Abstract
Leukemia inhibitory factor (LIF) has been recently identified as a p53 target gene, which mediates the role of p53 in maternal implantation under normal physiological conditions. Here, we report that LIF is a negative regulator of p53; LIF downregulates p53 protein levels and function in human colorectal cancer (CRC) cells. The downregulation of p53 by LIF is mediated by the activation of Stat3, which transcriptionally induces ID1. ID1 upregulates MDM2, a key negative regulator of p53, and promotes p53 protein degradation. LIF is overexpressed in a large percentage of CRCs. LIF overexpression promotes cellular resistance towards chemotherapeutic agents in cultured CRC cells and colorectal xenograft tumors in a largely p53-dependent manner. Overexpression of LIF is associated with a poor prognosis in CRC patients. Taken together, LIF is a novel negative regulator of p53, overexpression of LIF is an important mechanism for the attenuation of p53, which promotes chemoresistance in CRCs.
Introduction
p53 plays a critical role in tumor suppression 1, 2. p53 is the most frequently-mutated gene in human cancers; over 50% of all cancers harbor p53 mutations 3. As a transcription factor, p53 responds to stress signals and regulates the expression of its target genes, which leads to various cellular responses to prevent tumor formation 1. As a haplo-insufficient gene, a little decrease in p53 levels or activities (e.g. 2-fold difference) impacts greatly upon tumorigenesis 4–6. p53 is under tight regulation by a number of negative regulators, including MDM2, MDM4, Wip1, Pirh2, and Cyclin G1, to maintain its proper activities and function in cells 7–9. Many p53 negative regulators (e.g. MDM2, Wip1, Pirh2 and Cyclin G1) are p53 target genes, which form auto-regulatory negative feedback loops with p53. Overexpression and/or amplification of these negative regulators have been frequently observed in tumors, which leads to the attenuation of p53 function and promotes tumorigenesis 10, 11.
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the third leading cancer death in the United States 12. Around 50% of CRCs contain p53 mutations. Loss of p53 function plays a critical role in colorectal tumorigenesis by driving the progression of adenoma to carcinoma 13, 14. p53 is also critical for chemotherapeutic response in CRCs. Loss of p53 abolishes the apoptotic response to 5-flurorouracil (5-FU), a most commonly used chemotherapeutic agent for CRCs, in both cultured CRC cells and animal models 15, 16. In clinical, human CRCs with wild type p53 display a better response to 5-FU-based chemotherapy compared with CRCs with p53 mutations 17, 18. These findings demonstrate a critical role of p53 in tumor suppression and chemotherapeutic response in CRCs.
As a multi-functional protein, LIF plays different roles in a highly context-dependent manner. For instance, LIF induces the differentiation of murine myeloid leukemia cells, whereas inhibits the differentiation of murine embryonic stem cells 19, 20. LIF also plays a crucial role in embryonic implantation 21. LIF functions in autocrine and/or paracrine manners through binding to the LIF receptor complex composed of the LIF receptor (LIF-R) and gp-130, which in turn activates selective pathways, including the PI3K/AKT and JAK/Stat3 pathways, depending on the context 22–24.
Recently, we identified LIF as a novel p53 target gene. LIF is an important component of the p53 pathway, which mediates p53’s role in embryonic implantation 25–28. Considering the critical role of the p53 pathway in cancer, our findings suggest a potential role of LIF in cancer. However, to date, the role of LIF in tumorigenesis, especially CRC, is poorly understood. Here, we report that LIF has an important role in regulation of p53 function in tumor suppression; LIF negatively regulates p53 protein levels and function in human CRC cells. The negative regulation of p53 by LIF is through the activation of Stat3, which in turn induces the expression of ID1, the helix-loop-helix (HLH) protein inhibitor of differentiation and DNA binding. ID1 upregulates MDM2 expression, which leads to accelerated p53 protein degradation. LIF is overexpressed in a large percentage of human CRCs and is associated with a poor prognosis of CRC patients. Overexpression of LIF promotes chemoresistance in both cultured CRC cells and colorectal xenograft tumors in a largely p53-dependent manner.
Results
Overexpression of LIF in human CRCs
To study the potential role of LIF in CRCs, the expression of LIF was determined at both mRNA and protein levels in human CRC samples. The LIF mRNA levels were determined in 24 pairs of cDNA prepared from human CRC and their matched adjacent non-tumor tissues (Colorectal Cancer cDNA Array, Origene) by Taqman real-time PCR. LIF mRNA levels were significantly higher in CRCs than their adjacent non-tumor tissues (4.37-fold higher in average, p=6.6E-5; Fig. 1a). Furthermore, LIF protein levels were determined in 25 pairs of formalin-fixed and paraffin-embedded (FFPE) CRC specimen and their matched adjacent non-tumor tissues by Immunohistochemistry (IHC) staining. Consistently, a significantly higher percentage of CRC samples showed positive LIF staining (>10% cells are stained) compared with their matched adjacent non-tumor tissues (72% vs. 20%, p=0.005; Fig. 1b). These results demonstrated that LIF is overexpressed in a high percentage of CRCs, which suggests a potential role of LIF overexpression in colorectal tumorigenesis.
Figure 1. Overexpression of LIF in human CRCs.
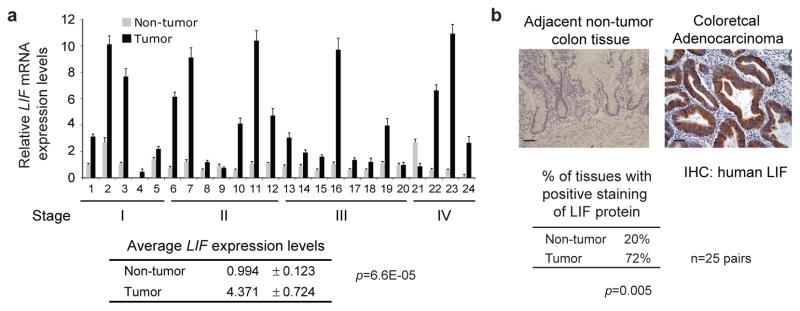
a. Human CRC tissues display significantly higher LIF mRNA expression levels compared with adjacent non-tumor tissues. cDNAs prepared from human CRC samples and their matched adjacent non-tumor tissues were provided by Origene. LIF mRNA expression levels were measured by Taqman real-time PCR and normalized with actin. LIF expression levels in a non-tumor tissue (#1) were designated as 1. Data were presented as mean±SD (n=3). p value was obtained by Student t-tests. b. Higher LIF protein levels in human CRC tissues. LIF protein levels were determined by IHC in human CRC samples and their matched adjacent non-tumor tissues (n=25). p value was obtained by Student t-tests. Representative IHC staining results for LIF in CRCs (right panel) and adjacent non-tumor colorectal tissues (left panel) are shown. Positive LIF staining: >10% cells stained with LIF. Scale bar: 50 μM.
LIF inhibits p53-mediaetd chemoresponse in human CRC cells
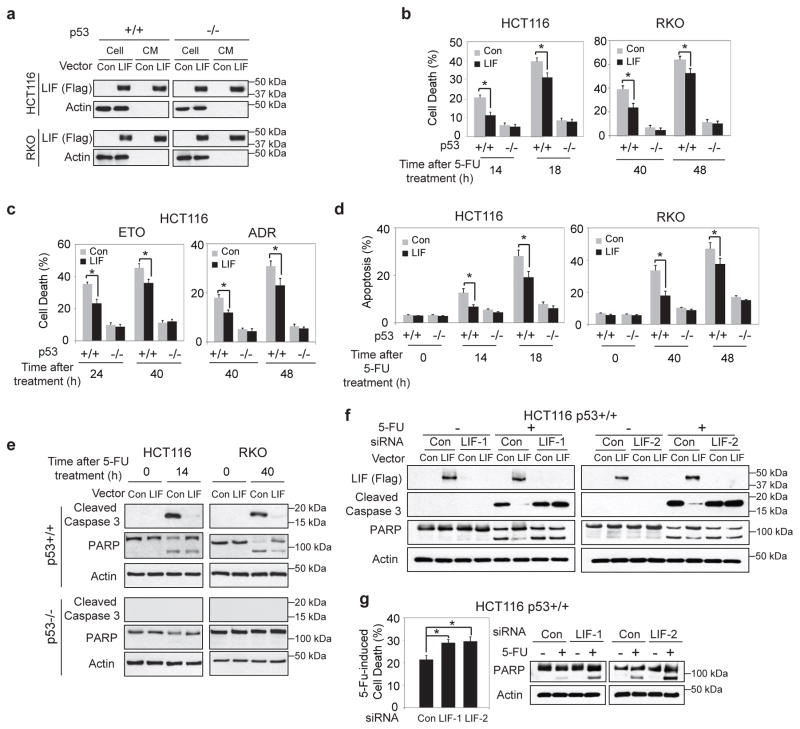
To investigate the potential role of LIF in CRCs, the effect of LIF on the cellular response towards chemotherapeutic agents was determined in a panel of human CRC cells. It has been well-established that p53 plays a central role in tumor suppression and is critical for an effective chemotherapeutic response in CRCs through induction of apoptosis. To this end, 3 pairs of isogenic human CRC cells with and without wild-type p53, including HCT116 p53+/+ & p53−/−, RKO p53+/+ & p53−/−, DLD-1 p53+/+ & p53−/− cells, were employed to establish stable cell lines with ectopic LIF expression. These cell lines were stably transduced with retroviral LIF expression vectors (pLPCX-LIF) (HCT116 p53+/+-LIF, HCT116 p53−/−-LIF, RKO p53+/+-LIF, RKO p53−/−-LIF, DLD-1 p53+/+-LIF and DLD-1 p53−/−-LIF). Cells transduced with empty vectors served as controls. The ectopic LIF expression in cells was confirmed at the protein level by Western-blot assays (Fig. 2a for HCT116 and RKO cells & Supplementary Fig. 1a for DLD-1 cells). The secretion of LIF into the conditioned medium was also confirmed (Fig. 2a). 5-FU clearly induced cell death in p53+/+ human CRC cells (HCT116 p53+/+, RKO p53+/+ and DLD-1 p53+/+), whereas had a very limited effect on cell death in their isogenic p53−/− cells. Notably, ectopic LIF expression markedly reduced 5-FU-induced cell death in p53+/+ CRC cells, but showed no apparent effect in isogenic p53−/− cells (Fig. 2b for HCT116 and RKO cells & Supplementary Fig. 1b for DLD-1 cells). A similar inhibitory effect of LIF on chemotherapeutic agents-induced cell death was observed in HCT116 p53+/+ cells treated with Etoposide (ETO) or Adriamycin (ADR), but not in HCT116 p53−/− cells that underwent same treatments (Fig. 2c). It has been well-established that 5-FU induces cell death in CRC cells mainly through p53-mediated apoptosis 15. The effect of LIF on 5-FU-induced apoptosis was determined in above-mentioned cells by employing Annexin V staining. 5-FU induced apoptosis in a highly p53-dependent manner; very limited apoptosis was observed in p53−/− cells treated with 5-FU (Fig. 2d), which is consistent with a previous report 15. Notably, ectopic LIF expression markedly decreased 5-FU-induced apoptosis in p53+/+ CRC cells (Fig. 2d for HCT116 and RKO cells & Supplementary Fig. 1c for DLD-1 cells). The inhibitory effect of LIF on p53-mediated apoptosis was further clearly demonstrated by the inhibition of activation and cleavage of caspase 3 and the subsequent proteolytic cleavage of poly (ADP ribose) polymerases (PARP) by LIF in p53+/+ but not p53−/− CRC cells treated with 5-FU (Fig. 2e for HCT116 and RKO cells & Supplementary Fig. 1d for DLD-1 cells). The inhibitory effect of LIF on p53-mediated apoptosis was also observed in HCT116 p53+/+ but not HCT116 p53−/− cells treated with ETO or ADR as determined by Annexin V staining and the levels of cleaved caspase 3 and PARP (Supplementary Fig. 2a & b). Knockdown of ectopic LIF in HCT116 p53+/+-LIF cells by siRNA oligos blocked the inhibitory effect of LIF on 5-FU-induced apoptosis (Fig. 2f). Consistently, knockdown of endogenous LIF by siRNA in HCT116 p53+/+ cells also clearly increased 5-FU-induced apoptosis (Fig. 2g). The knockdown of endogenous LIF was confirmed at both mRNA and protein levels (Supplementary Fig. 3). Taken together, these results demonstrated that LIF inhibits p53-mediated apoptosis, which in turn promotes chemoresistance in CRC cells.
Figure 2. LIF inhibits p53-mediated apoptosis in response to the treatments of chemotherapeutic agents in human CRC cells.
a. Two pairs of human CRC cell lines with and without p53 with stable ectopic LIF expression (HCT116 p53+/+-LIF, HCT116 p53−/−-LIF, RKO p53+/+-LIF and RKO p53−/−-LIF cells) and their control cells were established by transduction of LIF expression vectors (pLPCX-LIF) and control vectors, respectively. Ectopic LIF expression in cells was detected by Western-blot assays. The secretion of LIF from cells into the conditioned medium (CM) was also detected by Western-blot assays. CM was collected after serum-free medium was cultured overnight with 1/3 number of the cells used for Western-blot assays. b. LIF reduced 5-FU-induced cell death in a p53-dependent manner in cells. Above-mentioned cells with stable ectopic LIF expression and their control cells were treated with 5-FU (500 μM) for the indicated time periods. Cell viability was measured by Vi-CELL cell counter. c. LIF reduced Etoposide- and Adriamycin-induced cell death in a p53-dependent manner in HCT116 cells. HCT116 p53+/+-LIF, HCT116 p53−/−-LIF and their control cells were treated with Etoposide (ETO, 30 μM) or Adriamycin (ADR, 5 μM) for the indicated time periods. d&e. LIF reduced p53-meidated apoptosis induced by 5-FU in HCT116 and RKO cells. d. The percentage of apoptotic cells was determined by Annexin V staining. e. The levels of cleaved caspase 3 and PARP were determined by Western-blot assays. f. Knockdown of LIF in HCT116 p53+/+-LIF cells by two different siRNA oligos largely abolished the inhibitory effect of LIF on 5-FU-induced apoptosis. 5-FU-induced apoptosis was determined by measuring the levels of cleaved caspase 3 and PARP. g. Knockdown of endogenous LIF in HCT116 p53+/+ cells increased 5-FU-induced apoptosis. Cell viability (left panel) and apoptosis (right panel) were determined by Vi-CELL cell counter and the levels of cleaved PARP, respectively. For b, c, d & g, data are presented as mean ± SD (n=3). *: p<0.001 (Student t-test).
LIF decreases p53 protein levels and function in CRC cells
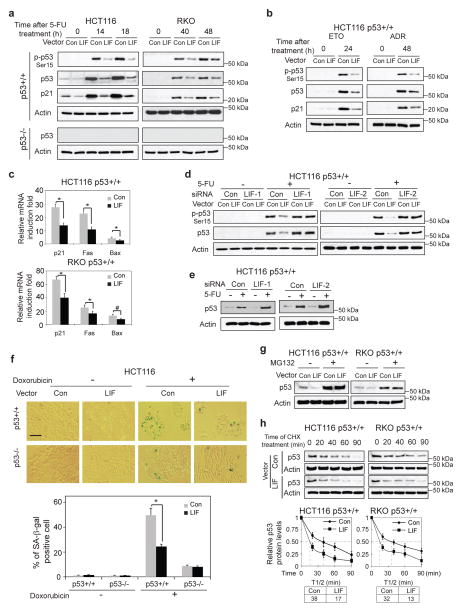
It has been well-established that in response to chemotherapeutic agents, including 5-FU, p53 is activated and its protein levels increase, which in turn initiates cellular responses, including apoptosis, to promote an efficient chemotherapeutic response 29. The levels of p53 protein are one of the most important determinants of its functions, including apoptosis. As shown in Supplementary Fig. 4, the degree of p53-meidated apoptosis is dependent upon the levels of p53 protein in H1299-Tet-on-p53 cells, which express p53 protein under the control of a Doxycycline-regulated promoter. While the cells did not express p53 protein in the absence of Doxycycline, low concentrations of Doxycycline (2.5 ng/ml, 50 ng/ml) induced p53 protein at a low level, and high concentrations of Doxycycline (1000 ng/ml) induced p53 protein at a much higher level. Furthermore, the levels of p53-mediated apoptosis were much lower in cells with low p53 protein levels than in cells with higher p53 levels (Supplementary Fig. 4). The observation that LIF inhibits chemotherapeutic agents-induced apoptosis in a largely p53-dependent manner raised the possibility that LIF may negatively regulate p53 protein levels and function. To test this possibility, HCT116 p53+/+-LIF, RKO p53+/+-LIF, DLD-1 p53+/+-LIF, and their control cells were treated with 5-FU. Their isogenic p53−/− cells with and without ectopic LIF expression were also employed. The phosphorylation of p53 at Ser15 (p-p53), which is the initial step of p53 activation in response to 5-FU treatment, and the accumulation of p53 and p21 proteins, a well-known p53 target, were measured by Western-blot assays. The induction of p-p53, p53 and p21 proteins in response to 5-FU was significantly lower in cells with ectopic LIF expression compared with control cells (Fig. 3a for HCT116 and RKO cells & Supplementary Fig. 5a for DLD-1 cells). The inhibitory effect of LIF on 5-FU-induced p53 protein accumulation was observed in the majority of cells as determined in HCT116 p53+/+ cells by immunofluorescence (IF) staining assays (Supplementary Fig. 5b). The inhibitory effect of LIF on 5-FU-induced p53 protein accumulation was also observed in non-apoptotic HCT116 p53+/+ cells that were sorted as cells negative for Annexin V staining by using a cell sorter (Supplementary Fig. 5c). These results demonstrated that the inhibitory effect of LIF on p53 existed in a major cell population. Similar results were observed in HCT116 p53+/+-LIF and control cells treated with ETO and ADR (Fig. 3b). LIF is a cytokine. The effect of exogenous LIF on p53 was also investigated. Exogenous LIF protein which was used to treat cells directly showed a similar inhibitory effect on the activation and accumulation of p53 protein, as well as the increase of p21 protein levels in response to 5-FU in HCT116 p53+/+ and RKO p53+/+ cells (Supplementary Fig. 5d).
Figure 3. LIF downregulates p53 protein levels and transcriptional activity through accelerating p53 protein degradation.
a. HCT116 p53+/+-LIF, HCT116 p53−/−-LIF, RKO p53+/+-LIF, RKO p53−/−-LIF and their control cells were treated with 5-FU. The levels of phosphorylated p53 at serine 15 (p-p53-Ser15), total p53 and p21 proteins were examined by Western-blot assays. b. HCT116 p53+/+-LIF and its control cells were treated with ETO (30 μM) or ADR (5 μM). The levels of p-p53-Ser15, p53 and p21 proteins were examined by Western-blot assays. c. HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells were treated with 5-FU (500 μM), the mRNA levels of p21, Fas and Bax were determined by Taqman real-time PCR and normalized with actin. d. Knockdown of LIF in HCT116 p53+/+-LIF cells largely blocked the inhibitory effect of LIF on the activation and accumulation of p53 protein in response to 5-FU treatment (500 μM). e. Knockdown of endogenous LIF in HCT116 p53+/+ cells increased p53 protein levels in response to 5-FU treatment. f. LIF negatively regulated p53 function in senescence. HCT116 p53+/+-LIF, HCT116 p53−/−-LIF and their control cells were treated with Doxorubicin (50 nM) for 3 days before senescent cells were detected by SA-β-gal staining. Upper panels: represented images of SA-β-gal staining of senescent cells. Scale bar: 100 μm. Lower panel: the percentage of SA-β-gal positive cells. g. Blocking proteasomal degradation by MG132 largely abolished the inhibitory effect of LIF on p53 protein levels in HCT116 p53+/+ and RKO p53+/+ cells. HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells were treated with MG132 (30μM) for 6 h. h. LIF decreased p53 protein half-life. HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells were treated with cycloheximide (CHX, 10 μg/ml) for the indicated time before being collected for Western-blot analysis. For c, f & h, data are presented as mean±SD (n=3). *: p<0.001 (Student t-test).
p53 functions as a transcription factor. The effect of LIF on p53 function in transcriptional activity was determined in HCT116 p53+/+-LIF, RKO p53+/+-LIF, DLD-1 p53+/+-LIF and their control cells. The mRNA levels of a group of well-known p53 target genes, including p21 (involved in cell cycle arrest), Fas and Bax (involved in apoptosis), were examined before and after 5-FU treatment by Taqman real-time PCR. 5-FU clearly induced the transcription of all these genes. Notably, the induction of these genes was significantly lower in cells with ectopic LIF expression compared with that in control cells (Fig. 3c for HCT116 p53+/+ and RKO p53+/+ cells & Supplementary Fig. 5e for DLD-1 p53+/+ cells). Knockdown of ectopic LIF by siRNA in HCT116 p53+/+-LIF cells largely abolished the inhibitory effect of LIF on 5-FU-induced activation and accumulation of p53 protein (Fig. 3d). Furthermore, knockdown of endogenous LIF clearly increased 5-FU-induced p53 protein accumulation in HCT116 p53 +/+ cells (Fig. 3e). The effect of LIF on p53 function in senescence was also determined in HCT116 p53+/+-LIF, HCT116 p53−/−-LIF and their control cells. Cells were treated with Doxorubicin, and senescent cells were detected by senescence associated β-galactosidase (SA-β-gal) assays. Doxorubicin effectively induced senescence in HCT116 cells in a largely p53-dependent manner (Fig. 3f). Notably, ectopic LIF expression clearly reduced Doxorubicin-induced senescence in HCT116 p53+/+ but not p53−/− cells (Fig. 3f).
p53 is mainly regulated at the post-translational level, and has a short protein half-life in cells under non-stressed conditions. Ectopic LIF expression did not clearly change p53 mRNA levels as determined by Taqman real-time PCR (Supplementary Fig. 6). p53 protein is mostly turned over through proteasomal degradation. Blocking proteasomal degradation by MG132, a proteasomal inhibitor, clearly increased p53 protein levels in HCT116 p53+/+ and RKO p53+/+ cells. Importantly, MG132 treatment largely abolished the inhibitory effect of LIF on the p53 protein level in both cell lines (Fig. 3g). Consistently, the half-life of p53 protein was clearly decreased in HCT116 p53+/+-LIF and RKO p53+/+-LIF cells (Fig. 3h). Taken together, these results strongly suggest that LIF down-regulates p53 protein levels and function through accelerating p53 protein degradation in cells.
LIF upregulates MDM2 to downregulate p53 through Stat3
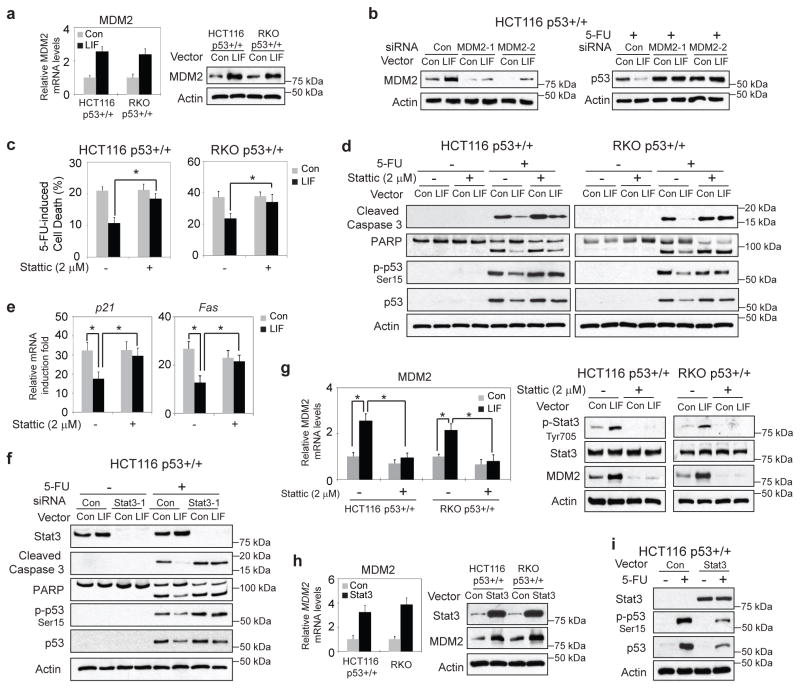
MDM2 is a key negative regulator of p53. As an E3 ubiquitin ligase, MDM2 binds to and ubiquitinates p53 for proteasomal degradation. Interestingly, we found that LIF increased the expression of MDM2 at both mRNA and protein levels (Fig. 4a). Knockdown of MDM2 in HCT116 p53+/+-LIF cells by siRNA oligos blocked the inhibitory effect of LIF on p53 protein accumulation in response to 5-FU treatment (Fig. 4b). Stat3 is a critical downstream effector of the LIF signaling in many types of cells and tissues 19, 23. To investigate whether the activation of Stat3 by LIF mediates the inhibitory effect of LIF on p53 protein levels and function in CRC cells, Stattic, a specific Stat3 inhibitor, was employed to block Stat3, and the effect of LIF on p53 was determined. HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells were treated with 5-FU along with or without Stattic treatment. Stattic greatly attenuated the inhibitory effect of LIF on 5-FU-induced cell death as well as the p53-mediated apoptosis induced by 5-FU (Fig. 4c&d). Importantly, Stattic treatment largely abolished the inhibitory effect of LIF on 5-FU-induced activation and accumulation of p53 protein in HCT116 p53+/+ and RKO p53+/+ cells (Fig. 4d). Stattic treatment also largely abolished the inhibitory effect of LIF on p53 transcriptional activity towards its target genes, including p21 and Fas, in HCT116 p53+/+ cells (Fig. 4e). Consistent results were obtained when the Stat3 function was blocked by using siRNA targeting Stat3. Knockdown of endogenous Stat3 by siRNA largely abolished the inhibitory effect of LIF on 5-FU-induced apoptosis, the activation and accumulation of p53 protein in HCT116 p53+/+ cells (Fig. 4f & Supplementary Fig. 7a). Importantly, the upregulation of MDM2 levels by LIF is largely mediated by Stat3; Stattic clearly blocked the increase of MDM2 levels in HCT116 p53+/+-LIF and RKO p53+/+-LIF cells (Fig. 4g). Consistently, ectopic Stat3 expression in HCT116 p53+/+ cells clearly increased MDM2 mRNA and protein levels (Fig. 4h). While Stat3 expression did not have an apparent effect on p53 mRNA levels (Supplementary Fig. 7b), it clearly decreased the activation and accumulation of p53 protein in response to 5-FU treatment (Fig. 4i). Collectively, these results demonstrated that LIF activates the Stat3 signaling pathway, which in turn upregulates MDM2 and downregulates p53 protein levels and function.
Figure 4. LIF upregulates MDM2 to downregulate p53 through activation of the Stat3 signaling.
a. LIF increased the expression levels of MDM2. The mRNA and protein levels of MDM2 were examined in HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells. b. Knockdown of MDM2 in HCT116 p53+/+-LIF cells largely blocked the inhibitory effect of LIF on 5-FU-induced p53 protein accumulation (right panel). The knockdown of MDM2 was confirmed at the protein levels (left panel). c.&d. HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells were treated with 5-FU (500 μM) along with or without Stattic (2 μM), a specific inhibitor of Stat3. c. Cell viability was measured by Vi-CELL cell counter. d. The levels of cleaved caspase 3, PARP, p-p53-Ser15 and p53 proteins were examined by Western-blot assays. e. HCT116 p53+/+-LIF and its control cells were treated with 5-FU along with or without Stattic. The mRNA levels of p21 and Fas were determined by Taqman real-time PCR and normalized with actin. f. HCT116 p53+/+-LIF and its control cells were transfected with siRNA targeting Stat3 (siRNA Stat3-1) or control siRNA, followed by 5-FU treatment. The levels of Stat3, cleaved caspase 3, PARP, p-p53-Ser15 and p53 proteins were determined by Western-blot assays. Similar results were obtained when a different Stat3 siRNA (siRNA Stat3-2) were used (see Supplementary Fig. 7a). g. The upregulation of MDM2 by LIF was largely abolished by Stattic. HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells were treated with Stattic. The MDM2 mRNA levels were determined by Taqman real-time PCR and normalized with actin (left panel). The levels of phosphorylated Stat3 at Tyrosine 705 (p-Stat3-Tyr705), Stat3 and MDM2 proteins were examined by Western-blot assays (right panels). h. Stat3 increased the expression levels of MDM2. The MDM2 mRNA and protein levels were examined in HCT116 p53+/+ and RKO p53+/+ cells transfected with Stat3 expression vectors or control vectors. i. Ectopic Stat3 expression reduced the activation and accumulation of p53 protein in response to 5-FU in HCT116 p53+/+ cells. For a., c., e. g. & h., data are presented as mean ± SD (n=3). *: p<0.001 (Student t-test).
Stat3 upregulates ID1 to mediate the effect of LIF on p53
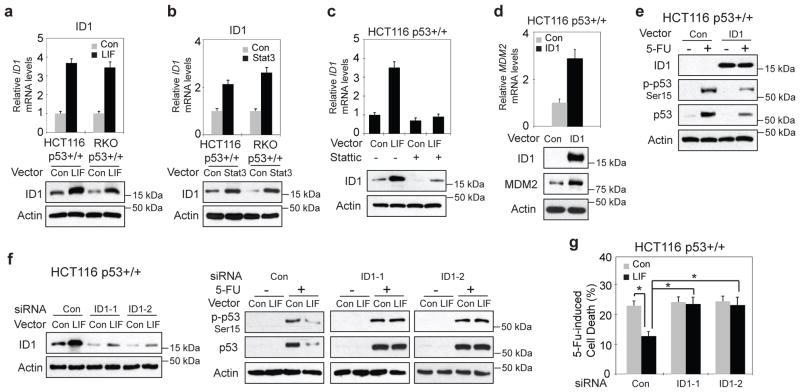
Stat3 is a transcription factor. Upon activation, Stat3 protein forms dimers that translocate to the nucleus to transcriptionally induce their target genes. ID1, the helix-loop-helix (HLH) protein inhibitor of differentiation and DNA binding, has been previously reported to upregulate MDM2 in human esophageal squamous cells 30. Interestingly, we found that ectopic LIF expression induced the expression of ID1 at both mRNA and protein levels in HCT116 p53+/+ and RKO p53+/+ cells (Fig. 5a). The induction of ID1 by LIF is mediated through Stat3; ectopic Stat3 expression induced ID1 at both mRNA and protein levels (Fig. 5b), whereas blocking Stat3 by Stattic largely abolished the induction of ID1 by LIF in HCT116 p53+/+ and RKO p53+/+ cells (Fig. 5c for HCT116 p53+/+ cells & Supplementary Fig. 8 for RKO p53+/+ cells). The induction of ID1 and MDM2 by ectopic LIF expression was also observed in HCT116 p53−/− and RKO p53−/− cells, suggesting that the induction of ID1 by LIF is p53 independent (Supplementary Fig. 9).
Figure 5. Stat3 induces the expression of ID1 to mediate the inhibitory effect of LIF on p53.
a. LIF induced ID1 expression in cultured cells. The mRNA and protein levels of ID1 were determined in HCT116 p53+/+-LIF, RKO p53+/+-LIF and their control cells. b. Stat3 induced ID1 expression in cultured cells. The mRNA and protein levels of ID1 were determined in HCT116 p53+/+ and RKO p53+/+ cells transfected with Stat3 expression vectors or control vectors. c. The induction of ID1 by LIF was largely blocked by Stattic. HCT116 p53+/+-LIF and their control cells were treated with 2 μM Stattic. The expression levels of ID1 were determined at both mRNA and protein levels. d. Ectopic ID1 expression by expression plasmid (pCMV-ID1) induced MDM2 expression as determined at both mRNA and protein levels in HCT116 p53+/+ cells. e. Ectopic ID1 expression decreased the activation and accumulation of p53 protein in response to 5-FU in HCT116 p53+/+ cells. f. Knockdown of endogenous ID1 by siRNA largely abolished the inhibitory effect of LIF on p53 activation and accumulation in response to 5-FU in HCT116 p53+/+ cells. The knockdown of ID1 was confirmed at the protein level (left panel). g. Knockdown of ID1 largely abolished the inhibitory effect of LIF on 5-FU-induced cell death in HCT116 p53+/+ cells. For a.–d. & g., data are presented as mean ± SD (n=3). *: p<0.001 (Student t-test).
To investigate whether ID1 mediates the inhibitory effect of LIF on p53, the effect of ID1 on p53 was determined in HCT116 p53+/+ cells. Ectopic ID1 expression clearly induced MDM2 at both mRNA and protein levels in HCT116 p53+/+ cells (Fig. 5d), which is similar to the observation in cells with ectopic expression of LIF or Stat3. Furthermore, ectopic ID1 expression reduced the activation and accumulation of p53 protein levels in response to 5-FU treatment (Fig. 5e). Importantly, knockdown of endogenous ID1 expression in HCT116 p53+/+ cells largely reduced the inhibitory effect of LIF on p53 protein levels and 5-FU-induced cell death (Fig. 5f & g). These data strongly suggest that Stat3 induces the expression of ID1 to mediate the inhibitory effect of LIF on p53 and p53-mediated apoptosis.
LIF promotes chemoresistance in colorectal xenograft tumors
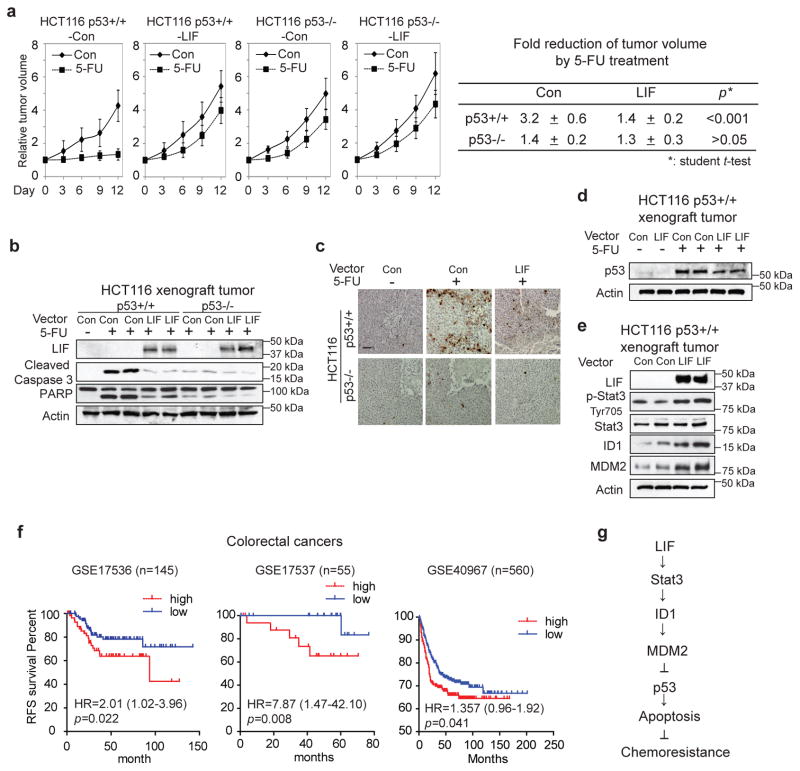
To investigate whether the downregulation of p53 by LIF promotes chemoresistance in vivo, the effect of LIF on the therapeutic response to 5-FU was determined in colorectal xenograft tumors. BALB/c nude mice were inoculated with HCT116 p53+/+-LIF, HCT116 p53−/−-LIF and their control cells. When tumors reached the size of ~ 200 mm3, mice were treated with 5-FU (i.p., 30mg/kg/daily) for 12 days. 5-FU inhibited the growth of xenograft tumors in a largely p53-dependent manner; 5-FU clearly inhibited the growth of HCT116 p53+/+ tumors and had much less pronounced effect on HCT116 p53−/− tumors (Fig. 6a). In response to 5-FU treatment, average HCT116 p53+/+-Con tumor sizes reduced by 3.2 fold while average HCT116 p53+/+-LIF tumor sizes only reduced by 1.4 fold (p<0.001, student’s t-test) (Fig. 6a). Notably, the promoting effect of LIF on the therapeutic resistance to 5-FU was greatly reduced in HCT116 p53−/− tumors (Fig. 6a). 5-FU induced p53-mediated apoptosis in HCT116 tumors; 5-FU treatment clearly increased the levels of cleaved caspase 3 and PARP in HCT116 p53+/+ tumors, but had a very limited effect in HCT116 p53−/− tumors. Ectopic LIF expression greatly reduced apoptosis induced by 5-FU in HCT116 p53+/+ tumors; the levels of cleaved caspase 3 and PARP were much lower in HCT116 p53+/+-LIF tumors compared with HCT16 p53+/+ control tumors in response to 5-FU treatment (Fig. 6b). In contrast, LIF displayed a very limited effect on 5-FU-induced apoptosis in HCT116 p53−/− tumors (Fig. 6b). These results were confirmed by IHC staining of cleaved caspase 3 in HCT116 xenograft tumors; the number of apoptotic cells with positive staining of cleaved caspase 3 were much lower in HCT116 p53+/+-LIF tumors compared with HCT116 p53+/+-Con tumors (Fig. 6c). Consistent with the observation made in in vitro cultured cells, the levels of basal p53 protein and the accumulation of p53 protein in response to 5-FU treatment were much lower in HCT116 p53+/+-LIF tumors than HCT116 p53+/+-Con tumors (Fig. 6d). Furthermore, LIF overexpression activated Stat3, increased ID1 and MDM2 protein levels in HCT116 p53+/+ xenograft tumors (Fig. 6e). These results suggest that the downregulation of p53 protein levels and function is an important mechanism by which LIF promotes chemoresistance in colorectal xenograft tumors.
Figure 6. LIF promotes chemoresistance in colorectal HCT116 xenograft tumors.
a. LIF promoted chemoresistance toward 5-FU in a largely p53-dependent manner in HCT116 xenograft tumors. HCT116 p53+/+-LIF, HCT116 p53−/−-LIF and their control cells were employed for xenograft tumor formation in nude mice. When tumor volumes reached 200 mm3, mice were treated with 5-FU (30 mg/kg/daily) or vehicle for 12 days. Relative tumor volumes are presented as mean±SD, n=8/group. The fold reduction of tumor volumes by 5-FU was much lower in HCT116 p53+/+-LIF tumors than HCT116 p53+/+-Con tumors, and no clear difference was observed between HCT116 p53−/−-LIF and HCT116 p53−/−-Con tumors. p value was obtained by ANOVA, followed by Student t-tests. b. & c. LIF inhibited 5-FU-induced apoptosis in a largely p53-dependent manner in HCT116 xenograft tumors. b. The levels of LIF, cleaved caspase 3 and PARP were examined by Western-blot assays in xenograft tumors. c. 5-FU-induced apoptosis was determined by IHC staining of cleaved caspase 3 in xenograft tumors. Scale bar: 50 μm. d. The levels of p53 protein were determined in HCT116 p53+/+-LIF and HCT116 p53+/+-Con tumors treated with and without 5-FU. e. LIF activated the Stat3 signaling, increased ID1 and MDM2 protein levels in HCT116 p53+/+ xenograft tumors. The levels of LIF, total and phosphorylated Stat3 (p-Stat3-Tyr705), ID1 and MDM2 proteins in xenograft tumors formed by HCT116 p53+/+-LIF and HCT1116 p53+/+-Con cells were examined by Western-blot assays. f. The correlation between higher LIF levels and a poor prognosis in relapse-free survival (RFS) of CRC patients. Kaplan-Meier curves indicate the RFS of CRC patients with different expression levels of LIF from three publically available datasets (GSE17536, GSE17537, and GSE40967). The patients whose tumors had higher LIF mRNA levels had significantly shorter RFS than patients whose tumors had lower LIF mRNA levels. p value was obtained by Kaplan-Meier statistics and log-rank (one tail) test. g. Schematic model depicting the negative regulation of p53 by LIF as an important mechanism that contributes to chemoresistance in human CRCs.
LIF is associated with a poor prognosis in CRC patients
To further evaluate the clinical importance of LIF in CRC, the prognostic value of LIF mRNA expression was analyzed in three publically available datasets of CRC patients (GSE17536, GSE17537 and GSE40967). Patients were divided into two groups according to the mean expression levels of LIF mRNA. As shown in Fig. 6f, there was a significant association of high LIF expression levels with a poor prognosis of relapse-free survival in all three cohorts tested. These results suggest that high LIF levels predict a poor prognosis of CRC patients, which supports our results from both cultured cells and mouse tumor models.
Taken together, these results clearly demonstrated that LIF attenuates p53 function, which in turn promotes the therapeutic resistance to 5-FU, and could be a prognostic marker for poor relapse-free survival in CRC patients.
Discussion
LIF is a multifunctional cytokine that plays a wide array of functions depending on cell and tissue types. LIF is essential for the maintenance of mouse embryonic stem cell pluripotency, regulation of inflammation, and also plays a critical role in blastocyst implantation 19, 21, 31. Our recent studies demonstrated that LIF is a p53 target and mediates p53 function in reproduction 25, 27, 28. LIF expression in uterus is under the control of p53, especially at the implantation stage, to ensure the proper embryonic implantation. Loss of p53 in female mice leads to impaired implantation 25. The deceased activity of the p53 pathway is associated with the decreased maternal fertility in human populations 27. While LIF functions as a downstream target of p53 to mediate p53 function in maternal reproduction, results from this study clearly demonstrated an additional important role of LIF in regulating p53 levels and function in tumor suppression in CRCs. LIF downregulates p53 protein levels and function, including transcriptional activity, apoptosis and senescence, which forms a negative feedback loop with p53. Results in this study further showed that LIF is overexpressed in a large percentage of human CRCs and is associated with a poor prognosis of CRC patients. Overexpression of LIF promotes chemoresistance in cultured CRC cells and colorectal xenograft tumors in a largely p53-dependent manner (Fig. 6g). Therefore, overexpression of LIF could be an important factor contributing to the chemoresistance in CRCs.
The pleiotropic effects of LIF signaling is mainly due to the selective activation of different kinases, including JAKs, MAPK, PI3K, and transcription factors, including Stat3, AP1, in a highly context-dependent manner. In this study, we investigated the mechanisms by which LIF negatively regulates p53. We found that the inhibitory effect of LIF on p53 is largely mediated by the activation of Stat3. Blocking the Stat3 pathway largely abolishes the inhibitory effect of LIF on p53. Stat3 is frequently activated in many types of human cancers, including CRC, and is crucial to the survival and growth of tumor cells 32–35. The regulation of mouse p53 by Stat3 has been suggested by a previous study showing that Stat3 binds to the promoter of mouse p53 gene and inhibits p53 expression at mRNA level in mouse cell lines 36. In this study, we found that Stat3 downregulates p53 protein levels but with no apparent effect on p53 mRNA levels in human cells. Furthermore, we found that Stat3 can upregulate MDM2, a key negative regulator of p53 protein, which is an unidentified mechanism by which Stat3 downregulates p53 in human CRC. Together, these findings suggest that Stat3 negatively regulates p53 in both mouse and human cell lines, but through different mechanisms.
We further identified that Stat3 transcriptionally induces ID1 to mediate its role in the negative regulation of p53. ID1 is often under the transcriptional regulation by the bone morphogenetic proteins (BMP)/Smad pathway. BMP activates Smad1/5, which binds to Smad4, then translocates to the nucleus and binds to Smad-binding elements in the promoter of ID1 to induce ID1 transcription 37, 38. In addition to the BMP/Smad pathway, ID1 can also be transcriptionally regulated by Scr, EGR-1 (early growth response protein 1) 39, 40. Results in this study demonstrated that Stat3 is an additional important regulator of ID1 expression. Consistently, analysis of a panel of supratentorial primitive neuroectodermal tumors (PNET) showed the activation of the Stat3 pathway along with the increased mRNA expression of ID1, suggesting the possible regulation of ID1 by the Stat3 pathway in PNETs 41. ID1 belongs to a family of HLH transcriptional regulatory factors which is involved in the regulation of gene expression. ID1 often functions as a transcription repressor through heterodimerization with the basic HLH transcription factors to inhibit their transcriptional function 42. ID1 has also been shown to upregulate the expression of some genes through different mechanisms 43–46. For example, ID-1 has been shown to induce VEGF transcription by stabilizing hypoxia-inducible factor 1α protein 43. ID1 can activate NF-κB signaling and upregulate its target genes, including Bcl-xL and ICAM-1 45. It has also been reported that ID1 upregulates MMP-2 and EGF-R 44, 46. ID1 plays an important role in development, stem cell self-renewal activity, senescence, differentiation, angiogenesis and migration 47. Overexpression of ID1 has been reported in a variety of human cancers, including CRC, and is often associated with poor prognosis and chemoresistance 48–51. In this study, we found that overexpression of ID1 clearly increases MDM2 mRNA and protein levels in human CRC cells, which in turn down-regulates p53 protein levels. Currently, it is still unclear how ID1 transcriptionally regulates MDM2, which needs further studies in the future. Importantly, knockdown of endogenous ID1 largely abolishes the effect of LIF on MDM2 and p53, demonstrating that downregulation of p53 by LIF/Stat3 signaling is largely mediated by ID1 (Fig. 6g).
The role of LIF in cancer and its underlying mechanisms are not well-understood. LIF has been shown to have a complex role in cancer from limited studies. LIF can induce the differentiation of murine myeloid leukemia cells and suppress their proliferation 52. Meanwhile, LIF displays a promoting effect on tumorigeneis in many solid tumors 53–55. LIF promotes proliferation of cancer cell lines, and is involved in the metastasis of cells from breast cancer, rhabdomyosarcoma, melanoma and nasopharyngeal carcinoma 54–57. The increased expression levels of LIF are associated with poor prognosis in patients of breast cancer and nasopharyngeal cancer 54, 55. Results in this study showed that LIF promotes chemoresistance towards a group of chemotherapeutic agents in CRC, and demonstrated that the downregulation of p53 by LIF is an important underlying mechanism. It is worth noting that majority of results in this study were obtained by using engineered cell lines with ectopic LIF expression or knockdown of endogenous LIF. Considering the limitation of engineered cell lines, it will be important to confirm the effect of LIF on chemoresistance and its underlying mechanisms in primary human tumors in future studies. In clinical, ~50% of CRC patients develop recurrent disease despite the use of surgical resection and chemotherapy. By employing several publically available datasets of CRC patients, we found that high LIF levels in tumors are associated with shorter recurrent-free survival of CRC patients.
In summary, this study demonstrates that LIF, which is overexpressed in a large percentage of human CRCs, is an important negative regulator of p53 through activating the Stat3/ID1/MDM2 pathway, which in turn promotes chemoresistance in CRC. Targeting LIF, Stat3 as well as ID1 to reactivate p53 is a potential therapeutic strategy to enhance chemosensitivity in CRC, especially in tumors with LIF overexpression.
MATERIAL AND METHODS
Cell culture and cell treatments
Human CRC HCT116 p53+/+, HCT116 p53−/−, RKO p53+/+, RKO p53−/−, DLD-1 p53+/+ and DLD-1 p53−/− cell lines were generous gifts from Dr. Bert Vogelstein at Johns Hopkins University. Cells with stable ectopic LIF expression were established by transduction of a retroviral LIF expression vector (pLPCX-LIF) and selected by puromycin. Cells with inducible p53 expression (H1299-Tet-on-p53) were established by stable transfection of a Tet-on p53 expression vector (Life technology) into p53-null human H1299 cells, which express p53 only in the presence of Doxycycline. Conditioned medium (CM) was collected after serum-free medium was cultured overnight with cells. Human Stat3 expression vector (EF.STAT3C.Ubc.GFP vector) was constructed by the group of Dr. L. Resar at Johns Hopkins University 58. pCMV-ID1 expression vector was constructed by amplifying ID1 cDNA using following primers: forward: 5′CCCAAGCTTATGAAAGTCGCCAGTGGCA-3′; reverse: 5′ CGGGATCCGCGACACAAGATGCGATCGT 3′, and inserting ID1 fragment into p3XFlag-CMV-14 vectors (Sigma). Expression plasmids were transfected into cells using lipofectamine 2000 (Invitrogen). For siRNA knockdown, 2 different siRNA oligos against LIF, Stat3 and ID1, respectively, were purchased from IDT. siRNA target LIF: siRNA-1: 5′ CAACAACCUGGACAAGCUAUGUGGC 3′; siRNA-2: 5′ GUCACAACCUCAUGAACCAGAT 3′. siRNA target Stat3: siRNA-1: 5′ UCCAGUUUCUUAAUUUGUUGACGGGUC 3′; siRNA-2: 5′ AUAGUCCUAUCUUCUAUUUGGAUGUCA 3′. siRNA target ID1: siRNA-1: 5′ AUAUUACAAUGAUCACCGACUGAAA 3′; siRNA-2: 5′ GGAAUUACGUGCUCUGUGGGUCUCC 3′. Two different siRNA oligos against MDM2 were purchased from Qiagen (Cat#SI00300846) and Dharmacon (Cat#M-003279-01). 5-FU, Etoposide, Adriamycin, MG132, cycloheximide (CHX), Doxorubicin and Stattic were purchased from Sigma.
Tissue samples
The Colon Cancer cDNA array that contains 24 pairs of cDNA prepared from human CRC samples and their matched adjacent non-tumor tissues was purchased from Origene. Twenty-five cases of FFPE primary CRC samples and their matched adjacent non-tumor tissues were collected at the University of Texas MD Anderson Cancer Center with approved IRB.
Western-blot assays
Standard Western-blot assays were used to analyze the levels of protein in cell lysates and CM. CM which was cultured with 1/3 number of the cells used for Western-blot assays was concentrated with an Amico Ultra-4 centrifugal filter device (Millipore) after a brief centrifugation to remove any cell debris. Antibodies against p53 (FL393, Santa Cruz; 1:1,000 dilution), anti-p-p53 (Ser15) (9284, Cell Signaling; 1:1,000 dilution), anti-LIF (AF-250-NA, R&D; 1:1,000 dilution), anti-PARP1/2 (H250, Santa Cruz; 1:1,000 dilution), anti-cleaved-caspase 3 (D175, Cell Signaling; 1:1,000 dilution), anti-MDM2 (2A10; 1:1,000 dilution), anti-p-Stat3 (Tyr705) (9131, Cell Signaling; 1:1,000 dilution), anti-Stat3 (C-20, Santa Cruz; 1:2,000 dilution), anti-ID1 (C-20, Santa Cruz; 1:2,000 dilution), anti-p21 (Ab-6, Calbiochem; 1:1,000 dilution, and anti–β-actin (A5441, Sigma; 1:125,000 dilution) antibodies were used in this study. The full blots are shown in Supplementary Fig. 10 – Supplementary Fig. 14.
Immunofluorescence staining assays
IF staining was performed as previously described 59. In brief, cells grown on slides were fixed with 4% paraformaldehyde for 30 min and treated with 0.5% TritonX-100 for 5 min, blocked with 1% bovine serum albumin for 30 min, and stained with anti-p53 (FL393; 1:300 dilution), and anti-Flag (1:200 dilution) antibodies overnight at 4°C to detect p53 and LIF-Flag, respectively. Slides were washed and then incubated with Alexa Fluor® 555 Goat Anti-Mouse IgG (H+L) (Invitrogen) and Alexa Fluor® 488 Goat Anti-Rabbit IgG (H+L) (Invitrogen). Nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI; Vector).
Taqman real-time PCR
Total RNA was prepared by using an RNeasy kit (Qiagen). All primers were purchased from Applied Biosystems. Real-time PCR was done in triplicate with TaqMan PCR mixture (Applied Biosystems). The expression of genes was normalized to the actin gene.
Assays for cell viability and Annexin V staining
The Vi-CELL cell counter (Beckman Coulter) that performs a trypan blue exclusion method was employed to determine cell viability. Apoptosis was measured by staining cells using Muse™ Annexin V & Dead Cell Assay Kit (Millipore) and analyzing cells in a bench flow cytometry, the Muse Cell Analyzer (Millipore) according to manufacturer’s instructions.
Cell sorting
HCT116 p53+/+-LIF and their control cells treated with 5-FU were trypsinized, washed with PBS and stained with Alexa Fluor® 488 Annexin V according to the manufacturer’s protocol (Invitrogen). After staining, cells were analyzed and cells negative for Annexin V staining were sorted by a BD Biosciences ultra-high-speed Influx cell sorter.
Cellular senescence analysis
Cells were treated with Doxorubicin (50 nM) for 3 days. Senescent cells were detected by senescence associated β-galactosidase assays using a Senescence β-Galactosidase Staining Kit (Cell Signaling).
5-FU treatment of colorectal xenograft tumors
The 7-week-old BALB/c nu/nu male athymic nude mice (Taconic) were inoculated (via s.c. injection) with HCT116 p53+/+ and HCT116 p53−/− cells with or without ectopic LIF expression (5 × 106 in 0.2 ml PBS) for xenograft tumor formation. When tumor volumes reach 200 mm3, mice were treated with 5-FU (i.p., 30mg/kg/daily) or vehicle for 12 days. Tumor volume and mouse weight were monitored 3 times/week. The mouse experiments were performed with the approval of the Institutional Animal Care and Use Committee of Rutgers State University of New Jersey.
IHC staining assays
IHC staining for LIF and Cleaved-caspase 3 was performed as previously described 60. In brief, tissue sections were deparaffinized in xylene and rehydrated with ethanol. After pre-incubation with 10% normal goat serum in PBS (pH 7.5), tissue sections were incubated with primary antibodies, including anti-LIF (AF-250-NA, R&D, 1:100 dilution) and anti-Cleaved caspase 3 antibodies (D175, cell signaling, 1:1,000 dilution), for overnight at 4°C. Tissue sections were then stained with biotinylated secondary antibody (Vector). Immunoreactivity was detected by using a Vectastain Elite ABC kit (Vector). Known positive controls were included in each experiment, and negative controls were obtained by omitting the primary antibody. The IHC results were scored according to the percentage of cells showing positive staining: -: 0%–<10%; +: ≥10%.
Datasets of colorectal cancer patients
Three cohorts of CRC patients with known LIF mRNA expression levels and relapse-free survival information were obtained from publically available datasets (GSE17536, GSE17537, and GSE40967) 61–63. Genome-wide expression levels of all tumor specimens were assessed on Affymetrix HGU133 Plus 2.0 arrays. To analyze the prognostic value of LIF expression levels, the patients were split into two groups according to the mean expression levels of LIF in tumors.
Statistical analysis
The data were expressed as mean±SD. The differences in xenograft tumor size in response to 5-FU treatment were analyzed for statistical significance by ANOVA, followed by Student’s t-tests using a GraphPad Prism software. Kaplan-Meier statistics and log-rank (one tail) test were performed to estimate the significance of differences in relapse-free survival of patients among different groups. All other p-values were obtained using two-tailed Student t-tests. The results were considered to be significant, when p value < 0.05.
Supplementary Material
Acknowledgments
We thank Arthur I. Roberts for his technical assistance in cell sorting analysis. W.H. is supported by the grants from NIH (1R01CA160558), DOD (W81XWH-10-1-0435), the Ellison Foundation, the New Jersey Health foundation, ACS (RSG-12-082-01-TBG) and the New Investigator Award of Rutgers Cancer Institute of New Jersey. Z.F. is supported by the grants from NIH (1R01CA143204) and NJCCR. This research was in part supported by the Flow Cytometry Core Facility of Rutgers Robert Wood Johnson Medical School, a Shared Resource of The Rutgers Cancer Institute of New Jersey (P30CA072720) and NIH Shared Instrumentation Grant (1 S10 RR025468-01).
Footnotes
Author Contributions
Z.F., W.H. designed experiments. H.Y., X.Y., Y. Z., X.L., L.W., C. Z., Z.L., K. L., Z.X., K. Y., J. L. performed experiments. H.Y., Z. S., Z. F., W.H. analyzed data. H.Y., Z.F., W.H. wrote the manuscript.
Authors declare no competing financial interests.
References
- 1.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004:247–270. [PubMed] [Google Scholar]
- 4.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 5.Hu W, et al. A single nucleotide polymorphism in the MDM2 gene disrupts the oscillation of p53 and MDM2 levels in cells. Cancer Res. 2007;67:2757–2765. doi: 10.1158/0008-5472.CAN-06-2656. [DOI] [PubMed] [Google Scholar]
- 6.Bond GL, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 7.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 8.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leng RP, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 10.Li J, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 11.Duan W, et al. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J Natl Cancer Inst. 2004;96:1718–1721. doi: 10.1093/jnci/djh292. [DOI] [PubMed] [Google Scholar]
- 12.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 13.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 14.Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat. 2003;21:271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- 15.Hwang PM, et al. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe SW, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 17.Elsaleh H, et al. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res. 2001;7:1343–1349. [PubMed] [Google Scholar]
- 18.Russo A, et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23:7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 19.Metcalf D. The unsolved enigmas of leukemia inhibitory factor. Stem cells. 2003;21:5–14. doi: 10.1634/stemcells.21-1-5. [DOI] [PubMed] [Google Scholar]
- 20.Williams RL, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 21.Stewart CL, et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi Y, et al. Leukemia inhibitory factor regulates trophoblast giant cell differentiation via Janus kinase 1-signal transducer and activator of transcription 3-suppressor of cytokine signaling 3 pathway. Mol Endocrinol. 2008;22:1673–1681. doi: 10.1210/me.2008-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinrich PC, et al. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. The Biochemical journal. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe S, et al. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25:2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 25.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 26.Hu W. The role of p53 gene family in reproduction. Cold Spring Harb Perspect Biol. 2009;1:a001073. doi: 10.1101/cshperspect.a001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang HJ, et al. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A. 2009;106:9761–9766. doi: 10.1073/pnas.0904280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Z, et al. Regulation of female reproduction by p53 and its family members. FASEB J. 2011;25:2245–2255. doi: 10.1096/fj.10-180166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunz F, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui CM, et al. Id-1 promotes proliferation of p53-deficient esophageal cancer cells. Int J Cancer. 2006;119:508–514. doi: 10.1002/ijc.21874. [DOI] [PubMed] [Google Scholar]
- 31.Gadient RA, Patterson PH. Leukemia inhibitory factor, Interleukin 6, and other cytokines using the GP130 transducing receptor: roles in inflammation and injury. Stem cells. 1999;17:127–137. doi: 10.1002/stem.170127. [DOI] [PubMed] [Google Scholar]
- 32.Lassmann S, et al. STAT3 mRNA and protein expression in colorectal cancer: effects on STAT3-inducible targets linked to cell survival and proliferation. Journal of clinical pathology. 2007;60:173–179. doi: 10.1136/jcp.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin L, et al. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226–7237. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bollrath J, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Niu G, et al. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Rovira T, Chalaux E, Massague J, Rosa JL, Ventura F. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J Biol Chem. 2002;277:3176–3185. doi: 10.1074/jbc.M106826200. [DOI] [PubMed] [Google Scholar]
- 39.Gautschi O, et al. Regulation of Id1 expression by SRC: implications for targeting of the bone morphogenetic protein pathway in cancer. Cancer Res. 2008;68:2250–2258. doi: 10.1158/0008-5472.CAN-07-6403. [DOI] [PubMed] [Google Scholar]
- 40.Tournay O, Benezra R. Transcription of the dominant-negative helix-loop-helix protein Id1 is regulated by a protein complex containing the immediate-early response gene Egr-1. Mol Cell Biol. 1996;16:2418–2430. doi: 10.1128/mcb.16.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phi JH, et al. Upregulation of SOX2, NOTCH1, and ID1 in supratentorial primitive neuroectodermal tumors: a distinct differentiation pattern from that of medulloblastomas. Journal of neurosurgery Pediatrics. 2010;5:608–614. doi: 10.3171/2010.2.PEDS1065. [DOI] [PubMed] [Google Scholar]
- 42.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 43.Lee TK, et al. Regulation of angiogenesis by Id-1 through hypoxia-inducible factor-1alpha-mediated vascular endothelial growth factor up-regulation in hepatocellular carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:6910–6919. doi: 10.1158/1078-0432.CCR-06-0489. [DOI] [PubMed] [Google Scholar]
- 44.Ling MT, et al. Id-1 expression induces androgen-independent prostate cancer cell growth through activation of epidermal growth factor receptor (EGF-R) Carcinogenesis. 2004;25:517–525. doi: 10.1093/carcin/bgh047. [DOI] [PubMed] [Google Scholar]
- 45.Ling MT, et al. Id-1 expression promotes cell survival through activation of NF-kappaB signalling pathway in prostate cancer cells. Oncogene. 2003;22:4498–4508. doi: 10.1038/sj.onc.1206693. [DOI] [PubMed] [Google Scholar]
- 46.Zigler M, et al. Expression of Id-1 is regulated by MCAM/MUC18: a missing link in melanoma progression. Cancer Res. 2011;71:3494–3504. doi: 10.1158/0008-5472.CAN-10-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fong S, Debs RJ, Desprez PY. Id genes and proteins as promising targets in cancer therapy. Trends in molecular medicine. 2004;10:387–392. doi: 10.1016/j.molmed.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Cheung HW, Ling MT, Tsao SW, Wong YC, Wang X. Id-1-induced Raf/MEK pathway activation is essential for its protective role against taxol-induced apoptosis in nasopharyngeal carcinoma cells. Carcinogenesis. 2004;25:881–887. doi: 10.1093/carcin/bgh087. [DOI] [PubMed] [Google Scholar]
- 49.Hu H, et al. The role of Id-1 in chemosensitivity and epirubicin-induced apoptosis in bladder cancer cells. Oncology reports. 2009;21:1053–1059. doi: 10.3892/or_00000323. [DOI] [PubMed] [Google Scholar]
- 50.O’Brien CA, et al. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell. 2012;21:777–792. doi: 10.1016/j.ccr.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 51.Meteoglu I, Meydan N, Erkus M. Id-1: regulator of EGFR and VEGF and potential target for colorectal cancer therapy. Journal of experimental & clinical cancer research: CR. 2008;27:69. doi: 10.1186/1756-9966-27-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gearing DP, et al. Molecular cloning and expression of cDNA encoding a murine myeloid leukaemia inhibitory factor (LIF) EMBO J. 1987;6:3995–4002. doi: 10.1002/j.1460-2075.1987.tb02742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fitzgerald JS, et al. Leukemia inhibitory factor triggers activation of signal transducer and activator of transcription 3, proliferation, invasiveness, and altered protease expression in choriocarcinoma cells. The international journal of biochemistry & cell biology. 2005;37:2284–2296. doi: 10.1016/j.biocel.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 54.Li X, et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget. 2014;5:788–801. doi: 10.18632/oncotarget.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu SC, et al. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J Clin Invest. 2013;123:5269–5283. doi: 10.1172/JCI63428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maruta S, et al. A role for leukemia inhibitory factor in melanoma-induced bone metastasis. Clinical & experimental metastasis. 2009;26:133–141. doi: 10.1007/s10585-008-9223-x. [DOI] [PubMed] [Google Scholar]
- 57.Wysoczynski M, et al. Leukemia inhibitory factor: a newly identified metastatic factor in rhabdomyosarcomas. Cancer Res. 2007;67:2131–2140. doi: 10.1158/0008-5472.CAN-06-1021. [DOI] [PubMed] [Google Scholar]
- 58.Hillion J, et al. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an achilles heel for hematopoietic malignancies? Cancer Res. 2008;68:10121–10127. doi: 10.1158/0008-5472.CAN-08-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang C, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng T, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nature communications. 2013;4:2996. doi: 10.1038/ncomms3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith JJ, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freeman TJ, et al. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. Gastroenterology. 2012;142:562–571. e562. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marisa L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS medicine. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.