Abstract

Cholesterol undergoes ozonolysis to afford a variety of oxysterol products, including cholesterol-5,6-epoxide (CholEp) and the isomeric aldehydes secosterol A (seco A) and secosterol B (seco B). These oxysterols display numerous important biological activities, including protein adduction; however, much remains to be learned about the identity of the reactive species and the range of proteins modified by these oxysterols. Here, we synthesized alkynyl derivatives of cholesterol-derived oxysterols and employed a straightforward detection method to establish secosterols A and B as the most protein-reactive of the oxysterols tested. Model adduction studies with an amino acid, peptides, and proteins provide evidence for the potential role of secosterol dehydration products in protein adduction. Hydrophobic separation methods—Folch extraction and solid phase extraction (SPE)—were successfully applied to enrich oxysterol-adducted peptide species, and LC-MS/MS analysis of a model peptide–seco adduct revealed a unique fragmentation pattern (neutral loss of 390 Da) for that species. Coupling a hydrophobic enrichment method with proteomic analysis utilizing characteristic fragmentation patterns facilitates the identification of secosterol-modified peptides and proteins in an adducted protein. More broadly, these improved enrichment methods may give insight into the role of oxysterols and ozone exposure in the pathogenesis of a variety of diseases, including atherosclerosis, Alzheimer’s disease, Parkinson’s disease, and asthma.
Introduction
Oxidative stress is a hallmark in numerous chronic and degenerative diseases associated with environmental factors. This stress can result in the formation and buildup of oxysterols (cholesterol-derived oxidation products) and other lipid-derived oxidation products. These lipid oxidation products exhibit toxicity and have been implicated in the pathogenesis of a variety of diseases, including atherosclerosis1,2 neurodegenerative disorders,3,4 and age-related macular degeneration.5,6 One significant source of oxidative stress is exposure to ozone, a main component of urban smog. Human exposure to biologically relevant levels of ozone is clearly linked to respiratory illnesses, such as asthma.7−9 Because ozone can travel in the air, both urban and rural populations can experience related adverse health effects.
Ozone is highly reactive with unsaturated lipids, such as cholesterol, which is present in the pulmonary surfactant lining the lungs.10−12 Ozone exposure studies in human pulmonary surfactant13 and bronchoalveolar lavage fluid and lung tissue of mice and rats10,14 show that the double bond of cholesterol is oxidized to afford various potentially reactive compounds, including epoxides and aldehydes. These resulting oxysterols exhibit diverse biological activities, including cytoxicity,13,15,16 inducing apoptosis,17,18 and altering membrane properties.19,20 Several of these ozone-derived oxidation products are electrophilic, meaning that they could react with the nucleophilic lysine, cysteine, and/or histidine residues of proteins to form covalent linkages. In fact, a number of studies demonstrated modification of specific proteins with cholesterol ozonolysis products.21−25
Covalent modification with electrophilic lipid oxidation products can affect protein function. For example, the cholesterol ozonolysis products secosterol A (seco A) and/or secosterol B (seco B) (both shown in Figure 1) modify β-amyloid,23 p53,26 apolipoprotein C-II,22 myelin basic protein,25 and antibody light chains,24 among other proteins. These modifications have been shown or are assumed to occur between the electrophilic aldehyde moiety of the secosterol and a nucleophilic lysine residue on the protein. The resulting secosterol adduction induces and accelerates amyloidogenesis in vitro(27) and may play a role in the initiation and progression of various diseases, including Alzheimer’s disease, cancer, atherosclerosis, and multiple sclerosis.
Figure 1.
Library of cholesterol-derived oxysterols and several alkynyl sterol analogs.
Covalent lipid modification increases the overall hydrophobicity of a modified protein; this added hydrophobicity can affect a protein’s localization. For example, palmitoylation can enable the lipidated proteins to associate with membranes and localize in lipid rafts/caveolae.28,29 Modification with oxysterols also confers significant hydrophobicity to the modified protein. Because of the added hydrophobicity, lipidated proteins can be challenging to study; however, exploiting hydrophobicity is central to many methods employed to isolate and study membrane proteins.30−32
We have developed a straightforward method to test a lipid’s ability to modify proteins: alkynyl lipid probes are allowed to react with protein, and any lipid-modified proteins are clicked with an azido-biotin reagent33 via Huisgen–Sharpless cycloaddition34,35 and subsequently detected by a fluorescent streptavidin conjugate.36 Here, we synthesized alkynyl analogs of several cholesterol-derived ozonolysis products and used these probes to determine the most protein-reactive oxysterol(s). Several model adduction studies with an amino acid, peptides, and proteins explored the identity of the reacting oxysterol, revealing the potential involvement of secosterol dehydration products in adduction. As prelude for studying more complex biological samples, hydrophobic methods were developed to enrich model systems in secosterol-modified peptides, and LC-MS/MS analyses elucidated a characteristic fragmentation pattern of secosterol–peptide adducts.
Materials and Methods
Synthetic Procedures
1H and 13C NMR spectra were collected on a 300, 400, or 600 MHz NMR. HRMS (high resolution mass spectrometry) analyses for small molecules were carried out at the University of Notre Dame. Purification by column chromatography was carried out on silica gel, and TLC plates were visualized with phosphomolybdic acid. The syntheses of alkynyl cholesterol (a-Chol) and alkynyl 7-ketocholesterol (a-7-ketoChol) have been previously reported.36 Full synthetic procedures for the remaining alkynyl lipids are described in the Supporting Information. CholEp (3:1 α to β),13 secosterols A and B,37 and their dehydration products37,38 were synthesized as previously described.
Alkynyl Oxysterol Treatment of Human Serum Albumin (HSA) and Biotinylation of Alkynyl (a) Lipid-Adducted Proteins
Solutions of HSA (1 mg/mL) and alkynyl (a)-oxysterol (20 μM) in 10 mM ammonium bicarbonate buffer (pH 7.4) were incubated at room temperature overnight with stirring. For the experiment testing the a-seco A concentration dependence of HSA adduction, a-seco A concentrations of 0–100 μM were assayed. The samples were reduced with sodium borohydride (5 mM, 1 h at room temperature) to stabilize any adducts that may have formed and neutralized with 10% HCl. The following click reagents were added to each of the samples: azido-biotin reagent (0.2 mM),33 tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) ligand (0.2 mM),39 copper sulfate (1 mM), and sodium ascorbate (1 mM), and the samples were vortexed and allowed to stir for 2 h at room temperature in the dark.
Analysis of Streptavidin-Labeled Proteins
Biotinylated proteins were resolved using 10% NuPAGE Novex BisTris gel (Life Technologies, Carlsbad, CA). Precision Plus Protein Kaleidoscope standards (10–250 kDa, Bio-Rad) were electrophoresed on the same gel for reference. The proteins were transferred electrophoretically to a polyvinylidene fluoride membrane (Life Technologies, Grand Island, NY) and probed with streptavidin conjugated with the Alexa Fluor 680 fluorophore (Life Technologies). Biotinylated proteins were visualized using the Odyssey Infrared Imaging System and Odyssey software according to the manufacturer (Licor, Lincoln, NE). Integrated intensities were obtained using the Odyssey software.
Modification of AcTpepX (Ac-AVAGXAGAR, X = K or H) or N-AcLys by Oxysterols
A solution of AcTpepX (1 mM) or N-AcLys (5 mM) and oxysterol (5 mM) in 50 mM phosphate buffer (pH 7.4):CH3CN (100 μL, 1:1 by volume) was incubated at 37 °C for 1–3 h. Samples containing secosterols were reduced with 2 M NaBH4 (10 μL) and then neutralized with 10% HCl for LC-MS analysis. Reverse-phase HPLC was performed on a Supelco Discovery C18 column (150 mm × 2.1 mm, 5 μm) using a mobile phase consisting of A: 0.05% TFA in H2O and B: 0.05% TFA in CH3CN. Although TFA sometimes suppresses ionization, resolution of the peaks was optimal with TFA as the additive. It did not significantly affect ionization in this case. The unreacted peptide and adducts were eluted with a gradient of 5% to 35% B over 20 min, then to 100% B over 5 min and held for 20 min, and back to 5% B over 5 min. The MS was operated in the positive-ion mode using electrospray ionization with conditions optimized for AcTpepK. For LC-MS/MS analysis, 35% normalized collision energy was used.
Modification of Cytochrome c (Cyt c) by Seco A and Secosterol Dehydration Products, MALDI-TOF Analysis, and Trypsinization
Cyt c (40 μM) was incubated in the presence of seco A, seco A–H2O, seco B–H2O (I), seco B–H2O (II), or seco B–2H2O (400 μM) in 10 mM NH4HCO3 buffer (pH 7.4) at room temperature overnight with stirring. Modified cyt c sample (1 μL) was spotted onto a MALDI target plate. A saturated solution of α-cyano-4-hydroxycinnamic acid (CHCA) matrix in H2O/CH3CN/TFA (50/50/0.1) (1 μL) was then spotted onto the same position of the MALDI plate. The modified cyt c and the matrix were immediately mixed by pipetting up and down twice and allowed to dry before sample analysis. MALDI-TOF-MS analyses were performed on a PerSeptive Biosystems Voyager-DE STR MALDI-TOF equipped with a pulsed N2 laser. Protein spectra were collected with an accelerating voltage of 20 kV in positive ion linear mode. Each spectrum was the accumulation of 1000 laser shots, with a laser intensity of 2200–2300 that was optimized for each spectrum to provide the best signal-to-noise ratio.
For the seco A-treated cyt c sample, to enable identification of sites of secosterol modification, the cyt c mixture was trypsinized using the following protocol: Trypsin (1:50 = w:w, trypsin:cyt c) was added, and the protein was allowed to digest for 4 h at 37 °C. Prior to LC-MS/MS analysis, either Folch extraction or solid phase extraction (SPE) (vide infra) was employed to enrich this solution in secosterol-modified peptides. The organic layer resulting from Folch extraction or the resulting SPE fractions were then analyzed by LC-MS/MS.
Folch Extraction
A mixture of unmodified and seco-modified peptides in 10 mM NH4HCO3 buffer (pH 7.4,100 μL) was diluted with chloroform and methanol in a ratio of 8:4:3 chloroform/methanol/buffer. The Folch solution was vortexed and centrifuged briefly to allow distinct organic and aqueous layers to separate. Each layer was subsequently analyzed by LC-MS using conditions described above for peptides.
Solid Phase Extraction (SPE) Fractionation
A C18 SPE cartridge (Sep-Pak Vac, 1 cm3, 50 mg sorbent, 55–105 μm particle size; Waters, Milford, MA) was conditioned with CH3CN/0.1% TFA (1 mL). The SPE cartridge was then flushed with 1 mL of water/0.1% TFA. The sample (e.g., seco-modified angiotensin II in 100 μL of buffer) was loaded onto the cartridge and separated using gradient elution (2 × 0.5 mL of each of the following): 10, 30, 50, and 90% CH3CN in H2O (0.1%TFA). Each 0.5 mL fraction was collected separately. Samples were either concentrated or analyzed directly using LC-MS/MS and/or MALDI-TOF-MS.
MALDI-TOF-MS Analysis of SPE Fractions
For SPE fractions containing <50% CH3CN, the sample (0.7 μL) was spotted on a MALDI target, followed by a saturated solution of CHCA matrix in H2O/CH3CN/TFA (50/50/0.1) (1 μL). The sample and matrix were immediately mixed by pipetting up and down twice and allowed to dry before sample analysis. Because of the volatility of organic solvents, sample preparation, specifically on-plate sample-matrix mixing, for MALDI analysis of samples containing significant amounts of organic solvent, can be difficult. For MALDI-TOF-MS analysis of SPE fractions containing ≥50% MeCN, the sample (0.7 μL) was spotted on a MALDI target, followed by a saturated solution of CHCA matrix in H2O/MeCN/TFA (50/50/0.1) (1 μL), followed by an additional 0.7 μL of the sample. The sample and matrix were immediately mixed by pipetting up and down twice and allowed to dry before sample analysis. This three-layer spotting technique ensured that the organic layer and matrix were sufficiently mixed to allow for analysis. Each MALDI-TOF-MS spectrum was the accumulation of 1000 laser shots, with a laser intensity of 1900–2100 that was optimized for each spectrum to provide the best signal-to-noise ratio.
HSA Modification with Seco A and Seco B
Refer to Scheme S1 for an outline of the general workflow for the experiments described here: HSA (1 mL, 4 mg/mL, 1X PBS) was incubated with seco A (1 mM) for 4 h at 37 °C. The reaction mixture was reduced with NaBH4 (20 mM) at room temperature for 1 h. Excess NaBH4 was removed using a Nanosep10K Omega (Pall) filter, and the volume of the reaction mixture was adjusted to 1 mL with 100 mM ammonium bicarbonate. DTT (10 mM) was added and incubated for 10 min at 50 °C, followed by iodoacetamide treatment (20 mM) in the dark for 10 min at RT. Trypsin (1:50 = w:w, trypsin:HSA) was added to the reaction mixture and incubated at 37 °C overnight. The digested peptides were loaded onto a preconditioned OASIS HLB (60 mg, 3 cm3) following the manufacturer’s protocol (Waters, Milford, MA). After a 1 mL H2O washing step, peptides were eluted and collected as described above in SPE fractionation. Collected fractions were evaporated to dryness in a SpeedVac concentrator and resuspended in 100 μL of H2O with 0.1% formic acid for LC-MS/MS analysis. The same procedure was followed for modification with seco B, except HSA was incubated with 0.2 mM seco B at 37 °C overnight.
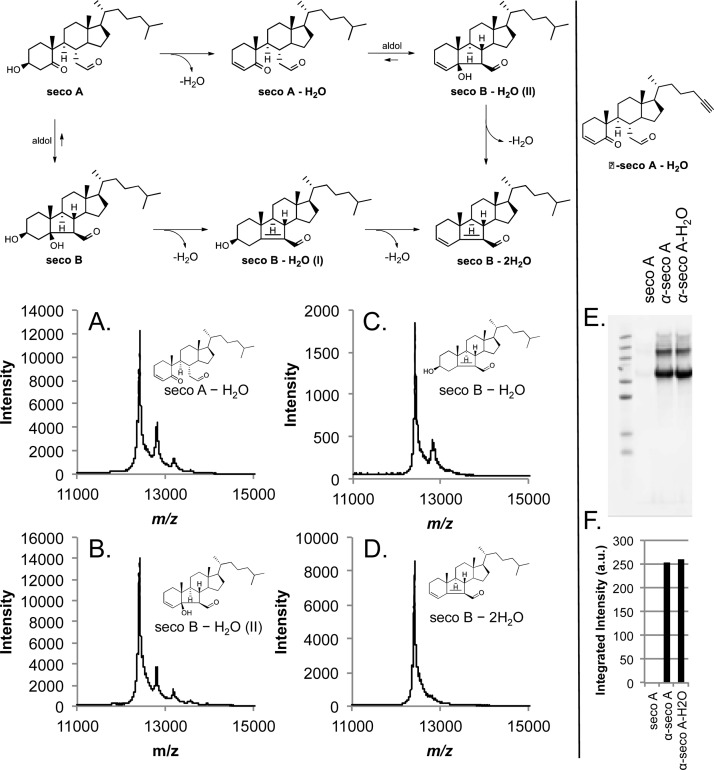
LC-MS/MS (Data Dependent Neutral Loss MS3) Analysis of Secosterol-Modified, Trypsin-Digested Proteins
The peptide solution (secosterol-treated, trypsinized protein) was separated using an Eksigent NanoLC Ultra HPLC and autosampler. The analytical column was packed with C18 resin (Jupiter, 3 μm, 300 Å, Phenomenex) in a capillary column (20 cm length, 360 μm O.D. × 100 μm I.D.). Peptides were eluted using a gradient as follows: 0–14.5 min, 2% B; 14.5–15 min, 2 to 5% B; 15–35 min, 5 to 35% B; 35–55 min, 35 to 99% B; 55–75 min, 99% B; and 75–90 min, 99 to 2% B at a flow rate of 500 nL/min (solvent A: 0.1% formic acid in H2O, solvent B: 0.1% formic acid in CH3CN). Eluting peptides were mass analyzed on an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) followed by a data dependent neutral loss (DDNL) MS3 method with dynamic exclusion enabled. Full-scan (m/z 350–2000) spectra were acquired with the Orbitrap mass analyzer (resolution 30,000), and the six most abundant ions in each MS scan were selected for fragmentation in the LTQ. An isolation width of 2 m/z units, activation time of 10 ms, and 35% normalized collision energy were used to generate MS2 spectra followed by MS3 neutral loss from MS2 with an isolation width of 2.2 m/z units. Neutral loss mass scans are enabled with the five most abundant in each MS2 with loss of m/z 390.349 (singly charged species), 195.175 (doubly charged species), and 130.120 (triply charged species). Dynamic exclusion settings allowed for a repeat count of 1 within a repeat duration of 15 s, and the exclusion duration time was set to 30 s. For identification of HSA peptides, tandem mass spectra were processed in BumberDash Tools,40 version 1.4.110, and searched by Myrimatch algorithm41 with a custom-built subset database originated from Homo_sapience_GRCh37-59_Cntm_r (2010_11-18). The sequence database was reversed so that each protein sequence appeared in both normal and reversed orientations, enabling false discovery rate estimation. MyriMatch was configured to have dynamic modifications of +57.021 for carbamidomethyl to Cys and 400.3342, 402.3498, 404.3654, 384.3342, and 386.3498 Da for [Cys,Lys,His]. MS3 spectra generated from DDNL were separately processed using ProteoWizard msConvert tool42 to convert to mzXML format, followed by Myrimatch search. In this case the configuration was consistent with dynamic modifications of +57.021 for carbamidomethyl to Cys and +12.00 for Lys residue. Protein assembly was handled by IDPicker algorithm, version 2.1.6710.43
Results
Synthesis of Alkynyl Lipids and Alkynyl Lipid Electrophiles
A series of cholesterol-derived oxysterols, several of which are established cholesterol ozonolysis products, were prepared according to known procedures13,37 along with their corresponding alkynyl analogs (Figure 1). Incorporation of the triple bond in place of the C24 isopropyl group of cholesterol introduces a minimal perturbation from the endogenous lipid structure. In previous studies, we demonstrated that alkynyl cholesterol (a-Chol) could be incorporated into Neuro2a cells and metabolized to the a-Chol fatty acid esters in a transformation identical to that of endogenous cholesterol. The immediate biosynthetic precursor to a-Chol, a-7-dehydrocholesterol, was also converted to a-Chol and its esters in cells, suggesting that alkynyl sterols are appropriate surrogates for the endogenous lipids in cell studies.36 The terminal alkyne moiety in our lipid probes allows for attachment of a biotin tag via click reaction with an azido-biotin reagent.33 The biotin tag can be used for visualization of proteins adducted with the oxysterols and/or enrichment techniques.
Human Serum Albumin Modification with Alkynyl Sterol Probes
With a set of alkynyl oxysterols in hand, we set up assays to determine the relative reactivity of their covalent attachment with protein nucleophiles. To survey our cholesterol-derived oxysterols for protein reactivity, buffer solutions of HSA were treated with each of the alkynyl oxysterol electrophiles shown in Figure 1. The adducts were stabilized by reduction with NaBH4 and then reacted with the azido-biotin reagent shown in Figure 2. Any alkynyl sterol-modified proteins can then be visualized by streptavidin-Alexa Fluor 680. A general scheme depicting the procedure is shown in Figure 2.
Figure 2.
General experimental workflow, including the azido-biotin reagent used for click biotinylation of alkynyl lipid-modified proteins.
The alkynyl secosterols (a-seco A and a-seco B) yielded the most protein adduction of the alkynyl oxysterols tested, as determined by streptavidin fluorophore intensity (Figure 3A). a-7-KetoChol gave no evidence of HSA adduction while a-CholEp afforded relatively low levels of protein modification. The relatively low level of a-CholEp–protein adduction found here agrees with a previous report that the 5,6-epoxides of endogenous cholesterol are remarkably unreactive toward nucleophiles.44 The reactivity of the cholesterol epoxides is in contrast to the epoxide derived from 7-dehydrocholesterol, an allylic epoxide that readily adducts to cellular proteins.36 Integrated intensities for the streptavidin fluorophore analyses of alkynyl lipid–HSA adduction indicate the extent of HSA modification by each probe (Figure 3B).
Figure 3.
HSA modification with alkynyl oxysterol electrophiles. (A) Streptavidin fluorophore visualization of HSA modification with alkynyl oxysterols (20 μM). [The higher weight protein (∼130 kDa) is HSA dimer.] (B) Integrated intensities from streptavidin fluorophore analysis of HSA modification with alkynyl oxysterols. (C) Streptavidin fluorophore visualization of HSA modification with different concentrations of a-seco A. [The higher weight protein (∼130 kDa) is HSA dimer.] (D) Integrated intensities from Western blot analysis of HSA modification with different concentrations of a-seco A. Alkynyl sterol-adducted protein was tagged with biotin via click reaction and labeled with the streptavidin-AlexaFluor 680 conjugate. The fluorophore was detected and integrated intensities were obtained using the Odyssey Infrared Imaging System and Application Software. For each streptavidin fluorophore analysis, the lane with the lowest integrated intensity was set to 0.
Figure 3C and D shows the dependence of protein modification on the concentration of the a-seco A probe. Thus, buffer solutions of HSA were treated with a-seco A at varying concentrations (0–100 μM), and biotinylated proteins were visualized using the streptavidin fluorophore (Figure 3C), with the integrated intensities (Figure 3D) showing that the amount of protein lipidation increases with increasing concentration of alkynyl lipid electrophile. The results shown in Figure 3B and D are extracted directly from a single gel.
Modification of a Peptide and an Amino Acid with Oxysterols: Model Studies
Having confirmed that the secosterols were protein-reactive, the chemistry of adduct formation was explored in greater detail. A peptide containing one nucleophilic site (Lys), Ac-Ala-Val-Ala-Gly-Lys-Ala-Gly-Ala-Arg (AcTpepK), was chosen as a model peptide. The peptide (1 mM) was incubated in the presence of the electrophile (5 mM) at 37 °C for 1 h. The reaction mixture was then reduced with NaBH4 to stabilize any adducts that form reversibly, and the solution was then neutralized and analyzed by LC-MS. While AcTpepK gave no evidence for formation of an adduct with CholEp and 7-ketoChol, both seco A and seco B gave rise to products with a mass corresponding to a peptide–oxysterol adduct (see Figure S1A–S1C for the LC-MS evidence). MS/MS analysis confirms that the product is a peptide with secosterol modification at the lysine residue (vide infra). This is likely the product that arises from reaction of the lysine amine side chain with the seco aldehyde. It is of some interest that the peptide–sterol adducts have chromatographic behavior on reverse-phase columns more like the free oxysterol than the unmodified peptide; both the peptide–sterol adducts and free oxysterols elute after the mobile phase reaches 100% organic solvent.
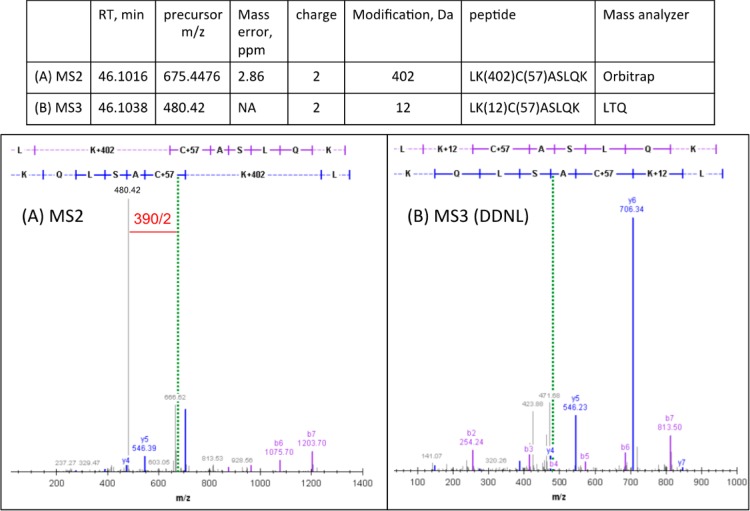
N-Acetyl lysine was chosen as a simple model nucleophile to investigate the fundamental stability and reactivity of the secosterols. In model studies, seco A and seco B were each incubated in the presence of N-acetyl lysine with subsequent NaBH4 reduction, and the adduction products before and after reduction were analyzed by LC-MS. Analyzing the product masses before and after reduction should shed light on the reactive moiety in the secosterol by considering the number of mass units added through reduction. Reaction of seco B with N-acetyl lysine afforded a product with m/z = 589 before reduction and m/z = 591 ([M + H]+) after reduction (Figure 4A and B, respectively), which likely results from the expected imine formation at the aldehyde with the amine side chain and subsequent reduction. Product analysis of the incubation of seco A with N-acetyl lysine was less straightforward. Before reduction, only one product with m/z = 589 was observed, suggesting lysine imine formation at the secosterol aldehyde (Figure S2A). However, there are four products observed after NaBH4 reduction (Figure S2B). One product corresponds to the reduced seco B-Lys adduct, resulting from seco A isomerization to seco B either prior to or following N-acetyl lysine modification. Two diastereomeric products have m/z = 593, a net addition of four hydrogen atoms resulting from complete reduction of the seco A-Lys imine adduct. The final product with m/z = 591 is a seco A-Lys adduct in which the ketone remains unreduced after treatment with NaBH4, suggesting that the ketone is resistant to reduction (confirmed in Figure S3A–C).
Figure 4.
LC-MS analysis of seco B-Lys products before and after reduction. (A) Products of seco B incubation with N-acetyl lysine, before reduction. (B) Products of seco B incubation with N-acetyl lysine, after reduction.
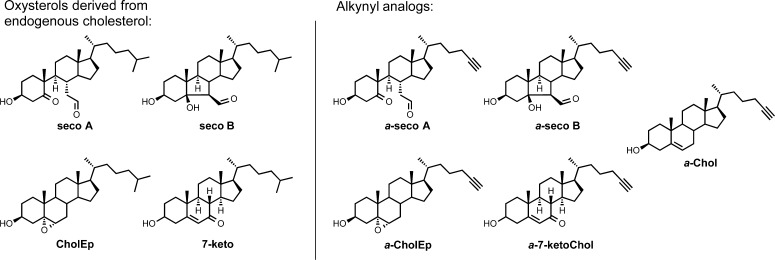
Using N-acetyl lysine as the limiting reagent allowed for analysis of the remaining unadducted secosterol, in addition to the Lys-secosterol adducts. After incubation in the presence of N-acetyl lysine, a significant amount of seco A was converted to seco B (Figure S4). Isomerization of seco A to seco B proceeded with continued incubation, as shown in the LC-MS analysis after 5 h (Figure S4D), eventually leading to seco B as the major isomer. This facile isomerization is well established; transient imine formation between the aldehyde of seco A and the amine groups of proteins has been shown to catalyze the conversion of seco A to seco B via an aldol reaction.21,37 There is also evidence for dehydration of seco A at longer incubation times (data not shown). These studies highlight an important consideration: the initial secosterol electrophile introduced into a system may not be the primary reactive electrophile; although we initially treat a system with seco A, the adduction results may better reflect the reactivity of seco B or some other electrophile to which seco A has converted over the course of the reaction.
Secosterol–Histidine Adduction on Cytochrome c (Cyt c)
Cyt c is an attractive model protein for adduction experiments due to its small size (12 kDa), which allows for straightforward monitoring of increases in molecular weight resulting from lipid electrophile covalent attachment. Incubation of cyt c and seco A in pH 7.4 buffer yielded significant protein modification that can be observed by MALDI-TOF-MS analysis (Figure 5, inset). High resolution FTMS measurement confirmed a nonreduced secosterol adduct to the cyt c tryptic peptide TGPNLHGLFGR by a mass difference of 6 ppm. The b and y ions in the LC-MS/MS analysis of the trypsinized protein suggested adduction of the secosterol at His-33, the first confirmed secosterol modification at a nonlysine residue (Figure 5 and Figure S5). This histidine residue has been shown to be a site of modification by other lipid electrophiles, namely 4-hydroxynonenal (HNE).45−47 However, unlike HNE, neither seco A nor seco B contains a Michael acceptor, a typical site of nucleophilic attack by histidine.
Figure 5.
High resolution FTMS analysis of cyt c tryptic peptide28TGPNLH(secoA-H2O)GLFGR (precursor m/z 784.9827 [M+2H]2+, mass error 1 ppm). Reaction mixture was not reduced with NaBH4 prior to trypsin digestion. Green dotted line denotes precursor ion. Singly and doubly charged b and y ions are highlighted with purple and blue, respectively. Significant water loss ions are observed at m/z 775 and 767 (−1H2O and −2H2O, respectively). Total loss of seco A is observed, displaying m/z 1168.7 [M + H]+ and 585.0 [M+2H]2+. b and y ions containing seco A, including b6 and y6, indicate that the seco A is adducted at the H-33 position. Inset: MALDI-TOF-MS analysis of secosterol-modified cyt c (intact modified protein). (FTMS/MS is provided in Figure S5A–C.)
While secosterols A and B have electrophilic aldehyde groups, histidine attack on these moieties is not expected; rather, lysine residues are common nucleophiles in reaction with aldehydes. Indeed, previous work had established Lys-22 of cyt c as the main site of seco B modification under SDS micellar conditions48 with no evidence of histidine adduction. In an SDS environment, the tertiary structure of cyt c is transformed to mimic membrane-bound cyt c;49 this alternative conformation may allow the secosterol to experience previously less accessible hydrophobic interactions around Lys-22 and modify cyt c at this site, whereas in the buffer solution used in this work, cyt c adopts a tertiary structure that may reduce access to Lys-22 and encourage the secosterol to react with the hydrophilic, solvent-exposed His-33. To explore the reactivity of seco A with His residues, a model peptide containing one nucleophilic site (His), Ac-Ala-Val-Ala-Gly-His-Ala-Gly-Ala-Arg (AcTpepH), was incubated with either seco A or seco A–H2O in pH 7.4 buffer. In both cases, LC-MS analysis of the product mixture showed evidence of a product with a mass corresponding to a peptide–seco A–H2O adduct (Figure S6A). LC-MS/MS analysis confirmed histidine as the site of modification, the most nucleophilic residue of N-terminal-protected AcTpepH (Figures S6C and S6D). After NaBH4 treatment, a mixture of dehydrated seco A and seco B adducts was observed with an addition of 2 and 4 mass units, respectively, indicating that the ketone is resistant to reduction, as we observed previously (Figure S6E and S6F). This observation suggests that seco A must undergo dehydration prior to adduction followed by aldol reaction.
The identification of a histidine residue (His-33) on cyt c as a site of secosterol modification forced a reconsideration of possible electrophiles that may be formed from seco A. Seco A can undergo cyclization via an aldol reaction to form seco B,37 and both seco A and seco B can lose one50 or two water molecules, affording the dehydration products shown in Figure 6. Dehydration products seco A–H2O and seco B–H2O (I) do contain an α,β-unsaturated carbonyl, a prototypical Michael acceptor that might explain the observation of secosterol covalent attachment at cyt c His-33.
Figure 6.
Secosterol dehydration products and their reactivity. (top scheme) Secosterols and their dehydration products. MALDI-TOF-MS analysis of cyt c modification with secosterol dehydration products. (A) Modification with seco A–H2O. (B) Modification with seco B–H2O (II). (C) Modification with seco B–H2O (I). (D) Modification with seco B–2H2O. (E) Streptavidin fluorophore visualization of HSA modification with a-seco A compared to a-seco A–H2O. [The higher weight protein (∼130 kDa) is HSA dimer.] (F) Integrated intensities from streptavidin fluorophore analysis of HSA (1 mg/mL) modification with a-seco A (20 μM) compared to a-seco A–H2O (20 μM). Alkynyl sterol-adducted protein was tagged with biotin via click reaction and labeled with the streptavidin–AlexaFluor 680 conjugate. The fluorophore was detected and integrated intensities were obtained using the Odyssey Infrared Imaging System and Application Software. For each streptavidin fluorophore analysis, the lane with the lowest integrated intensity was set to 0.
The dehydration products seco A–H2O, seco B–H2O (I), seco B–H2O (II), and seco B–2H2O were prepared according to literature procedures.37,38 MALDI-TOF-MS analysis indicated protein adduction of cyt c with all of the compounds except for the double dehydration product seco–2H2O (Figure 6A–D). These experiments all suggest that electrophiles generated by dehydration play a major role in the protein adduction, and this was confirmed by studies of the alkynyl analog, a-seco A–H2O. Both a-seco A and a-seco A–H2O were prepared, and each was incubated with HSA, as previously described following the protocol outlined in Figure 2. The resulting solutions were reduced with NaBH4 and treated with the azido-biotin reagent, and alkynyl sterol-modified proteins were visualized with streptavidin Alexa Fluor 680. The relative intensities for the fluorescent signal indicated the extent of HSA modification by each probe (Figure 6E and F). Under these reaction conditions, the dehydration product of a-seco A (a-seco A–H2O) produced a comparable amount of protein adduction to the parent secosterol (a-seco A) itself. This result supports the notion that secosterol dehydration products contribute to the reactivity of secosterols with proteins. Because dehydrated a-seco A contains an α,β-unsaturated ketone, nucleophilic protein residues may react with the electrophilic secosterol through either Schiff base formation at the aldehyde or Michael addition. Considering dehydration products containing a Michael acceptor in the overall scheme of secosterol reactivity introduces the previously unrecognized possibility of protein modification at histidine side chains.
Enrichment Methods and MSn Analysis of Secosterol–Peptide Adducts
Sterols and the seco-sterols are hydrophobic, and methods were investigated to separate sterol-modified peptides from unmodified peptides based on hydrophobicity. Thus, a mixture of secosterol-adducted AcTpepK and unmodified AcTpepK was submitted to Folch extraction,51 and the resulting organic and aqueous layers were analyzed by LC-MS (Figure 7). This analysis showed that the majority of the secosterol-modified peptide was extracted into the organic phase and suggests that hydrophobic separations may find use in the enrichment of sterol-adducted peptides and proteins. Solid phase extraction with a reverse-phase (C18) SPE cartridge on a mixture of unmodified angiotensin II (Asp-Arg-Val-Tyr-Ile-His-Pro-Phe) and angiotensin II modified with seco A also showed a complete separation of the modified peptide from the unmodified peptide (Figure 8). This model study demonstrates the potential use of SPE as a preparative enrichment method for peptides modified with oxysterol electrophiles.
Figure 7.

Folch extraction of seco B-adducted AcTpepK. (A–C) LC-MS analysis. (A) Seco B-adducted AcTpepK. (B) Aqueous phase of Folch-extracted mixture. (Additional AcTpepK was spiked into initial mixture prior to Folch extraction.) (C) Organic phase of Folch-extracted mixture.
Figure 8.
MALDI-TOF-MS analysis of reverse-phase SPE of a secosterol-modified peptide. (A) Mixture of unmodified and secosterol-modified angiotensin II (M = Asp-Arg-Val-Tyr-Ile-His-Pro-Phe). (B) SPE fractions eluted with 10% CH3CN in water with 0.1% TFA contain unmodified angiotensin II. (C) SPE fractions eluted with 50–100% CH3CN in water with 0.1% TFA contain secosterol-modified angiotensin II.
The MS/MS fragmentation of a model peptide modified with a secosterol was investigated to establish characteristic fragmentation associated with secosterol-modified peptides (e.g., neutral loss of the secosterol) to provide a basis for identifying secosterol-modified peptides in a complex proteomic analysis. In a preliminary study of the AcTpepK–seco B lysine imine adduct, the dominant peaks observed were due to loss of water ([M + 2H – H2O]2+ and [M + 2H – 2H2O]2+) (Figure S7). The borohydride-reduced peptide–seco B adduct gave a dominant ion at m/z = 427.9 in the MS2 spectrum (Figure 9B). Apparently, this fragment ion is due to cleavage at the C–C bond between the secosterol core and the reduced imine group. Cleavage in this position should result in a neutral loss of 390 Da (most of the secosterol), and the ability to identify seco B-modified AcTpepK via an LC-MS/MS/MS neutral loss experiment was confirmed, exhibiting fragment ions with +12 m/z relative to the ions observed from the parent peptide (Figure 9A and C). This unique fragmentation may be useful in identifying secosterol-modified peptides during proteomic analysis.
Figure 9.
Fragmentation of AcTpepK–seco B adduct after NaBH4 reduction. (A) MS2 of unmodified AcTpepK (m/z 421.8 [M + 2H]2+) using 30% normalized collision energy. (B) MS2 of AcTpepK + seco B (=SB) adduct (m/z 622.9 [M + SB + 2H]2+). Characteristic neutral loss (m/z 390 = @) fragmentation ion of seco B-modified AcTpepK after reduction is observed at m/z 427.9. 321.7 = [y7 + SB – @ + 2H]2+, 371.4 = [y8 + SB – @ + 2H]2+, 374.2 = y4, 427.9 = [M + SB −@ + 2H]2+, 514.4 = y5 + SB – @, 571.4 = y6 + SB – @, 642.5 = y7 + SB – @, 741.6 = y8 + SB – @, 842.6 = M. (C) DDNL MS3 of m/z 427.9 [M + SB – @ + 2H]2+ using 35% normalized collision energy aided identification of covalent modification to peptides. 212.9 = b2, 284.0 = b3, 303.1 = y3, 321.8 = [y7 + SB – @ + 2H]2+, 371.4 = [y8 + SB – @ + 2H]2+, 374.2 = y4, 481.4 = b5 + SB – @, 514.4 = y5 + SB – @, 571.4 = y6 + SB – @, 642.5 = y7 + SB – @, 741.5 = y8 + SB – @.
Proteomics Assay of Seco B-Modified Human Serum Albumin
HSA was exposed to seco B as previously described, and the protein was trypsin-digested, with the resulting peptides fractionated by SPE and subjected to LC-MS/MS (Scheme S1). Spectra were analyzed using the automated algorithm search described in the Materials and Methods section.
A total of 3600 peptide spectra were filtered under stringent criteria, covering 91% of HSA. A majority (75%) of the filtered spectra were eluted in the 10–30% CH3CN SPE fractions (Table S1), which contained mostly unadducted peptides. Most of the seco B–peptide adducts identified were contained in the 60% CH3CN SPE fraction and are listed in Table 1. All the precursor masses of seco B-modified peptides have an accuracy of <10 ppm mass error. K-223 seems to be the most frequently modified position by oxysterols, based on the number of spectral counts.
Table 1. Combined List of HSA–Secosterol Adduction Sites Analyzed by High Resolution FTMSa.
| sequence | sites | precursor, m/z | charge | mass error, ppm |
|---|---|---|---|---|
| 1DAHK*SEVAHR | K4 | 517.9285 | 3 | 6.5 |
| 187DEGK*ASSAK | K190 | 647.8977 | 2 | 1.1 |
| 182LDELRDEGK*ASSAK | K190 | 641.0491 | 3 | 3.7 |
| 191ASSAK*QR | K195 | 575.3854 | 2 | 2.7 |
| 191ASSAK&QR | K195 | 566.3806 | 2 | 3.6 |
| 196QRLK*C#ASLQK | K199 | 817.5283/545.3542 | 2/3 | 3.4/2.7 |
| 198LK*C#ASLQK | K199 | 675.4467/450.6332 | 2/3 | 1.6/0.9 |
| 198LK&C#ASLQK | K199 | 444.6304 | 3 | 2.5 |
| 414K*VPQVSTPTLVEVSR | K414 | 681.4367 | 3 | 4.0 |
| 433VGSK*C#C#K | K436 | 620.8777 | 2 | –5.5 |
| 525K*QTALVELVK | K525 | 766.0295 | 2 | 2.3 |
| 525KQTALVELVK*HKPK | K534 | 506.0964 | 4 | 6.8 |
HSA was treated with either seco A or seco B followed by NaBH4 reduction. SPE fractionation was performed Supporting Information Scheme 1). The majority of unmodified peptides were contained in the 30% MeCN fraction (Supporting Information Table 1). All the identified adducted peptides were found in the 60% MeCN fraction. Based on MS/MS fragmentation patterns, the final form of secosterol was identified as seco B or its dehydrated products. A separate list of adducted peptides is shown in Supporting Information Table 2. The same sites have been modified by both electrophiles repeatedly. Identified peptides were further confirmed by DDNL MS3. The mass accuracy and peptide fragmentation assignments are in good agreement with their covalent modification and sites. K* represents a seco B imine adduct on lysine. K& is a dehydrated adduct on lysine. C# is a carbamidomethyl modification on cysteine.
As observed in our model studies, the seco B-Lys adducts of tryptic serum albumin peptides yielded a diagnostic fragmentation pattern in the MS2 spectra that corresponds to the loss of 390 Da (the seco B core) (Figure 10 and Figure S8). This neutral loss can be used as a signature fragment for seco B–Lys adducts. Furthermore, this characteristic neutral loss ion can be incorporated into DDNL52 LC-MSn analysis to identify seco B–Lys adducts in more complex mixtures of proteins/peptides. The residual peptide modification of m/z = +12 Da (due to the partial fragmentation of seco B) was observed in the resulting MS3 spectra. All MS3 spectra were extracted separately using an MS Converter42 to generate mzXML for Myrimatch searching; the peptides are indeed identified as the same peptide sequences identified by the MS2 spectra (Table S2).
Figure 10.
Characteristic MS2 of seco B-Lys adduct and DDNL MS3 spectra. HSA-seco B adducted peptide after NaBH4 reduction. The (A) and (B) spectra are summarized in the head table. Green dotted lines denote precursor ions of the adducted peptides. b and y ions are in purple and blue, respectively. (A) MS2 spectrum of LK(402)C(57)ASLQK, which is modified with seco B on Lys-199. As illustrated in Figure 9, the characteristic 390 neutral loss fragment ion is observed at m/z = 480.42 ([peptide +2H + seco B – 390]2+). (B) The subsequent DDNL MS3 spectrum of m/z 480.42 provides further evidence of seco B adduction at Lys-199 in the same sequence with a shift of +12 m/z units after loss of 390 [LK(12)C(57)ASLQK].
Seco A-Modified Human Serum Albumin
The picture of seco A adduction has the potential to be more complicated than that of seco B, as described previously and outlined in Figure 6. Nevertheless, a study of HSA–seco A adduction was performed, evaluating SPE as a method of enrichment to aid in probing the sites of seco A modification. Seco A–protein adduction was carried out overnight at room temperature at 5-fold lower concentration than the aforementioned seco B–protein adduction. Whether seco A adduction occurs prior to aldol isomerization or vice versa, the identified modified peptides correspond to seco B–Lys adducts exhibiting ions in the MS2 spectra resulting from the signature 390 neutral loss, as shown in Figure 10. MS3 data generated from DDNL match the peptide sequences identified with the MS2 spectra. There are no other significant adducts observed except the dehydrated form of previously identified modified peptides (Table S2). This experiment verifies SPE as an effective method to enrich complex mixtures in sterol-modified peptides and also confirms the characteristic fragmentation pattern of seco B–Lys adducts.
Discussion
Because of the strong biological activity and electrophilic nature of several cholesterol ozonolysis products, we synthesized alkynyl analogs of several of these cholesterol-derived oxysterols and investigated their reactivity with proteins. The trend in protein reactivity was the following: a-seco A ≈ a-seco B ≫ a-CholEp > a-7-ketoChol, with alkynyl secosterols A and B yielding significantly more protein adduction than the other oxysterols tested. These results agree with previous work demonstrating secosterol modification of various specific proteins.21−26 Also supporting our results is a prior study that reported low reactivity of 5,6-cholesterol epoxides with amine and thiol nucleophiles.44
The dehydration product seco A–H2O has been identified in atherosclerotic arteries, along with seco A and seco B, but the reactivity of seco A–H2O with nucleophiles remains unexplored.37 Five pieces of experimental evidence suggest the potential involvement of secosterol dehydration products in the overall scheme of secosterol reactivity: (1) Seco A loses a water molecule in the presence of catalytic N-acyl lysine, forming a dehydration product upon incubation at 37 °C. (2) Incubation of seco A in the presence of cyt c results in secosterol adduction at a histidine residue of cyt c (His-33); however, reaction of histidine with seco A was unexpected, as seco A does not contain a typical histidine-reactive electrophile (e.g., a Michael acceptor). (3) Seco A–H2O and seco B–H2O (I and II) modify cyt c after incubation at 37 °C for 24 h. (4) Both seco A and seco A–H2O modify model peptide AcTpepH at the histidine residue, and the MS2 spectra of the resulting adducts show the same fragment ions and isotope distributions. (5) The alkynyl analog a-seco A–H2O afforded a comparable amount of protein modification to the precursor secosterol a-seco A when using HSA as a model protein. Taken together, these results demonstrate that secosterols can dehydrate under mild conditions, and the resulting dehydration products are protein reactive, presumably via the α,β-unsaturated carbonyl. The presence of this Michael acceptor expands the reactivity of cholesterol-derived secosterols, enabling reaction with histidine nucleophiles. We note that a study of cyt c modification with trans,trans-2,4-decadienal identified His-33 adducts, affording mass increases indicative of Schiff base formation rather than Michael addition.53 Because there are multiple mechanisms that could account for the histidine–secosterol adduction we observed, we are performing additional experiments to unambiguously identify both the structure of the reactive secosterol(s) and the histidine-reactive site(s) of the identified secosterol(s).
As demonstrated by our LC-MS analyses, modification with an oxysterol conveys considerable hydrophobicity to the adducted peptide, causing the peptide–secosterol adduct to behave chromatographically more like the free oxysterol than the unmodified peptide. Methods exploiting this hydrophobicity, Folch extraction and SPE, were employed to successfully enrich a simple mixture in (nonalkynyl) secosterol-modified peptide(s). SPE enrichment was also applied to identify sites of secosterol modification in HSA, effectively removing the majority of the unmodified peptides with ≤30% CH3CN in water. Because SPE is typically one of the final sample cleanup steps before proteomic analysis, hydrophobic enrichment by SPE may be easily incorporated into the sample preparation workflow.
Utilizing hydrophobic enrichment in conjunction with proteomic analysis incorporating the characteristic fragmentation of secosterol–adducted peptides (neutral loss of 390 Da) may improve the likelihood of the identification of secosterol-modified peptides and proteins in complex mixtures. Additionally, developing enrichment methods that use hydrophobicity, rather than tagged analogs of the oxysterols of interest, will permit the analysis of samples of endogenous sterol oxidation products. These tools may help shed light on the link between ozone exposure and diseases associated with environmental factors.
In conclusion, a-seco A and a-seco B were the most protein-reactive cholesterol ozonolysis products in our study. These oxysterols can dehydrate (under mild conditions) to afford products containing Michael acceptors, which expands the recognized reactivity of secosterols. Furthermore, secosterol modification of peptides and proteins conveys sufficient lipophilicity to allow for hydrophobic enrichment of secosterol–peptide adducts in complex mixtures. Employing a hydrophobic enrichment method coupled with proteomic analysis incorporating the unique fragmentation of secosterol–peptide adducts may ultimately provide a more comprehensive map of proteins that are modified with secosterols. It should be noted, however, that the samples tested here are not representative of protein mixtures found in tissues and fluid samples. Experiments that extend the methods described here for single peptides and proteins to endogenous sources will undoubtedly pose additional challenges. Experiments that apply these approaches to cells in culture are ongoing and will be reported in due course.
Glossary
Abbreviations:
- CholEp
cholesterol-5,6-epoxide
- seco A
secosterol A
- seco B
secosterol B
- SPE
solid phase extraction
- HSA
human serum albumin
- THPTA
tris(3-hydroxypropyltriazolylmethyl)amine
- cyt c
cytochrome c
- CHCA
α-cyano-4-hydroxycinnamic acid
- DTT
dithiothreitol
- DDNL
data dependent neutral loss
- a-Chol
alkynyl cholesterol
- a-seco A
alkynyl secosterol A
- a-seco B
alkynyl secosterol B
- a-CholEp
alkynyl cholesterol 5,6-epoxide
- a-7-ketoChol
alkynyl 7-ketocholesterol
- HNE
4-hydroxynonenal
Supporting Information Available
Synthesis of secosterols and NMR spectra and additional LC-MS/MS and DDNLMS3 spectra of model and tryptic HSA peptides–secosterol adducts. This material is available free of charge via the Internet at http://pubs.acs.org.
The National Institutes of Health (P01 NIEHS ES013125) supported this work. The Ph.D. scholarship of Thiago C. Genaro-Mattos was supported by São Paulo Research Foundation (FAPESP, 2012/16145-5).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jessup W.; Kritharides L.; Stocker R. (2004) Lipid oxidation in atherogenesis: an overview. Biochem. Soc. Trans. 32, 134–138. [DOI] [PubMed] [Google Scholar]
- Shao B.; Heinecke J. W. (2009) HDL, lipid peroxidation, and atherosclerosis. J. Lipid Res. 50, 599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre L. M.; Perry G.; Smith M. A. (2008) Oxidative Stress and Neurotoxicity. Chem. Res. Toxicol. 21, 172–188. [DOI] [PubMed] [Google Scholar]
- Reed T. T. (2011) Lipid peroxidation and neurodegenerative disease. Free Radical Biol. Med. 51, 1302–1319. [DOI] [PubMed] [Google Scholar]
- Ebrahimi K. B.; Handa J. T. (2011) Lipids, Lipoproteins, and Age-Related Macular Degeneration. J. Lipids 802059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopitz J.; Holz F. G.; Kaemmerer E.; Schutt F. (2004) Lipids and lipid peroxidation products in the pathogenesis of age-related macular degeneration. Biochimie 86, 825–831. [DOI] [PubMed] [Google Scholar]
- Peden D. B. (2011) The role of oxidative stress and innate immunity in O(3) and endotoxin-induced human allergic airway disease. Immunol. Rev. 242, 91–105. [DOI] [PubMed] [Google Scholar]
- Muller L.; Jaspers I. (2012) Epithelial cells, the ″switchboard″ of respiratory immune defense responses: effects of air pollutants. Swiss Med. Wkly. 142, w13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivian L. (2011) Outdoor air pollution and asthma in children. J. Asthma 48, 470–481. [DOI] [PubMed] [Google Scholar]
- Pryor W. A.; Wang K.; Bermúdez E. (1992) Cholesterol ozonation products as biomarkers for ozone exposure in rats. Biochem. Biophys. Res. Commun. 188, 618–623. [DOI] [PubMed] [Google Scholar]
- Pryor W. A.; Squadrito G. L.; Friedman M. (1995) The cascade mechanism to explain ozone toxicity: The role of lipid ozonation products. Free Radical Biol. Med. 19, 935–941. [DOI] [PubMed] [Google Scholar]
- Santrock J.; Gorski R. A.; O’Gara J. F. (1992) Products and mechanism of the reaction of ozone with phospholipids in unilamellar phospholipid vesicles. Chem. Res. Toxicol. 5, 134–141. [DOI] [PubMed] [Google Scholar]
- Pulfer M. K.; Murphy R. C. (2004) Formation of Biologically Active Oxysterols during Ozonolysis of Cholesterol Present in Lung Surfactant. J. Biol. Chem. 279, 26331–26338. [DOI] [PubMed] [Google Scholar]
- Pulfer M. K.; Taube C.; Gelfand E.; Murphy R. C. (2005) Ozone Exposure in Vivo and Formation of Biologically Active Oxysterols in the Lung. J. Pharm. Exp. Ther. 312, 256–264. [DOI] [PubMed] [Google Scholar]
- Sevanian A.; Berliner J.; Peterson H. (1991) Uptake, metabolism, and cytotoxicity of isomeric cholesterol-5,6-epoxides in rabbit aortic endothelial cells. J. Lipid Res. 32, 147–155. [PubMed] [Google Scholar]
- Sathishkumar K.; Murthy S. N.; Uppu R. M. (2007) Cytotoxic effects of oxysterols produced during ozonolysis of cholesterol in murine GT1–7 hypothalamic neurons. Free Radical Res. 41, 82–88. [DOI] [PubMed] [Google Scholar]
- Sathishkumar K.; Haque M.; Perumal T. E.; Francis J.; Uppu R. M. (2005) A major ozonation product of cholesterol, 3β-hydroxy-5-oxo-5,6-secocholestan-6-al, induces apoptosis in H9c2 cardiomyoblasts. FEBS Lett. 579, 6444–6450. [DOI] [PubMed] [Google Scholar]
- Kosmider B.; Loader J. E.; Murphy R. C.; Mason R. J. (2010) Apoptosis induced by ozone and oxysterols in human alveolar epithelial cells. Free Radical Biol. Med. 48, 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtel E.; Bach D.; Epand R. F.; Tishbee A.; Epand R. M. (2006) A Product of Ozonolysis of Cholesterol Alters the Biophysical Properties of Phosphatidylethanolamine Membranes. Biochemistry 45, 1345–1351. [DOI] [PubMed] [Google Scholar]
- Bach D.; Wachtel E.; Miller I. R. (2009) Kinetics of Schiff base formation between the cholesterol ozonolysis product 3β-hydroxy-5-oxo-5,6-secocholestan-6-al and phosphatidylethanolamine. Chem. Phys. Lipids 157, 51–55. [DOI] [PubMed] [Google Scholar]
- Bosco D. A.; Fowler D. M.; Zhang Q.; Nieva J.; Powers E. T.; Wentworth P. Jr.; Lerner R. A.; Kelly J. W. (2006) Elevated levels of oxidized cholesterol metabolites in Lewy body disease brains accelerate alpha-synuclein fibrilization. Nat. Chem. Biol. 2, 249–253. [DOI] [PubMed] [Google Scholar]
- Stewart C. R.; Wilson L. M.; Zhang Q.; Pham C. L. L.; Waddington L. J.; Staples M. K.; Stapleton D.; Kelly J. W.; Howlett G. J. (2007) Oxidized Cholesterol Metabolites Found in Human Atherosclerotic Lesions Promote Apolipoprotein C-II Amyloid Fibril Formation. Biochemistry 46, 5552–5561. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Powers E. T.; Nieva J.; Huff M. E.; Dendle M. A.; Bieschke J.; Glabe C. G.; Eschenmoser A.; Wentworth P.; Lerner R. A.; Kelly J. W. (2004) Metabolite-initiated protein misfolding may trigger Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 101, 4752–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J.; Shafton A.; Altobell L. J.; Tripuraneni S.; Rogel J. K.; Wentworth A. D.; Lerner R. A.; Wentworth P. (2008) Lipid-Derived Aldehydes Accelerate Light Chain Amyloid and Amorphous Aggregation. Biochemistry 47, 7695–7705. [DOI] [PubMed] [Google Scholar]
- Cygan N. K.; Scheinost J. C.; Butters T. D.; Wentworth P. (2011) Adduction of Cholesterol 5,6-Secosterol Aldehyde to Membrane-Bound Myelin Basic Protein Exposes an Immunodominant Epitope. Biochemistry 50, 2092–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J.; Song B.-D.; Rogel J. K.; Kujawara D.; Altobel L. III; Izharrudin A.; Boldt G. E.; Grover R. K.; Wentworth A. D.; Wentworth P. Jr. (2011) Cholesterol Secosterol Aldehydes Induce Amyloidogenesis and Dysfunction of Wild-Type Tumor Protein p53. Chem. Biol. 18, 920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieschke J.; Zhang Q.; Powers E. T.; Lerner R. A.; Kelly J. W. (2005) Oxidative Metabolites Accelerate Alzheimer’s Amyloidogenesis by a Two-Step Mechanism, Eliminating the Requirement for Nucleation. Biochemistry 44, 4977–4983. [DOI] [PubMed] [Google Scholar]
- Aicart-Ramos C.; Valero R. A.; Rodriguez-Crespo I. (2011) Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta Biomembr. 1808, 2981–2994. [DOI] [PubMed] [Google Scholar]
- Linder M. E.; Deschenes R. J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell. Biol. 8, 74–84. [DOI] [PubMed] [Google Scholar]
- Qoronfleh M. W.; Benton B.; Ignacio R.; Kaboord B. (2003) Selective Enrichment of Membrane Proteins by Partition Phase Separation for Proteomic Studies. J. Biomed. Biotechnol. 2003, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. (2011) Strategies for the Purification of Membrane Proteins, In Protein Chromatography (Walls D., and Loughran S. T., Eds.) pp 485–496, Humana Press. [Google Scholar]
- Seddon A. M.; Curnow P.; Booth P. J. (2004) Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta 1666, 105–117. [DOI] [PubMed] [Google Scholar]
- Vila A.; Tallman K. A.; Jacobs A. T.; Liebler D. C.; Porter N. A.; Marnett L. J. (2008) Identification of Protein Targets of 4-Hydroxynonenal Using Click Chemistry for ex Vivo Biotinylation of Azido and Alkynyl Derivatives. Chem. Res. Toxicol. 21, 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. (2002) A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem., Int. Ed. Engl. 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- Thirumurugan P.; Matosiuk D.; Jozwiak K. (2013) Click Chemistry for Drug Development and Diverse Chemical–Biology Applications. Chem. Rev. 113, 4905–4979. [DOI] [PubMed] [Google Scholar]
- Windsor K.; Genaro-Mattos T. C.; Kim H. Y.; Liu W.; Tallman K. A.; Miyamoto S.; Korade Z.; Porter N. A. (2013) Probing lipid-protein adduction with alkynyl surrogates: application to Smith-Lemli-Opitz syndrome. J. Lipid Res. 54, 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth P. Jr.; Nieva J.; Takeuchi C.; Galve R.; Wentworth A. D.; Dilley R. B.; DeLaria G. A.; Saven A.; Babior B. M.; Janda K. D.; Eschenmoser A.; Lerner R. A. (2003) Evidence for ozone formation in human atherosclerotic arteries. Science 302, 1053–1056. [DOI] [PubMed] [Google Scholar]
- Gan C.; Fan L.; Cui J.; Huang Y.; Jiao Y.; Wei W. (2012) Synthesis and in vitro antiproliferative evaluation of some ring B abeo-sterols. Steroids 77, 1061–1068. [DOI] [PubMed] [Google Scholar]
- Hong V.; Presolski S. I.; Ma C.; Finn M. G. (2009) Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem., Int. Ed. Engl. 48, 9879–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman J. D., Ma Z.-Q., and Tabb D. L. (2012) Identifying proteomic LC-MS/MS data sets with Bumbershoot and IDPicker. Current Protocols in Bioinformatics, Chapter 13, Unit 13.17, Wiley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D. L.; Fernando C. G.; Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M. Y.; Paulse C.; Creasy D.; Flashner L.; Kani K.; Moulding C.; Seymour S. L.; Nuwaysir L. M.; Lefebvre B.; Kuhlmann F.; Roark J.; Rainer P.; Detlev S.; Hemenway T.; Huhmer A.; Langridge J.; Connolly B.; Chadick T.; Holly K.; Eckels J.; Deutsch E. W.; Moritz R. L.; Katz J. E.; Agus D. B.; MacCoss M.; Tabb D. L.; Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z. Q.; Dasari S.; Chambers M. C.; Litton M. D.; Sobecki S. M.; Zimmerman L. J.; Halvey P. J.; Schilling B.; Drake P. M.; Gibson B. W.; Tabb D. L. (2009) IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 8, 3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillasse M. R.; Saffon N.; Gornitzka H.; Silvente-Poirot S.; Poirot M.; de Medina P. (2012) Surprising unreactivity of cholesterol-5,6-epoxides towards nucleophiles. J. Lipid Res. 53, 718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom A. L.; Barnes S.; Wilson L.; Kirk M.; Coward L.; Darley-Usmar V. (2004) Modification of Cytochrome c by 4-hydroxy-2-nonenal: evidence for histidine, lysine, and arginine-aldehyde adducts. J. Am. Soc. Mass Spectrom. 15, 1136–1147. [DOI] [PubMed] [Google Scholar]
- Tang X.; Sayre L. M.; Tochtrop G. P. (2011) A mass spectrometric analysis of 4-hydroxy-2-(E)-nonenal modification of cytochrome c. J. Mass Spectrom. 46, 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. V.; Wishnok J. S.; Tannenbaum S. R. (2007) Covalent adducts arising from the decomposition products of lipid hydroperoxides in the presence of cytochrome c. Chem. Res. Toxicol. 20, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Appolinário P. P.; Mugnol K. C. U.; Bloch C. Jr; Nantes I. L.; Di Mascio P.; Miyamoto S. (2013) Covalent Binding and Anchoring of Cytochrome c to Mitochondrial Mimetic Membranes Promoted by Cholesterol Carboxyaldehyde. Chem. Res. Toxicol. 26, 1536–1544. [DOI] [PubMed] [Google Scholar]
- Mugnol K. C.; Ando R. A.; Nagayasu R. Y.; Faljoni-Alario A.; Brochsztain S.; Santos P. S.; Nascimento O. R.; Nantes I. L. (2008) Spectroscopic, structural, and functional characterization of the alternative low-spin state of horse heart cytochrome C. Biophys. J. 94, 4066–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomono S.; Miyoshi N.; Shiokawa H.; Iwabuchi T.; Aratani Y.; Higashi T.; Nukaya H.; Ohshima H. (2011) Formation of cholesterol ozonolysis products in vitro and in vivo through a myeloperoxidase-dependent pathway. J. Lipid Res. 52, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J.; Lees M.; Stanley G. H. S. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509. [PubMed] [Google Scholar]
- Villen J.; Beausoleil S. A.; Gygi S. P. (2008) Evaluation of the utility of neutral-loss-dependent MS3 strategies in large-scale phosphorylation analysis. Proteomics 8, 4444–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigolo C. A.; Di Mascio P.; Medeiros M. H. (2007) Covalent modification of cytochrome c exposed to trans,trans-2,4-decadienal. Chem. Res. Toxicol. 20, 1099–1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.