Abstract
Resistance to chemotherapy is a major cause of mortality in advanced cancer patients. In this study, digital karyotyping was used to search for genomic alterations in liver metastases that were clinically resistant to 5-fluorouracil (5-FU). In two of four patients, we identified amplification of an ≈100-kb region on 18p11.32 that was of particular interest because it contained the gene encoding thymidylate synthase (TYMS), a molecular target of 5-FU. Analysis of TYMS by fluorescence in situ hybridization identified TYMS gene amplification in 23% of 31 5-FU-treated cancers, whereas no amplification was observed in metastases of patients that had not been treated with 5-FU. Patients with metastases containing TYMS amplification had a substantially shorter median survival (329 days) than those without amplification (1,021 days, P <0.01). These data suggest that genetic amplification of TYMS is a major mechanism of 5-FU resistance in vivo and have important implications for the management of colorectal cancer patients with recurrent disease.
Since its introduction over four decades ago, 5-fluorouracil (5-FU) has become a staple of treatment for many cancers. In particular, it is the mainstay of chemotherapeutic regimens for colorectal cancers, both in metastatic and adjuvant settings (1). Metabolites of 5-FU and other fluoropyrimidines irreversibly inhibit thymidylate synthase (TYMS, Online Mendelian Inheritance in Man reference no. 188350), the enzyme normally responsible for conversion of deoxyuridine monophosphate to deoxythymidine monophosphate (2). As this process generates the sole de novo source of thymidylate, an essential precursor to DNA synthesis, inhibition of TYMS leads to DNA damage and blocks DNA replication and repair. In addition to its effects on DNA, metabolites of 5-FU can be incorporated into RNA, thereby disrupting normal RNA processing and function.
Although many colorectal cancer patients initially respond to 5-FU-based therapies, most develop recurrences and are usually retreated with 5-FU in combination with other drugs such as oxaliplatin (3, 4) or irinotecan (5). A subset of patients respond to such therapy, but in a variable and unpredictable manner. Despite significant research on the effects of 5-FU on cancer cells in vitro (2, 6-9), the molecular mechanisms underlying the development of 5-FU resistance in patients remain largely unknown. High TYMS protein or mRNA levels in tumors as determined by immunohistochemistry or RT-PCR have tended to be associated with a worse response to 5-FU in patients (10, 11). However, some reports have shown the opposite, i.e., that patients with high TYMS protein expression have improved outcome compared to those with low expression when treated with 5-FU (12). Additionally, measurement of TYMS protein expression in primary tumors does not aid in predicting outcome or response to 5-FU at sites of metastatic disease (13, 14). Alterations in levels of enzymes affecting 5-FU metabolism, including thymidine phosphorylase (TP) and dihydropyrimidine dehydrogenase (DPD), have also been postulated to affect 5-FU resistance. Overexpression of TP protein has been reported to increase sensitivity to 5-FU (15), whereas elevated levels of DPD mRNA have been associated with resistance (16). However, these correlations are also controversial, as some studies have shown that increased levels of TP mRNA were found in tumors that were less likely to respond to 5-FU (17), whereas others have reported that DPD and TP protein levels have no effect on patient survival (18).
Although levels of protein and RNA expression can provide clues to causal events during tumorigenesis, gene expression is difficult to measure accurately for technical reasons and may be affected by complex regulatory circuits specific to each tumor's environment. In contrast, genetic alterations can provide unambiguous information about pathogenetic mechanisms. For example, genetic alterations of the p53 gene provided critical clues to its pathogenic role that were not anticipated from prior measurements of p53 protein expression levels (19). Similarly, genetic mutations and gene amplification of the BCR/ABL gene in patients refractory to therapy with Gleevec have provided unique insights into the mechanisms underlying resistance to this tyrosine kinase inhibitor (20, 21). Unfortunately, previous genetic studies on 5-FU resistance have been limited to analyses of the development of chemoresistance in vitro (2, 6-9) or to a small number of patient case reports (22, 23).
Based on the above genetic precedents and the lack of a systematic study of 5-FU resistance in human cancer, we have undertaken a comprehensive genomic analysis of 5-FU resistance in colorectal cancer by using digital karyotyping (DK) (24). DK permits high-resolution analyses of copy number alterations on a genomewide scale. The approach involves isolation and high-throughput analysis of short (21-bp) sequence tags from ≈800,000 specific loci distributed throughout the genome. Analysis of sequence tag densities in sliding windows throughout each chromosome allows identification of potential amplifications and deletions at high resolution. This analysis permits a systematic genetic examination of resistance to 5-FU in vivo, convincingly identifying TYMS gene amplification as a major determinant of 5-FU chemoresistance in human cancers.
Materials and Methods
Tissue Samples. Tissue samples, including normal tissues, primary tumors, and metastases were obtained from colorectal cancer patients undergoing surgery at The Johns Hopkins Hospital between 1990 and 2002. A diagnosis of colorectal cancer was established by histological examination of surgical specimens, and clinical information was retrospectively retrieved from patient records. Acquisition of tissue specimens and examination of clinical records was approved by an institutional review board and was performed in accordance with Health Insurance Portability and Accountability Act regulations. Metastatic samples were obtained from complete resections, debulking, or biopsies of metastatic lesions.
Tumor Cell Purification of Liver Metastases. Tumor cells were purified from liver metastases as described (25). Briefly, tissues were obtained immediately after surgical removal and digested with 1 mg/ml collagenase for 1 h at 37°C. Single cell suspensions were obtained by sequential filtering through nylon mesh of 400, 50, and 25 μm. Epithelial cells were isolated by binding to anti-BerEP4 immunomagnetic beads (Dynal, Oslo), and the purified cells were immediately frozen at -80°C.
DK Library Construction and Analysis. DK libraries were constructed as described (24). Briefly, genomic DNA was isolated by using a DNeasy kit (Qiagen, Chatsworth, CA). For each sample, 1 μg of genomic DNA was sequentially digested with mapping enzyme SacI (New England Biolabs), ligated to 20-40 ng of biotinylated linkers (Integrated DNA Technologies, Coralville, IA), and digested with the fragmenting enzyme NlaIII (New England Biolabs). DNA fragments containing biotinylated linkers were isolated by binding to streptavidin-coated magnetic beads (Dynal). Captured DNA fragments were ligated to linkers containing MmeI recognition sites, and tags were released with MmeI (New England Biolabs). Tags were self-ligated to form ditags, which were then further ligated to form concatemers and cloned into pZero (Invitrogen). Clones were sequenced by using Big Dye terminators (Applied Biosystems) and analyzed with a 384-capillary automated sequencing apparatus (Spectrumedix, State College, PA) or a 96-capillary ABI3700 instrument at Agencourt Biosciences (Beverly, MA). DK sequence files were trimmed by using phred sequence analysis software (CodonCode, Dedham, MA), and 21-bp genomic tags were extracted by using the SAGE2000 software package (Invitrogen). Tags were matched to the human genome (University of California Santa Cruz human genome assembly, June 2002 freeze), and tag densities were evaluated by using the DK software package. Genomic densities were calculated as the ratio of experimental tags to the number of virtual tags present in a fixed window. Sliding windows of sizes ranging from 100 to 300 virtual tags were used to identify regions of increased and decreased genomic density. Chromosomal regions were considered to contain an amplification if maximal genomic densities were more than six genome copies per diploid genome. DK protocols and software for extraction and analysis of genomic tags are available at www.digitalkaryotyping.org.
Quantitative PCR. Genome content differences between metastatic tumors and normal liver cells were determined by quantitative real-time PCR using an iCycler apparatus (Bio-Rad) as described (24). DNA content was normalized to that of Line-1, a repetitive element for which copy numbers per haploid genome are similar among all human cells. PCR primers with the following sequences, TYMS-F, 5′-TTTTCGAAGAATCCTGAGCTTTG-3′ and TYMS-R, 5′-CACTCTCGATCTGTGCAAGAGAA-3′, were used to amplify a portion of the TYMS gene located at chromosome 18 position 988,801 to 989,046 base pair (University of California Santa Cruz human genome assembly, June 2002 freeze).
PCRs for each sample were performed in triplicate, and threshold cycle numbers were calculated by using ICYCLER 2.3 software (Bio-Rad).
Fluorescence in Situ Hybridization (FISH). Formalin-fixed paraffin-embedded tissue array sections 4 μm in thickness were analyzed by FISH as described (26, 27). Bacterial artificial chromosome (BAC) clone RP11-806L2 (located on chromosome 18p, 0.8-1.0 Mb from the telomere) and RP11-151D11 (located on chromosome 18p, 13.0-13.2 Mb from the telomere) were obtained from Bacpac Resources (Children's Hospital, Oakland, CA) and used as probes for the TYMS gene and a reference region on chromosome 18, respectively. RP11-806L2 and RP11-151D11 were labeled by nick translation with biotin-dUTP and digoxigenin-dUTP, respectively. To detect biotin-labeled and digoxigenin-labeled signals, slides were first incubated with FITC-avidin (Vector Laboratories) and an anti-digoxigenin mouse antibody (Molecular Biochemicals). The slides were subsequently incubated with a biotinylated anti-avidin antibody (Vector Laboratories) and tetramethylrhodamine B isothiocyanate (TRITC)-conjugated rabbit anti-mouse antibody (Sigma), then finally incubated with FITC-avidin and TRITC-conjugated goat anti-rabbit antibody (Sigma). Slides were counterstained with 4′,6′-diamidino-2-phenylindole stain (Sigma).
FISH signals were evaluated with a Nikon fluorescence microscope E800 by two individuals who were blinded to the treatment history of each patient. Separate narrow band pass filters were used for the detection of tetramethylrhodamine B isothiocyanate, FITC, and 4′,6′-diamidino-2-phenylindole stain signals. Using ×40 objective lens, ≈100 tumor cells were examined for each specimen, and the number of fluorescent signals within tumor cells from the TYMS gene BAC probe and chromosome 18 reference BAC probe was recorded. Amplification of the TYMS gene was defined as a ratio of TYMS BAC probe signals to chromosome 18 reference BAC probe signals 2:1 or more.
Statistical Analysis. Overall survival was calculated from the date of the surgical excision of the metastasis to the date of death or last follow-up and computed by the Kaplan-Meier method. Data were censored when patients were lost to follow-up.
Results
DK of Colorectal Cancer Metastases. DK was used to evaluate genomic DNA from liver metastases of four different colorectal cancer patients that had previously received 5-FU-based adjuvant chemotherapy (FU-M1-4). As controls, two liver metastases from colorectal cancer patients that had not previously received 5-FU (M1-2) were also analyzed. In each case, tumor epithelial cells were immunopurified from the metastases by using antibody-conjugated magnetic beads (25). This purification was essential to obtain DNA templates that were free of significant contamination from non-neoplastic cells within the metastatic lesions.
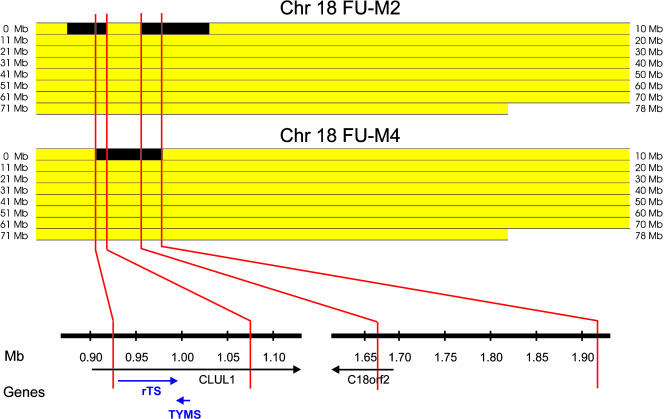
A total of ≈200,000 genomic tags were obtained from each sample, permitting analysis of loci spaced at an average distance of ≈30 kb throughout the genome. Computation of genomic tag densities identified distinct subchromosomal regions of amplification and deletion on several chromosomes. Each of the alterations occurred in individual tumors with the exception of a region of amplification on chromosome 18. This amplification, located at 18p11.32, was observed in two of the four 5-FU-resistant metastases (FU-M2 and FU-M4), but not in the two metastases from untreated patients, suggesting that this region could be related to 5-FU resistance.
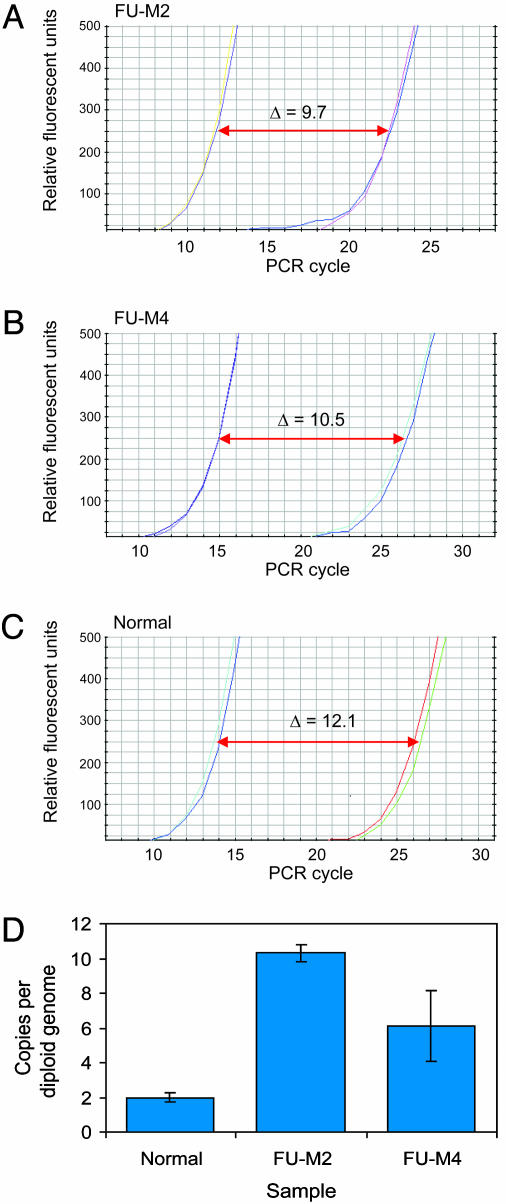
In tumor FU-M2, two separate, but closely spaced, amplicons were identified at 18p11.32, whereas a single continuous amplicon was found in tumor FU-M4 in the same region. Detailed analyses of these amplicons showed two common regions of amplification, one 0.92-1.06 Mb from the telomere and the other 1.66-1.92 Mb from the telomere (Fig. 1). Examination of genome databases identified two genes that were completely contained within the first region, TYMS and the HSRTSβ gene (rTS), whereas no known or predicted genes were present in the second. Amplification of TYMS was of particular interest because (i) the region containing this gene had a higher tag density within the overlapping amplicons; (ii) TYMS expression has been correlated with 5-FU resistance in some studies (10, 11), and (iii) TYMS amplification has been documented to develop in cancer cell lines that became resistant to 5-FU after exposure to this drug in vitro (28, 29). Quantitative PCR analyses of genomic DNA using primers specific to the TYMS locus confirmed that TYMS was amplified to levels of 10 and 6 gene copies per diploid genome in FU-M2 and FU-M4, respectively (Fig. 2).
Fig. 1.
Overlapping regions of amplification on chromosome 18p identified by DK. Bitmap views comprised of 18,431 pixels representing tag density values at the chromosomal position of each virtual tag on chromosome 18. Yellow regions indicate tag densities that were not amplified, and black regions represent areas with genomic tag densities indicating amplification. Genomic tag densities were determined as described in Materials and Methods and had maximal values of 10 and 6 copies per diploid genome for the amplifications in FU-M2 and FU-M4, respectively. Genes present within overlapping amplified regions are indicated below on a high-resolution map. Only TYMS and rTS were entirely contained within the regions that were amplified in both FU-M2 and FU-M4.
Fig. 2.
Quantitative PCR analysis of genomic DNA from colorectal metastases. Quantitive PCR analysis of TYMS (right curves) and LINE element control (left curves) performed on genomic DNA from colorectal cancer metastases FU-M2 (A) and FU-M4 (B) and normal (nontumor) DNA (C). (D) Differences in threshold cycle numbers between LINE element and TYMS confirm that TYMS is present at increased gene copy numbers in colorectal metastases.
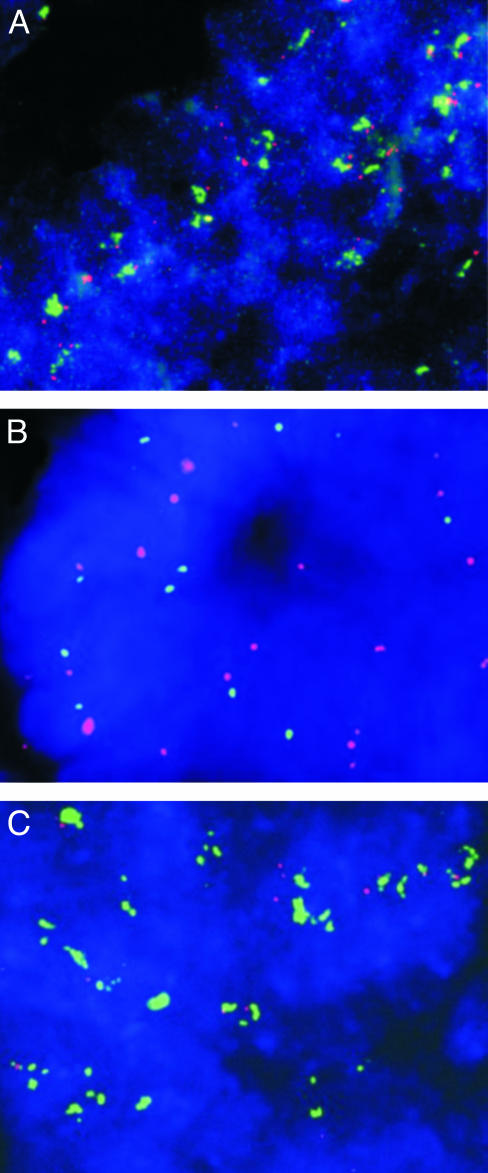
FISH Analysis of TYMS Amplification. To further evaluate the role of TYMS in 5-FU resistance, we analyzed TYMS gene copy number by using dual-color FISH. A total of 89 colorectal cancers embedded in tissue microarrays were assessed. These comprised 53 metastases derived from liver, lung, and brain tissues, including the four metastases originally analyzed by DK, and 36 primary colorectal cancers. Thirty one of the analyzed lesions were from patients that had received 5-FU therapy before tumor resection. Biotinylated DNA from a BAC containing the TYMS gene was used as probe, and sections were cohybridized with digoxigenin-labeled DNA from a BAC containing sequences from 18p11.21, 12 Mb closer to the centromere. Two probes from the same chromosome are necessary to distinguish chromosome duplications from true amplification events, the latter involving relatively small amplicons (30). Using FISH, multiple copies of the TYMS gene were detected in interphase nuclei in seven lesions, including the two metastases previously detected by DK (Table 1 and examples in Fig. 3 A and C). All seven lesions were derived from patients who had been treated with 5-FU: six were metastatic lesions of patients who had previously been treated with 5-FU-based therapy, and one was a primary colorectal cancer from a familial adenomatous polyposis (FAP) patient who had been treated with 5-FU before colectomy. In contrast, none of the 58 cancers from patients that had not been treated with 5-FU showed increased copies of the TYMS gene (Table 1, P < 0.001, χ2 test). To examine the temporal relationship between TYMS amplification and 5-FU treatment, we analyzed a primary colorectal cancer from a patient that later developed metastases with TYMS amplification. This primary cancer was removed before the initiation of 5-FU therapy and did not contain amplified TYMS genes when studied by FISH (data not shown). Similarly, TYMS amplification in the FAP patient noted above was only present in the resected cancer and was not observed in adenoma tissue obtained before 5-FU treatment (Fig. 3B).
Table 1. Prevalence of TYMS amplification in colorectal cancers.
| Previous treatment
|
|||
|---|---|---|---|
| TYMS status | 5-FU | No 5-FU | Total |
| Amplified | 7 | 0 | 7 |
| Not amplified | 24 | 58 | 82 |
| Total | 31 | 58 | 89 |
Fig. 3.
TYMS amplification assessed by interphase FISH. Analysis of interphase nuclei from a colorectal cancer metastasis to the liver after 5-FU treatment (A). Matched colorectal adenoma obtained before 5-FU treatment (B) and colorectal cancer obtained after 5-FU neoadjuvant treatment (C) from a patient with familial adenomatous polyposis. Nuclei are visualized with 4′,6′-diamidino-2-phenylindole stain (blue); TYMS probe (located on chromosome 18p, 0.8-1.0 Mb from the telomere) is visualized by using FITC-avidin (green), and chromosome 18 control probe (located on chromosome 18p, 13.0-13.2 Mb from the telomere) is visualized by using tetramethylrhodamine B isothiocyanate-conjugated antibodies (red). Increased TYMS gene copy number was observed only in patients previously treated with 5-FU (A and C). (Magnification: ×600.)
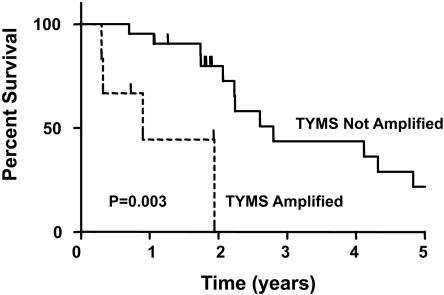
TYMS Amplification and Survival. The results described above show that TYMS amplification is exclusively found in cancer lesions of patients who had been treated with 5-FU. These data are consistent with the idea that the exposure to 5-FU had selected for cells with amplified TYMS genes, and that such cells would be resistant to 5-FU treatment. A corollary of this idea is that such patients would fare worse than those without TYMS amplification, as postsurgical therapy of patients with metastatic cancers often involves retreatment with 5-FU plus other agents (see Introduction). To evaluate this possibility, we compared the survival of patients with metastatic lesions that had previously been treated with 5-FU, segregated according to TYMS amplification status. Although the average age, stage at initial diagnosis, and metastasis size and location were similar between patients with and without increased TYMS gene copies, the median overall survival after surgical removal of metastases among patients with TYMS gene amplification was 329 days, as compared with 1,021 days for patients without TYMS amplification (Table 2, P < 0.01, t test). Using a proportional hazard model, patients with TYMS amplification showed a relative risk of death that was 3.5-fold higher (relative risk 1.06-11.4, 95% confidence interval, P < 0.05) than patients without TYMS amplification. These differences were also significant in Kaplan-Meier analyses (Fig. 4). In particular, this analysis showed that no patient with TYMS amplification has survived >2 years, whereas over a quarter of the patients without TYMS amplification survived >4 years (Fig. 4, P < 0.01, log rank test).
Table 2. Clinical characteristics of 5-FU treated patients with and without TYMS amplification.
| Patient group | TYMS status in metastasis | Previous treatment | Age, years | Stage at diagnosis | Metastasis size, cm | Metastasis location | Median survival after metastasis removal (survival range) |
|---|---|---|---|---|---|---|---|
| Group A (n = 6) | Amplified | 5-FU | 65 | I-0% | 2.2 | Liver-67% | 329 days (109-708 days) |
| II-0% | Lung-0% | ||||||
| III-33% | Brain-33% | ||||||
| IV-50% | |||||||
| ND-17% | |||||||
| Group B (n = 21) | Not amplified | 5-FU | 63 | I-5% | 3.2 | Liver-80% | 1,021 days (255-3,790 days) |
| II-10% | Lung-10% | ||||||
| III-29% | Brain-10% | ||||||
| IV-43% | |||||||
| ND-14% |
Fig. 4.
Five-year survival curve for patients with and without TYMS amplification.
Discussion
Our results demonstrate a significant association between treatment with 5-FU, amplification of the TYMS gene, and survival after surgical excision of metastatic lesions. These observations have important implications for basic and clinical aspects of human cancer.
Drug resistance is a major cause of treatment failure and death in cancer patients. But the basis for drug resistance in such tumors is generally unknown. There have been numerous reports of mechanisms underlying the development of drug resistance in cell culture systems. For example, expression of multidrug resistance genes can confer resistance to drugs in vitro (31, 32), but the relationship of such expression to the development of resistance in vivo remains conjectural. Similarly, the dihydrofolate reductase gene is commonly found to be amplified after treatment of cultured cells with methotrexate (33), but there are only a few case reports of gene amplification occurring in the tumors of cancer patients after exposure to conventional chemotherapeutic agents in vivo (22, 34, 35).
There are two examples of gene amplification developing in patients treated with targeted therapies, and both are informative with respect to 5-FU. The first involves androgen receptor mutations in prostate cancer patients treated with antiandrogens (36). The second involves genetic alterations (often amplification) of the BCR/ABL fusion gene in chronic myelogenous leukemia (CML) patients treated with Gleevec (20, 21). Gleevec inhibits several tyrosine kinases in addition to BCR/ABL (37). However, the specific genetic alterations of BCR/ABL that develop during treatment unambiguously point to the BCR/ABL gene as the central drug target in CML. Similarly, many potential mechanisms of action of 5-FU have been suggested (8, 38, 39). But the amplification of the TYMS gene after 5-FU treatment in vivo provides compelling evidence that TYMS is a major target of this drug in human cancer patients.
The fact that TYMS amplification was observed only in patients after treatment with 5-FU suggests that the cancers must pass through a bottleneck that effectively kills the vast majority of cancer cells (those without TYMS gene amplification) in these patients. Our observations, coupled with the experimental demonstration that engineered overexpression of TYMS in cultured cells can cause 5-FU resistance (40), provide compelling evidence that TYMS amplification is responsible for a significant fraction of 5-FU resistance.
The considerably worse survival of patients with TYMS amplification compared to similar patients without TYMS gene amplification (Table 2 and Fig. 4) is consistent with the conclusion the amplification of this gene is responsible for 5-FU resistance. Although the reasons for the reduced survival are not known with certainty, patients whose tumors recur after metastectomy are generally treated with regimens containing 5-FU. In our patient population, the majority of patients received 5-FU alone or in combination with other chemotherapeutic agents after removal of metastases (data not shown). The 5-FU component of such regimens would not likely be of benefit in patients with TYMS gene amplification but would be expected to cause the same degree of systemic toxicity observed in patients without TYMS gene amplification, potentially explaining the worse survival of these patients.
In addition to TYMS amplification, it is possible that other genetic mechanisms of resistance are present in patients with clinical resistance to 5-FU. It is important to note that not all patients without TYMS amplification had longer survival times, and in fact several patients had extremely short survival periods after surgical resection. Such mechanisms of resistance could include genetic modification of other members of the pathway, including TP or dihydropyrimidine dehydrogenase, or candidate genes within previously described loci affected by 5-FU in vitro (8, 41).
Although larger prospective studies will be important to confirm the present findings, our results have clear implications for the management of colorectal cancer patients. In particular, our data suggest that recurrences in patients whose biopsies show TYMS gene amplification should not be treated with 5-FU. Many of the newer second-line therapies undergoing clinical trials involve combinations of 5-FU with other agents. In patients with TYMS gene amplification, 5-FU would likely add toxicity without efficacy. TYMS gene amplification is straightforward to detect by using the probes and methods described in this article and can be performed on routinely fixed and paraffin-embedded samples. In addition to eliminating the 5-FU from regimens that would ordinarily include it, these results should stimulate efforts to develop compounds that specifically target cancers with amplified TYMS genes (42) and provide an ideal subset of patients in which to test such agents.
Acknowledgments
This work was supported by the Benjamin Baker Scholarship Fund, the Cancer Genome Anatomy Project, and National Institutes of Health Grants CA43460, CA57345, and CA62924.
Abbreviations: DK, digital karyotyping; 5-FU, 5-fluorouracil; TYMS, thymidylate synthase; TP, thymidine phosphorylase; FISH, fluorescence in situ hybridization; BAC, bacterial artificial chromosome.
References
- 1.Moertel, C. G. (1994) N. Engl. J. Med. 330, 1136-1142. [DOI] [PubMed] [Google Scholar]
- 2.Longley, D. B., Harkin, D. P. & Johnston, P. G. (2003) Nat. Rev. Cancer 3, 330-338. [DOI] [PubMed] [Google Scholar]
- 3.Giacchetti, S., Perpoint, B., Zidani, R., Le Bail, N., Faggiuolo, R., Focan, C., Chollet, P., Llory, J. F., Letourneau, Y., Coudert, B., et al. (2000) J. Clin. Oncol. 18, 136-147. [DOI] [PubMed] [Google Scholar]
- 4.de Gramont, A., Figer, A., Seymour, M., Homerin, M., Hmissi, A., Cassidy, J., Boni, C., Cortes-Funes, H., Cervantes, A., Freyer, G., et al. (2000) J. Clin. Oncol. 18, 2938-2947. [DOI] [PubMed] [Google Scholar]
- 5.Saltz, L. B., Cox, J. V., Blanke, C., Rosen, L. S., Fehrenbacher, L., Moore, M. J., Maroun, J. A., Ackland, S. P., Locker, P. K., Pirotta, N., et al. (2000) N. Engl. J. Med. 343, 905-914. [DOI] [PubMed] [Google Scholar]
- 6.Peters, G. J., Backus, H. H., Freemantle, S., van Triest, B., Codacci-Pisanelli, G., van der Wilt, C. L., Smid, K., Lunec, J., Calvert, A. H., Marsh, S., et al. (2002) Biochim. Biophys. Acta 1587, 194-205. [DOI] [PubMed] [Google Scholar]
- 7.Plasencia, C., Rooney, P. H., Taron, M., Martinez-Balibrea, E., McLeod, H. L. & Abad, A. (2003) Int. J. Oncol. 22, 945-953. [DOI] [PubMed] [Google Scholar]
- 8.Maxwell, P. J., Longley, D. B., Latif, T., Boyer, J., Allen, W., Lynch, M., McDermott, U., Harkin, D. P., Allegra, C. J. & Johnston, P. G. (2003) Cancer Res. 63, 4602-4606. [PubMed] [Google Scholar]
- 9.Wang, W., Marsh, S., Cassidy, J. & McLeod, H. L. (2001) Cancer Res. 61, 5505-5510. [PubMed] [Google Scholar]
- 10.Johnston, P. G., Lenz, H. J., Leichman, C. G., Danenberg, K. D., Allegra, C. J., Danenberg, P. V. & Leichman, L. (1995) Cancer Res. 55, 1407-1412. [PubMed] [Google Scholar]
- 11.Lenz, H. J., Hayashi, K., Salonga, D., Danenberg, K. D., Danenberg, P. V., Metzger, R., Banerjee, D., Bertino, J. R., Groshen, S., Leichman, L. P. & Leichman, C. G. (1998) Clin. Cancer Res. 4, 1243-1250. [PubMed] [Google Scholar]
- 12.Edler, D., Glimelius, B., Hallstrom, M., Jakobsen, A., Johnston, P. G., Magnusson, I., Ragnhammar, P. & Blomgren, H. (2002) J. Clin. Oncol. 20, 1721-1728. [DOI] [PubMed] [Google Scholar]
- 13.Findlay, M. P., Cunningham, D., Morgan, G., Clinton, S., Hardcastle, A. & Aherne, G. W. (1997) Br. J. Cancer 75, 903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston, P. G., Benson, A. B., 3rd, Catalano, P., Rao, M. S., O'Dwyer, P. J. & Allegra, C. J. (2003) J. Clin. Oncol. 21, 815-819. [DOI] [PubMed] [Google Scholar]
- 15.Evrard, A., Cuq, P., Robert, B., Vian, L., Pelegrin, A. & Cano, J. P. (1999) Int. J. Cancer 80, 465-470. [DOI] [PubMed] [Google Scholar]
- 16.Salonga, D., Danenberg, K. D., Johnson, M., Metzger, R., Groshen, S., Tsao-Wei, D. D., Lenz, H. J., Leichman, C. G., Leichman, L., Diasio, R. B. & Danenberg, P. V. (2000) Clin. Cancer Res. 6, 1322-1327. [PubMed] [Google Scholar]
- 17.Metzger, R., Danenberg, K., Leichman, C. G., Salonga, D., Schwartz, E. L., Wadler, S., Lenz, H. J., Groshen, S., Leichman, L. & Danenberg, P. V. (1998) Clin. Cancer Res. 4, 2371-2376. [PubMed] [Google Scholar]
- 18.Ikeguchi, M., Makino, M. & Kaibara, N. (2002) Langenbecks Arch. Surg. 387, 240-245. [DOI] [PubMed] [Google Scholar]
- 19.Baker, S. J., Fearon, E. R., Nigro, J. M., Hamilton, S. R., Preisinger, A. C., Jessup, J. M., vanTuinen, P., Ledbetter, D. H., Barker, D. F., Nakamura, Y., et al. (1989) Science 244, 217-221. [DOI] [PubMed] [Google Scholar]
- 20.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, P. N. & Sawyers, C. L. (2001) Science 293, 876-880. [DOI] [PubMed] [Google Scholar]
- 21.Shah, N. P., Nicoll, J. M., Nagar, B., Gorre, M. E., Paquette, R. L., Kuriyan, J. & Sawyers, C. L. (2002) Cancer Cell 2, 117-125. [DOI] [PubMed] [Google Scholar]
- 22.Clark, J. L., Berger, S. H., Mittelman, A. & Berger, F. G. (1987) Cancer Treat. Rep. 71, 261-265. [PubMed] [Google Scholar]
- 23.Gorlick, R., Metzger, R., Danenberg, K. D., Salonga, D., Miles, J. S., Longo, G. S., Fu, J., Banerjee, D., Klimstra, D., Jhanwar, S., et al. (1998) J. Clin. Oncol. 16, 1465-1469. [DOI] [PubMed] [Google Scholar]
- 24.Wang, T. L., Maierhofer, C., Speicher, M. R., Lengauer, C., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Proc. Natl. Acad. Sci. USA 99, 16156-16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha, S., Bardelli, A., Buckhaults, P., Velculescu, V. E., Rago, C., St. Croix, B., Romans, K. E., Choti, M. A., Lengauer, C., Kinzler, K. W. & Vogelstein, B. (2001) Science 294, 1343-1346. [DOI] [PubMed] [Google Scholar]
- 26.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623-627. [DOI] [PubMed] [Google Scholar]
- 27.Bardelli, A., Saha, S., Sager, J., Romans, K. E., Xin, B., Markowitz, S. D., Lengauer, C., Velculescu, V. E., Kinzler, K. W. & Vogelstein, B. (2003) Clin. Cancer Res. 9, 5607-5615. [PubMed] [Google Scholar]
- 28.Jenh, C. H., Geyer, P. K., Baskin, F. & Johnson, L. F. (1985) Mol. Pharmacol. 28, 80-85. [PubMed] [Google Scholar]
- 29.Berger, S. H., Jenh, C. H., Johnson, L. F. & Berger, F. G. (1985) Mol. Pharmacol. 28, 461-467. [PubMed] [Google Scholar]
- 30.Brodeur, G. M. & Hogarty, M. D. (1998) in The Genetic Basis of Human Cancer, eds. Kinzler, K. W. & Vogelstein, B. (McGraw-Hill, New York), Vol. 1, pp. 161-179. [Google Scholar]
- 31.Kartner, N., Riordan, J. R. & Ling, V. (1983) Science 221, 1285-1288. [DOI] [PubMed] [Google Scholar]
- 32.Gros, P., Ben Neriah, Y. B., Croop, J. M. & Housman, D. E. (1986) Nature 323, 728-731. [DOI] [PubMed] [Google Scholar]
- 33.Schimke, R. T. (1984) Cell 37, 705-713. [DOI] [PubMed] [Google Scholar]
- 34.Horns, R. C., Jr., Dower, W. J. & Schimke, R. T. (1984) J. Clin. Oncol. 2, 2-7. [DOI] [PubMed] [Google Scholar]
- 35.Carman, M. D., Schornagel, J. H., Rivest, R. S., Srimatkandada, S., Portlock, C. S., Duffy, T. & Bertino, J. R. (1984) J. Clin. Oncol. 2, 16-20. [DOI] [PubMed] [Google Scholar]
- 36.Koivisto, P., Visakorpi, T. & Kallioniemi, O. P. (1996) Scand. J. Clin. Lab. Invest. Suppl., 226, 57-63. [PubMed] [Google Scholar]
- 37.Druker, B. J. (2002) Cancer Cell 1, 31-36. [DOI] [PubMed] [Google Scholar]
- 38.Scherf, U., Ross, D. T., Waltham, M., Smith, L. H., Lee, J. K., Tanabe, L., Kohn, K. W., Reinhold, W. C., Myers, T. G., Andrews, D. T., et al. (2000) Nat. Genet. 24, 236-244. [DOI] [PubMed] [Google Scholar]
- 39.Hwang, P. M., Bunz, F., Yu, J., Rago, C., Chan, T. A., Murphy, M. P., Kelso, G. F., Smith, R. A., Kinzler, K. W. & Vogelstein, B. (2001) Nat. Med. 7, 1111-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saga, Y., Suzuki, M., Mizukami, H., Kohno, T., Takei, Y., Fukushima, M. & Ozawa, K. (2003) Int. J. Cancer 106, 324-326. [DOI] [PubMed] [Google Scholar]
- 41.Rooney, P. H., Stevenson, D. A., Marsh, S., Johnston, P. G., Haites, N. E., Cassidy, J. & McLeod, H. L. (1998) Cancer Res. 58, 5042-5045. [PubMed] [Google Scholar]
- 42.Neuteboom, S. T., Karjian, P. L., Boyer, C. R., Beryt, M., Pegram, M., Wahl, G. M. & Shepard, H. M. (2002) Mol. Cancer Ther. 1, 377-384. [PubMed] [Google Scholar]