Abstract
The tRNA m1A58 methyltransferase is composed of two subunits encoded by the essential genes TRM6 and TRM61 (formerly GCD10 and GCD14). The trm6-504 mutation results in a defective m1A methyltransferase (Mtase) and a temperature-sensitive growth phenotype that is attributable to the absence of m1A58 and consequential tRNAiMet instability. We used a genetic approach to identify the genes responsible for tRNAiMet degradation in trm6 cells. Three recessive extragenic mutations that suppress trm6-504 mutant phenotypes and restore hypomodified tRNAiMet to near normal levels were identified. The wild-type allele of one suppressor, DIS3/RRP44, encodes a 3′-5′ exoribonuclease and a member of the multisubunit exosome complex. We provide evidence that a functional nuclear exosome is required for the degradation of tRNAiMet lacking m1A58. A second suppressor gene encodes Trf4p, a DNA polymerase (pol σ) with poly(A) polymerase activity. Whereas deletion of TRF4 leads to stabilization of tRNAiMet, overexpression of Trf4p destabilizes the hypomodified tRNAiMet in trm6 cells. The hypomodified, but not wild-type, pre-tRNAiMet accumulates as a polyadenylated species, whose abundance and length distribution both increase upon Trf4p overexpression. These data indicate that a tRNA surveillance pathway exists in yeast that requires Trf4p and the exosome for polyadenylation and degradation of hypomodified pre-tRNAiMet.
Keywords: Transfer RNA, exosome, RNA processing, RNA turnover, tRNA modification
The relatively unstable nature of messenger RNAs fueled the discovery of pathways that control the degradation of normal and abnormal mRNAs in the nucleus and cytoplasm (Hilleren and Parker 1999; Mitchell and Tollervey 2001; Wilusz et al. 2001; Maquat 2002; Moore 2002; Long and McNally 2003). Two general pathways of mRNA decay have been characterized in the yeast Saccharomyces cerevisiae, and homologs of most of the yeast proteins involved in mRNA turnover have been identified in metazoans. The first pathway initially requires shortening of the mRNA polyadenylate tail, followed by removal of the 5′ cap structure (Wilusz et al. 2001), which leaves the body of the mRNA susceptible to 5′-3′ exonucleolytic degradation by Xrn1p. The second pathway involves deadenylation of mRNAs and the 3′-5′ degradation of the body of the mRNA by the exosome (Jacobs et al. 1998; Burkard and Butler 2000; van Hoof et al. 2000b; van Hoof and Parker 2002; Mitchell and Tollervey 2003).
The exosome is a multisubunit complex of proteins with multiple functions in the processing, degradation, and retention of stable and unstable RNAs in the nucleus and cytoplasm. The cytoplasmic exosome directly interacts with Ski7p (Araki et al. 2001) and recruits the Ski2p, Ski3p, and Ski8p complex to the 3′ end of a deadenylated mRNA (Brown et al. 2000) or an mRNA that is stalled on the ribosome because it lacks a stop codon (Jacobs et al. 1998; van Hoof et al. 2000b), and in turn each is degraded in a 3′-to-5′ direction. In the nucleus, the exosome has been implicated in elimination of by-products of rRNA processing (ETS sequence). The nuclear exosome possesses an exonuclease, Rrp6p, not found in the cytoplasmic form (Allmang et al. 1999b). A specialized function of Rrp6p and the nuclear exosome appears to be in retaining mRNAs incorrectly processed at their 3′ ends at the site of transcription to prevent their release into the cytoplasm (Hilleren et al. 2001; Libri et al. 2002). Thus far, the exosome has not been implicated in the destruction of stable RNAs that are rendered unstable due to mutations or defects in processing.
In contrast to mRNA, tRNA and rRNA are highly stable RNA species with half-lives measured in days. The longevity of tRNA and rRNA can be disrupted by mutations in the RNA themselves (Eisenberg and Yarus 1980; Colby et al. 1981; Yuo and Weiner 1989; Eschenlauer et al. 1993; Johansson and Bystrom 2002) or by inactivation of a protein required for the processing or modification of a normally stable RNA (Anderson et al. 1998). Recent studies have shown that precursor tRNAs (pre-tRNAs) rendered unstable because of a mutation activate a surveillance mechanism in Escherichia coli that results in 3′ adenylation and degradation of the precursor form of the mutant tRNA. This degradation mechanism requires poly(A) polymerase and the exoribonuclease PNpase (Li et al. 2002). In E. coli, the degradation of mRNA also involves, but is not limited to, poly(A) polymerase and PNpase (Kushner 2002). The fact that poly(A) polymerase and PNpase play an active role in mRNA decay and tRNA degradation suggests that aberrant pre-tRNA degradation and mRNA turnover share similar mechanisms in E. coli.
The processing of eukaryotic pre-tRNA is a multistep pathway consisting of 5′ end cleavage by RNaseP, intron removal, 3′ end-trimming by endo- and/or exonucleases, CCA addition to the 3′ end, and ribose or nucleotide modification (Hopper and Phizicky 2003). One such nucleotide modification is the addition of a methyl group to the N1 position of adenosine at position 58 of the T ψ C loop. The yeast 1-methyladenosine 58 methyltransferase (m1A58 Mtase) is composed of two proteins encoded by genes essential for cell viability (Garcia-Barrio et al. 1995; Cuesta et al. 1998). Mutation of the genes TRM6 (formerly named GCD10) or TRM61 (formerly named GCD14) encoding the m1A Mtase lead to tRNAiMet instability, supporting the postulate that m1A58, along with several other modified nucleosides found in tRNA from all three kingdoms, is important for tRNA structure or function (Bjork 1995). Apart from the requirement for aminoacylation of some tRNAs prior to nucleocytoplasmic transport (Lund and Dahlberg 1998; Sarkar et al. 1999; Grosshans et al. 2000), there are no known surveillance mechanisms to ensure the structural or functional integrity of nuclear tRNA processing events prior to nucleocytoplasmic transport.
We previously showed that the defect in tRNAiMet expression in trm6 or trm61 mutants due to a lack of (m1A58) is overcome by increasing the copy number of IMT4, encoding tRNAiMet, showing that increasing the level of tRNAiMet transcription is one mechanism of suppression. We also found that increasing the copy number of LHP1, encoding the tRNA 3′-end processing factor Lhp1p, augmented the synthesis of tRNAiMet or stabilized pre-tRNAiMet during processing (Anderson et al. 1998; Calvo et al. 1999). The instability of pre-tRNAiMet lacking m1A58 is unique in that no other tRNAs exhibit instability in strains possessing an inactivated m1A Mtase (Anderson et al. 1998). The threedimensional structure of tRNAiMet from S. cerevisiae has been solved by X-ray crystallography (Basavappa and Sigler 1991). Those authors showed that adenosines at positions 20, 54, and 60 (rarely found in noninitiator tRNAs in eukaryotes) along with m1A58 in tRNAiMet form a substructure that is predicted to be crucial for the maintenance of D- and T-loop interactions. Based on these data, we postulated that the instability of tRNAiMet occurs from a weakened tertiary structure through disruption of normal D- and T-loop interactions, which stem from the absence of 1-methyl on adenosine 58 (Anderson et al. 1998). Because the substructure described is unlikely to be represented in elongator tRNAs from S. cerevisiae, the initiator tRNAMet is unique in exhibiting instability in the absence of m1A58. Having determined that the absence of m1A58 from pre-tRNAiMet leads to its degradation, we reasoned that impairing the ability of the cell to degrade pre-tRNAiMet lacking m1A58 should restore normal levels of mature hypomodified tRNAiMet in trm6 cells. In this report, we outline a pathway in yeast for the degradation of tRNAiMet lacking m1A58 by identifying mutations in TRF4 and DIS3/RRP44 as spontaneous suppressors of the trm6-504 Ts- phenotype. Trf4p was first identified as a DNA polymerase, along with its redundant homolog Trf5p, in S. cerevisiae (Castano et al. 1996b; Wang et al. 2000). DIS3/RRP44 was first identified as a homolog of Schizosaccharomyces pombe DIS3 (Noguchi et al. 1996), and later as a component of a purified exosome fraction from S. cerevisiae (Mitchell et al. 1997). Dis3p/Rrp44p is a member of the RNR superfamily of exoribonucleases (Zuo and Deutscher 2001), and the purified protein possesses 3′-5′ riboexonuclease activity in vitro (Mitchell et al. 1997). Our results show that mutation of TRF4 or DIS3/RRP44 restores mature hypomodified tRNAiMet to near normal levels. By blocking degradation of tRNAiMet lacking m1A58, we found that the hypomodified molecule accumulates as an adenylated precursor, whose abundance and size distribution increase upon Trf4p overexpression. These findings establish that Trf4p and the exosome are functioning together in the nucleus to degrade the aberrant pre-tRNAiMet lacking m1A58.
Results
Extragenic suppressors of trm6-504
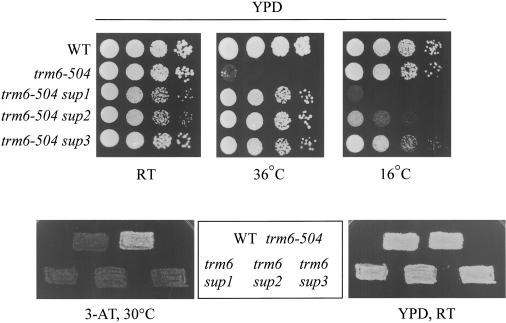
We took a genetic approach to identify the molecular pathway responsible for the degradation of pre-tRNAiMet lacking m1A58 by isolating second-site mutations that suppress the growth defect associated with the trm6-504 mutation in the m1A Mtase. This mutation confers a temperature-sensitive (Ts-) growth phenotype at 36°C, and resistance to the drug 3-aminotriazole (3ATr; Harashima and Hinnebusch 1986). 3-AT is an inhibitor of the His3p protein; in this context 3-AT simulates amino acid starvation and induces the general control pathway through activation of the Gcn2p kinase (Wolfner et al. 1975), and the attendant induction of GCN4 mRNA translation (Hinnebusch 1997). The trm6-504 mutant bypasses the requirement for Gcn2p to induce the general control pathway because of reduced tRNAiMet levels, and thus a trm6-504 mutant exhibits a constitutive 3-AT resistant (3-ATr) phenotype in the absence of Gcn2p function (Harashima and Hinnebusch 1986). Increasing the level of mature hypomodified tRNAiMet in a trm6 gcn2 mutant reverses the 3-ATr phenotype to yield a 3-AT-sensitive phenotype (3-ATs; Anderson et al. 1998). We isolated 150 spontaneous revertants of the Ts- and 3ATr growth phenotypes of the trm6-504 gcn2-101 mutant, of which 51 confer a cold-sensitive (Cs-) phenotype at 16°C. Initial genetic characterization revealed that all 51 Cs- revertants contained recessive suppressor mutations. Upon tetrad dissection of revertants back-crossed to the trm6-504 gcn2-101 parental strain, the Cs- phenotype always cosegregated 2:2 with suppression of trm6-504, indicating that suppression is linked to the Cs- phenotype and due to mutation of a single suppressor gene. We assigned the 51 Cs- suppressors to three complementation groups, designated sup1 to sup3, and the phenotypes of representatives of each complementation group are shown in Figure 1. We ruled out the possibility that suppression was due to intragenic reversion of TRM6, by showing that a wild-type copy of TRM6 on a single-copy plasmid failed to complement the recessive Cs- phenotype of each complementation group (data not shown).
Figure 1.
Mutations of sup1, sup2, or sup3 suppress the mutant phenotypes of trm6-504. TRM6, trm6-504 and representative trm6-504, sup- strains all harboring gcn2-101 were grown to saturation in YPD medium, and serial 10-fold dilutions were spotted onto YPD plates and incubated at 36°C, 16°C, and 26°C. The same strains were grown on YPD at 26°C and replica-plated to synthetic complete (SC) plates lacking histidine and supplemented with 30 mM 3-aminotriazole (3-AT) or YPD and incubated at 30°C and 26°C, respectively.
Identification of suppressor genes
We used the cold-sensitive phenotypes of stable representatives of sup1 and sup2 to clone the corresponding wild-type alleles of the suppressor genes by complementation using a yeast genomic DNA library constructed in a single-copy shuttle vector (Rose et al. 1987). The ends of the genomic DNA inserts in the complementing plasmids were sequenced, and the DNA sequences were used to query the Saccharomyces Genome Database to identify all predicted open reading frames (ORFs) within the identified genomic intervals. So far, we have been unable to clone the wild-type allele of sup3 due to its weak Cs- phenotype.
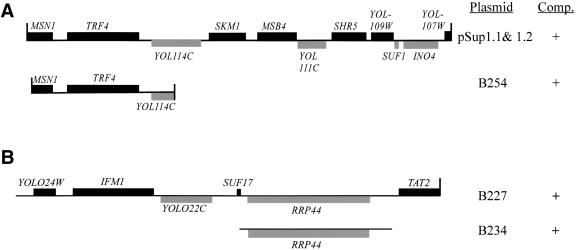
The sequence of two independent plasmids that complemented sup1 (pSup1.1 and pSup1.2) had identical fragments ∼12 kbp in length from Chromosome XV (coordinates 100202-112210 bp) containing nine complete ORFs (Fig. 2A). By constructing subclones and testing them for complementation of trm6-504, we identified TRF4 as the suppressor gene (Fig. 2A,C). Because TRF4 is a nonessential gene, we were able to confirm its identity as SUP1+ by crossing a trm6-504 mutant with a trf4Δ strain and finding that trf4Δ suppresses the Ts- phenotype of trm6-504 (data not shown; see Materials and Methods). Moreover, we determined that TRF4 is allelic to sup1 using marker rescue (Materials and Methods).
Figure 2.
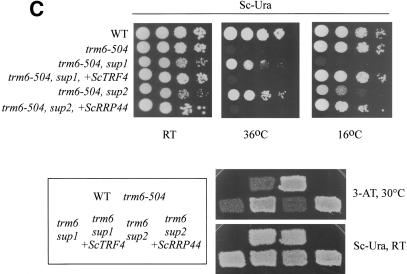
Identification of SUP1+ and SUP2+ as TRF4 and RRP44, respectively. (A) Schematic representation of genomic DNA inserts present in YCp50 (pSUP1.1 and pSUP1.2) and the single-copy plasmid subclone derived from pSUP1.1 (B254). All three plasmids complemented the Cs- and 3-ATr phenotypes of the sup1, trm6-504, gcn2-101 strain. (B) Schematic representation of the largest genomic DNA insert that complemented sup2 Cs- phenotype (B227) and the single-copy plasmid subclone derived from B227 (B234). Both plasmids complemented the Cs- and 3-ATr phenotypes of the sup1, trm6-504, gcn2-101 strain (see Materials and Methods for details). The chromosomal coordinates for all three genomic clones containing DIS3/RRP44 (B227, B228, and B229) are given in Table 1. Black boxes, ORFs encoded on the Watson strand; stippled boxes, ORFs encoded on the Crick strand; vertical lines, incomplete ORFs. (C) Growth of strains wild type, trm6-504, sup1, trm6-504, and sup2, trm6-504 all harboring gcn2-101 transformed with YCpLac33 (rows 1-3, 5), with scTRF4 (B254, row 4) or scRRP44 (B234, row 6). Cultures of each strain grown in SC-Ura and serial 10-fold dilutions were spotted onto SC-Ura and incubated at 36°C, 16°C, and 26°C (RT). The same strains were grown as patches on SC-Ura plates and replica-plated to SC-Ura, His medium supplemented with 30mM 3-AT or SC-Ura and incubated at 30°C and 26°C, respectively.
We identified three independent plasmids from the single-copy genomic library that complemented the sup2 Cs- phenotype. DNA sequence analysis and searches of the S. cerevisiae genome database revealed that all three genomic clones were derived from a single chromosomal region (Fig. 2B; Table 1). Only DIS3/RRP44 and SUF17, encoding tRNAGCCGly, were represented as complete ORFs in all three clones, and we found that the DIS3/RRP44 gene subcloned into a single-copy plasmid lacking SUF17 (Fig. 2B) complemented the sup2 Cs- phenotype and restored the Ts- and 3ATr phenotypes of the trm6-504 gcn2-101 strain (Fig. 2C). We confirmed the identity of SUP2+ as DIS3/RRP44 by conducting linkage analysis after directing integration of URA3 near the DIS3/RRP44 locus in sup2 (see Materials and Methods). From these results, we concluded that mutation of either TRF4 or DIS3/RRP44 can suppress the growth phenotypes of a trm6-504 mutant. Throughout the remainder of this report, DIS3/RRP44 will be referred to as RRP44.
Table 1.
Plasmids used
| Name | Description | Reference |
|---|---|---|
| B176 | YCp50: CEN4, URA3 cloning vector | Rose et al. 1987 |
| B181 | YCpLac33: CEN4, URA3 cloning vector | Gietz and Sugino 1988 |
| B184 | YEpLac 195: URA3 high copy vector | Gietz and Sugino 1988 |
| B187 | YIpLac211: URA3 integrative vector | Gietz and Sugino 1988 |
| B234 | 3.9 kb XhoI/XmaI PCR fragment of RRP44/DIS3 in SalI/XmaI-digested YCpLac33 | This study |
| B251 | 3.9 kb XhoI/XmaI product of RRP44/DIS3 in SalI/XmaI YEpLac195 | This study |
| B254 | 3.0 kb SspI/PmeI fragment pSup1 containing TRF4 into SmaI-digested YCpLac33 | This study |
| B256 | 3.1 kb SacI/PstI fragment of TRF4 from B254 into SacI/PstI YEpLac195 | This study |
| B269 | 3.4 kb XbaI/EcoRI fragment of RRP6 in XbaI/EcoRI YCpLac33 | Briggs et al. 1998 |
| CB432 | 3.8 kb SmaI/HindIII genomic fragment containing TRF4 into SmaI/HindIII pRS306 | Sadoff et al. 1995a |
| B227 | YCp50 containing chromosome XV DNA coordinates 280049 to 286635 | This study |
| B228 | YCp50 containing chromosome XV DNA coordinates 280689 to 287099 | This study |
| B229 | YCp50 containing chromosome XV DNA coordinates 276767 to 287099 | This study |
| B285 | 1.1 kb EcoRI fragment of IFM1 cloned into EcoRI-digested YIpLac211 | This study |
| B145 | A 3.0 kb XbaI/XhoI fragment containing TRM6 into SalI/XbaI-digested YIpLac211 | This study |
| a pSup 1.1 | YCp50 containing chromosome XV DNA coordinates 100202-112210 | This study |
| AKYOL114C | 2.5 kb YOL114C HindIII fragment from CB432 into HindIII-digested YCpLac33 | This study |
| p2704 | 3.0 kb XbaI/XhoI fragment containing TRM6 into XbaI/SalI-digested YCplac33 | Anderson et al. 1998 |
Genomic DNA contained in pSup1.1 and pSup1.2 is identical.
TRF4 was originally discovered in a genetic screen to identify mutants that require topoisomerase I for growth (Sadoff et al. 1995a). It has been shown that Trf4p exhibits DNA polymerase activity with β-polymerase-like characteristics and is required for sister-chromatid cohesion (Wang et al. 2000). Trf4p is an ortholog of S. pombe Cid13p, a cytoplasmic poly(A) polymerase that is required for the stability of ribonucleotide reductase mRNA by maintaining its polyadenylated state (Saitoh et al. 2002). It was further demonstrated that both Cid13p and Trf4p exhibit poly(A) polymerase activity in vitro (Saitoh et al. 2002). Rrp44p is a core component of the yeast exosome, a complex of 10 predicted 3′-5′ exohydrolases or phosphorylases involved in numerous RNA processing events in the nucleus and cytoplasm of yeast (van Hoof and Parker 1999). Rrp44p has been classified as a member of the RNR superfamily of exoribonuclease based on its 3′-5 hydrolytic exonuclease activity (Mitchell et al. 1997; Zuo and Deutscher 2001).
tRNAiMet lacking m1A58 is stabilized by mutation of TRF4, RRP44, or sup3
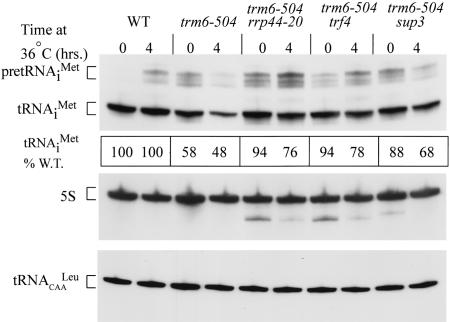
Previously, we established that trm6-504 confers selective degradation of pre-tRNAiMet due to the absence of m1A58 (Anderson et al. 1998), and that genetic suppression of trm6-504 can occur by increasing the level of hypomodified tRNAiMet (Anderson et al. 1998). We sought to determine whether mutations in TRF4 or RRP44 suppress trm6-504 by this same mechanism. Total RNA was isolated from the isogenic strains TRM6, trm6-504, trm6-504 sup1 (trf4-20), trm6-504 sup2 (rrp44-20), and trm6-504 sup3 grown at 26°C and following a shift to 36°C, the nonpermissive temperature for trm6-504 (Anderson et al. 1998). Northern blot analysis of the steady-state levels of tRNAiMet revealed a significant increase in the level of mature tRNAiMet in all three suppressor-containing strains compared to the parental trm6-504 mutant (Fig. 3). Quantification of mature tRNAiMet after normalization to 5S rRNArRNA revealed that the levels of mature tRNAiMet in the trm6-504 single mutant were 58% and 48% of the wild-type levels in the TRM6 strain at 26°C and 36°C, respectively (Fig. 3), in agreement with our previous findings (Anderson et al. 1998). Importantly, the levels of mature tRNAiMet in the suppressor strains were elevated to 88%-94% of the wild-type level at 26°C and to 68%-78% of wild type at 36°C (Fig. 3). The fact that the level of mature tRNAiMet in the suppressor strains was significantly lower than that in wild type cells may reflect degradation of tRNAiMet by a Trf4p- and Rrp44p-independent pathway that is revealed only at elevated temperatures. Alternatively, the suppressor mutations may not completely inactivate the functions of Trf4p and Rrp44p involved in degradation of hypomodified tRNAiMet. The high level of pre-tRNAiMet in the trm6-504 mutant compared to the TRM6 strain at 26°C (Fig. 3 TRM6 vs. trm6-504, 0 h) indicates a reduced efficiency of pre-tRNAiMet processing in the absence of m1A58. Consistent with the idea that processing of pre-tRNAiMet is slowed in a trm6 mutant (Calvo et al. 1999) and that mutant pre-tRNAiMet is the target for degradation at elevated temperatures (Anderson et al. 1998), the steady-state level of pre-tRNAiMet in trm6-504 is reduced at 36°C compared to wild type (Fig. 3). In the trm6-504 suppressor strains, pre-tRNAiMet is more stable at 36°C and its accumulation varies between suppressors at 26°C, suggesting that pre-tRNAiMet is more stable in the suppressors strains at 36°C and that processing rates of pre-tRNAiMet vary significantly between individual suppressors (Fig. 3). To rule out the remote possibility that increased levels of mature tRNAiMet in the suppressors was due to an increase in m1A production, we measured the level of m1A in total tRNA from TRM6, trm6-504, and the trm6-504 trf4-20, rrp44-20, or sup3 double mutants by HPLC analysis (Table 2). As anticipated, m1A was nearly undetectable in the parental trm6-504 strain, and the lowlevel of m1A detected in tRNA from trm6-504 cells was unchanged in the two suppressor strains. We conclude from these results that the mechanism of suppression is most likely due to defects in the machinery that normally degrades hypomodified pre-tRNAiMet.
Figure 3.
Mutations in TRF4, RRP44, and sup3 increase the steady-state level of tRNAiMet in a trm6-504 background. Northern blot analysis of total RNA isolated from wild-type, trm6-504, or trm6-504 trf4-20, rrp44-20 or sup3 double mutants grown at a permissive (26°C) or nonpermissive (36°C) temperature. Total RNA (10 μg) was separated by electrophoresis on a 6% denaturing acrylamide (19:1) gel cast in 8 M urea. Hybridization with probes JA11 (tRNAiMet), JA99 (5S rRNA), and JA151 (tRNACAALeu) were performed as described in Materials and Methods. tRNAiMet was visualized by autoradiography and quantified by Phosphor-Imager analysis using Image Quant software; normalized to 5S rRNA or tRNACAALeu and then expressed as a percentage of tRNAiMet in wild type at the same temperature. Normalization of tRNAiMet to 5S rRNA or tRNACAALeu yielded similar results, and the quantity of tRNAiMet shown is after normalizing to 5S rRNA.
Table 2.
Detection of m1A in total tRNA by HPLC analysis
| Strains | m1A as % of WT |
|---|---|
| WT | 100 |
| trm6-504 | 7.5 |
| trm6-504 rrp44-20 | 7.2 |
| trm6-504 trf4-20 | 7.8 |
| trm6-504 sup3 | 3.0 |
m1A levels normalized to m1G are shown; normalization to m5C and Ψ gave similar results.
Degradation of tRNAiMet occurs in the nucleus
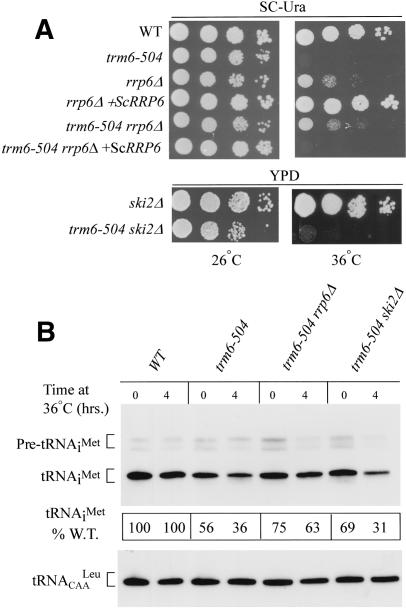
Based on results from previous work, we hypothesized that the precursor form of tRNAiMet lacking m1A58 is rapidly degraded whereas the mature form of the hypomodified tRNAiMet is stable upon reaching the cytoplasm (Anderson et al. 1998). The exosome exists in two forms, with the nuclear form containing an additional nonessential 3′-5′ exonuclease, Rrp6p. If degradation of hypomodified pre-tRNAiMet occurs in the nucleus, then it should be affected by deletion of RRP6. To test this prediction, we created a trm6-504 rrp6Δ double mutant by genetic cross (see Materials and Methods) and examined its phenotype. The results in Figure 4A suggest that the rrp6Δ mutation suppressed the Ts- phenotype of trm6-504. This conclusion was confirmed by showing that a single-copy plasmid bearing RRP6 (Briggs et al. 1998) complemented the ability of rrp6Δ to suppress trm6-504 by restoring the original Ts- phenotype of trm6-504 (Fig. 4A). Because trm6-504 suppression by trf4-20, rrp44-20, and sup3 were accompanied by an increase in the steady-state level of mature tRNAiMet, we predicted the same would be true for suppression by rrp6Δ. To test this possibility, we determined the steadystate level of tRNAiMet in the trm6-504 rrp6Δ mutant at 26°C and 36°C by Northern blot analysis. As expected, suppression of the trm6-504 Ts- phenotype was accompanied by an increase in mature tRNAiMet level compared to the trm6-504 mutant (Fig. 4B, trm6-504 vs. trm6-504 rrp6Δ). Taken together, these results strongly support the conclusion that degradation of hypomodified pre-tRNAiMet does occur in the nucleus, although they do not exclude the possibility that mature tRNAiMet degradation also occurs in the cytoplasm.
Figure 4.
(A) Degradation of hypomodified tRNAiMet occurs in the nucleus. Wild-type (Y200), trm6-504 (Y190), rrp6Δ (F23), and trm6-50 rrp6Δ (Y298) strains were transformed with YCpLac33 (B181, rows 1-3, 5) or ScRRP6 (B269, rows 4,6), or untransformed strains ski2Δ (F24) and trm6-504 ski2Δ (Y299) were tested for growth. Transformants or ski2Δ (F24) and trm6-504 ski2Δ (Y299) were grown to saturation in SC-Ura or YPD, respectively, and serial 10-fold dilutions were spotted on SC-Ura or YPD plates and incubated at 26°C and 36°C. (B) Northern blot analysis of total RNA (10μg) isolated from wild-type (F27), trm6-504 (Y190), trm6-504 rrp6Δ (Y298), and trm6-504 ski2Δ (Y299) strains grown in YPD at permissive (26°C) and after shift to the nonpermissive temperature, 36°C. RNAs separated on a 6% polyacrylamide (19:1) 8 M urea gel and transferred to a membrane were probed with JA11 to detect tRNAiMet and JA151 to detect tRNACAALeu. tRNAiMet and tRNACAALeu were visualized by autoradiography and quantified by PhosphorImager analysis using Image Quant software. tRNAiMet was normalized to the amount of tRNACAALeu in the same sample and is expressed as percentage of wild type at the same temperature.
To address whether the cytoplasmic exosome might also play a role in the degradation of aberrant tRNAiMet, we created a trm6-504 ski2Δ double mutant by genetic cross (see Materials and Methods). Ski2p is an accessory factor to the exosome that is required for the degradation of mRNA in the cytoplasm (Jacobs et al. 1998; van Hoof et al. 2002), but thus far has not been shown to function in the nuclear exosome (Jacobs et al. 1998; van Hoof et al. 2000a). The trm6-504 ski2Δ double mutant showed no significant difference in growth at 36°C compared to trm6-504, and the mature tRNAiMet level at 36°C in the double mutant was not significantly different than that seen in trm6-504 (Fig. 4A,B). From these results we conclude that it is unlikely that the cytoplasmic exosome is involved in the degradation of hypomodified tRNAiMet; however, we cannot rule out the possibility that the cytoplasmic exosome can degrade the aberrant tRNAiMet independently of the accessory factor Ski2p.
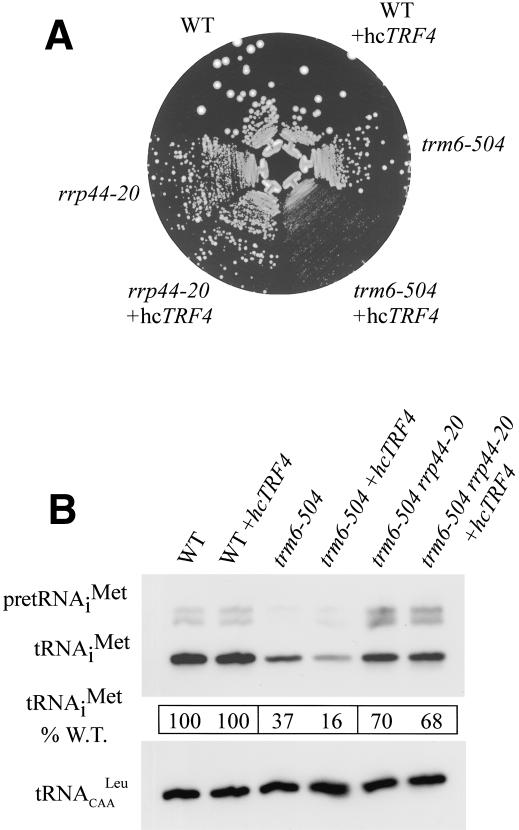
Overexpression of TRF4 exacerbates the slow growth phenotype of trm6-504 and results in a further reduction of tRNAiMet levels
Our results demonstrated that the exosome and Trf4p are required for the degradation of pre-tRNAiMet lacking m1A58 in trm6-504 cells. Next, we wished to address whether Trf4p and the exosome function independently or in conjunction to bring about degradation of hypomodified pre-tRNAiMet. Toward this end, we asked whether overexpression of Trf4p would accelerate the degradation of tRNAiMet lacking m1A58 in a manner dependent on the exosome. This was tested by introducing a multicopy-number plasmid containing wild-type TRF4 (hcTRF4) into trm6-504, TRM6, and trm6-504 rrp44-20 haploid strains. We found that the presence of hcTRF4 exacerbated the slow growth of the trm6-504 mutant at 26°C or 30°C while having no effect on the growth of TRM6 cells (Fig. 5A). This suggested that high levels of Trf4p might further destabilize pre-tRNAiMet lacking m1A58 in the trm6-504 mutant while having no affect on wild-type tRNAiMet. This idea was borne out by the results of Northern blot analysis. The tRNAiMet level in a trm6-504 mutant bearing hcTRF4 was reduced to 43% of the level found in the trm6-504 containing an empty plasmid (Fig. 5B). Thus, the level of tRNAiMet in the trm6-504 mutant was 37% of that seen in TRM6 cells but only 16% of wild type in the trm6-504+hcTRF4 cells (Fig. 5B). The reduction in the mature tRNAiMet levels in the presence of excess Trf4p requires that the tRNAiMet lacks m1A58, because the TRM6+hcTRF4 and TRM6 cells had indistinguishable amounts of mature tRNAiMet (Fig. 5B). As might be expected for a protein residing in a multisubunit complex such as the exosome, similar experiments overexpressing RRP44 in trm6-504 or TRM6 strains did not result in a measurable change in growth at 26°C or 30°C (data not shown).
Figure 5.
Overexpression of Trf4p enhances degradation of hypomodified tRNAiMet in an exosome-dependent manner. (A) Wild type (Y200), trm6-504 (Y190), and trm6-504 rrp44-20 were transformed with a high-copy-number plasmid YEpLac195 (B184) or a high-copy-number plasmid bearing TRF4 (B256), and single transformants were streaked to SC-Ura and incubated for 4 d at 30°C. (B) Northern blot analysis of total RNA (10μg) isolated from the same strains described in A grown at 30°C. Detection of precursor and mature tRNAiMet was done by hybridization with probes JA11 (tRNAiMet) and JA151 (tRNACAALeu), followed by autoradiography or PhosphorImager analysis and quantification using Image Quant software. tRNAiMet was normalized to the amount of tRNACAALeu in the same sample and is expressed as percentage of wild type bearing the same plasmid.
Trf4p function is required prior to exosome-mediated degradation of aberrant tRNAiMet
To determine whether a functional exosome is required for Trf4p-mediated degradation of tRNAiMet lacking m1A58, we asked whether overexpression of Trf4p in a strain bearing rrp44-20 would affect the growth rate or levels of mature tRNAiMet in a trm6-504 background. If exosome function is not required for the accelerated pre-tRNAiMet degradation mediated by Trf4p, then overexpression of Trf4p should reduce the growth rate of the trm6-504 rrp44-20 double mutant. At odds with this prediction, we found that hcTRF4 had no effect on the growth rate (Fig. 5A) or levels of tRNAiMet (Fig. 5B) in the trm6-504 rrp44-20 double mutant. Thus, it appears that Trf4p mediates degradation of pre-tRNAiMet lacking m1A58 via the exosome.
Polyadenylation of hypomodified precursor tRNAiMet
Exosome-mediated degradation of hypomodified pre-tRNAiMet occurs in the nucleus, and the direct involvement of Trf4p (also a nuclear protein) in the degradative process suggested that this mechanism might require the poly(A) polymerase function of Trf4p to polyadenylate pre-tRNAiMet prior to degradation. This would be reminiscent of what has been shown for degradation of a mutant tRNATrp in E. coli (Li et al. 2002). Thus, we reasoned that if polyadenylation of hypomodified pre-tRNAiMet occurs in yeast, it could signify the existence of a tRNA surveillance mechanism similar to that identified in bacterial cells. We predicted that polyadenylated hypomodified tRNAiMet would migrate in gel electrophoresis as a heterogeneous smear above the pre-tRNAiMet from trm6-504 cells. However, after long exposures, Northern blots of total RNA from trm6-504 cells failed to provide evidence that hypomodified pre-tRNAiMet was polyadenylated (data not shown).
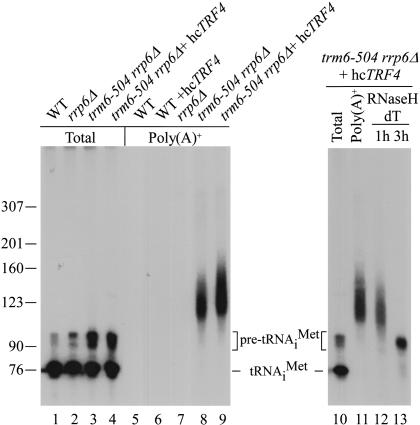
Because rrp6Δ suppresses trm6-504 and previous reports showed that deletion of RRP6 led to accumulation of polyadenylated pre-sno and snRNAs (Allmang et al. 1999a; van Hoof et al. 2000a), we reasoned that by combining rrp6Δ and trm6-504 in the same strain we might increase the abundance of polyadenylated pre-tRNAiMet by inactivating Rrp6p. Similarly, we reasoned that if Trf4p influences the polyadenylation of pre-tRNAiMet, then overexpression of Trf4p may cause a greater accumulation of the polyadenylated pre-tRNAiMet in the trm6-504 rrp6Δ mutant. Northern blot analysis of total RNA from trm6-504 rrp6Δ under conditions that favor hybridization of precursor tRNAiMet (Materials and Methods) and extended autoradiography revealed a small amount of slower migrating, heterogeneously sized tRNAiMet (data not shown). In order to enrich for the presumptive poly(A)+ pre-tRNAiMet, we selected poly(A)+ RNA from wild-type, rrp6Δ, trm6-504 rrp6Δ, and trm6-504 rrp6Δ +hcTRF4 total RNA using oligo-d(T) cellulose (Materials and Methods). We conducted a Northern blot analysis of equivalent proportions of the resulting poly(A)+ RNAs after correcting for the different input amounts of total RNA used in the oligo-d(T) selection. As predicted, we detected a smear of tRNAiMet molecules longer than pre-tRNAiMet in poly(A)+ RNA from trm6-504 rrp6Δ that was not present in the corresponding sample from wild type or rrp6Δ, indicating that hypomodified but not wild-type tRNAiMet was polyadenylated (Fig. 6, lanes 5-9). Significantly, the abundance of polyadenylated tRNAiMet increased, and it displayed a wider length distribution, in the poly(A)+ RNAs isolated from trm6-504 rrp6Δ +hc-TRF4 versus trm6-504 rrp6Δ cells (Fig. 6, lanes 8,9), demonstrating that Trf4p stimulates polyadenylation of hypomodified tRNAiMet. Pretreatment of the poly(A)+ RNA from trm6-504 rrp6Δ +hcTRF4 with oligo-d(T) and RNaseH converted the smear of polyadenylated tRNAiMet to a single species similar in size to pre-tRNAiMet (Fig. 6, lane 13) These data strongly support our model that hypomodified pre-tRNAiMet is polyadenylated prior to degradation by the nuclear exosome.
Figure 6.
Pre-tRNAiMet lacking m1A58 is polyadenylated. RNA isolated from the indicated strains cultured at 30°C was subjected to Northern blot analysis using total RNA (lanes 1-4, 5 μg; lane 10, 1 μg) or poly(A)+ RNA (lanes 5,6, 1 μg; lanes 7-9, 2 μg; lanes 11-13, 1 μg) after separation by denaturing polyacrylamide gel electrophoresis and transfer to a membrane. Hybridization with probe JA11 (tRNAiMet) was done as described in Materials and Methods with the exception that the hybridizing temperature was 60°C, and the tRNAiMet was visualized by autoradiography. Pretreatment of 1 μg of poly(A)+ RNA from the trm6-504 rrp6Δ +hcTRF4 strain with oligo-d(T18) and RNaseH was done as described in Materials and Methods; the reactions were terminated after 1 or 3 h (lanes 12,13, respectively). The standards used in size determination were radiolabeled pBR322 digested with MspI.
Discussion
We previously determined that TRM6 and TRM61 (formerly GCD10 and GCD14) comprise the essential tRNA m1A58 methyltransferase. Mutations in TRM6 or TRM61 lead to the selective instability of tRNAiMet due to the absence of the m1A58 modification, conferring temperature sensitivity and resistance to 3-AT in a gcn2 background (Harashima and Hinnebusch 1986; Garcia-Barrio et al. 1995; Anderson et al. 1998). Here we used genetic suppressor analysis to establish that mutation of the exosome subunit Rrp44p, or inactivation of Trf4p, suppresses the Ts- and 3-ATr phenotypes of a trm6-504 mutant. Our molecular analysis showed that the rrp44 and trf4 mutations suppress trm6-504 by increasing the steady-state amounts of hypomodified tRNAiMet to nearly wild-type levels. Deletion of another exosome subunit gene, RRP6, also suppressed the Ts- phenotype conferred by trm6-504, providing strong evidence that impairment of exosome function is the mechanism of suppression by the rrp44-20 mutation. Because Rrp6p is associated only with the nuclear form of the exosome, we conclude that the bulk of the degradation of hypomodified pre-tRNAiMet in trm6-504 cells occurs in the nucleus. Consistent with this, inactivation of Ski2p, an accessory factor for the cytoplasmic form of the exosome, did not suppress the Ts- trm6-504 mutant phenotype.
Overexpression of Trf4p exacerbated the reduction in hypomodified tRNAiMet levels in trm6-504 cells in a manner that was completely suppressed by the rrp44-20 mutation. Thus, we conclude that Trf4p is required for efficient degradation of unmodified tRNAiMet by the nuclear exosome. Trf4p belongs to the family of β-polymerase-like nucleotidyltransferases (Aravind and Koonin 1999) that was also shown to have poly(A) polymerase activity (Saitoh et al. 2002). Below, we propose a working model to account for the function of Trf4p in exosomemediated degradation of hypomodified tRNAiMet.
The exosome is required for degradation of tRNAiMet lacking m1A58
The role of the exosome in the degradation of tRNAiMet lacking m1A58 represents a novel function for this multisubunit RNA processing machinery. The exosome facilitates the 3′-to-5′ degradation of mRNA and the processing of many small stable RNAs, and it possesses other functions related to RNA metabolism in both the nucleus and cytoplasm (Mitchell et al. 1997; Jacobs et al. 1998; van Hoof et al. 2000a). The ability of rrp6Δ to suppress trm6-504 (Fig. 4A,B) clearly demonstrated that the nuclear exosome is heavily involved in the degradation of tRNAiMet lacking m1A58. This is consistent with the localization of Trf4p to the nucleus (Walowsky et al. 1999) and our previous finding that pre-tRNAiMet but not mature tRNAiMet lacking m1A is subject to degradation (Anderson et al. 1998). Thus, we conclude that the degradation of hypomodified tRNAiMet is carried out in the nucleus.
A proposed pathway for the surveillance and degradation of tRNAiMet lacking m1A58
TRF4 was originally identified in a genetic screen for mutations that lead to synthetic lethality in combination with a topoisomerase I mutant, top1-7 (Sadoff et al. 1995a). This finding was followed by several studies that provided strong support for a model where Trf4p functions as a DNA polymerase, termed pol σ, which may couple DNA replication with the formation of sites of sister chromatid cohesion during mitosis (Castano et al. 1996a; Wang et al. 2000, 2002). Additional genetic studies uncovered TRF5, encoding a highly related protein Trf5p, which is 55% identical to Trf4p (Castano et al. 1996b). More recently, TRF4 and TRF5 were found to physically and functionally interact with DNA polymerase ε (Edwards et al. 2003).
How does the loss of Trf4p function lead to an increased level of tRNAiMet lacking m1A58? As pol σ, Trf4p might function to repress transcription of the IMT genes during mitosis. This possibility is suggested by the failure of a trf4 top1 double mutant to condense rDNA repeats during mitosis (Castano et al. 1996a) and to repress ACT1 transcription after cells reach stationary phase (Sadoff et al. 1995b). If true, then loss of Trf4p function should lead to increased IMT gene transcription and higher steady-state levels of pre-tRNAiMet in the trm6-504 mutant at all growth temperatures, analogous to the effect of increased IMT4 dosage (Anderson et al. 1998). However, the trf4 mutation did not produce a significant increase in the steady-state level of pre-tRNAiMet in the trm6-504 mutant at the permissive temperature (Fig. 3), nor did it affect the steady-state levels of other tRNAs (data not shown). Instead, we favor a model whereTrf4p is directly involved in the detection and degradation of aberrant tRNA independently of its function as a DNA polymerase. This hypothesis is supported by our finding that overexpression of Trf4p in trm6-504 exacerbated its slow growth phenotype and reduced the level of tRNAiMet lacking m1A58. Because Trf4p overexpression in the trm6-504 rrp44-20 mutant did not decrease tRNAiMet levels or reduce growth (Fig. 5), we conclude that Trf4p promotes the degradation of tRNAiMet lacking m1A58 via the exosome. It is important to note that overexpression of Trf4p reduced the level of tRNAiMet lacking m1A58, but had no effect on wild-type tRNAiMet expression (Fig. 5B). We propose that this selective degradation results from an abnormal structure of the mutant tRNAiMet that is recognized as aberrant and targeted for degradation, rather than being a specific response to the absence of m1A58. This argument is supported by the unique tertiary structure of tRNAiMet involving m1A58 (Basavappa and Sigler 1991) and the fact that tRNAiMet is the only known tRNA that is degraded in the absence of m1A.
Mutation of TRF4 affects rRNA processing as well as tRNAiMet levels in trm6-504 cells (Fig. 3; S. Kadaba and J. Anderson, unpubl.), suggesting that the activity of Trf4p may be required for the exosome to efficiently process rRNA precursors. By extending this idea, Trf4p could be required for all functions of the exosome. In this regard, Trf4p could be functioning as a poly(A) polymerase, like the related Cid13p in S. pombe (Saitoh et al. 2002) to stabilize one or more mRNAs in the cytoplasm. The predicted target of Trf4p would be one or more mRNAs encoding an exosome subunit. In this view, loss of Trf4p function would result in destabilization of an mRNA encoding an exosome subunit, with attendant reduction in exosome function and subsequent stabilization of tRNAiMet lacking m1A58. This hypothesis is inconsistent with the finding that a GFP-Trf4p fusion was found localized to the nucleus of yeast (Walowsky et al. 1999), although a small amount of GFP-Trf4p could be undetectable in the cytoplasm under these conditions and might be enough to carry out polyadenylation of a limited number of mRNAs.
A more direct role for Trf4p in promoting the degradation of unmodified tRNAiMet is suggested by the mechanism recently established for degradation of a mutant tRNA in bacteria (Saitoh et al. 2002). A mutation in E. coli tRNATrp renders it unstable and results in temperature-sensitive growth. Introduction of mutations in poly(A) polymerase and polynucleotide phosphorylase stabilized the mature mutant tRNATrp and caused its precursor to accumulate (Li et al. 2002). Subsequently, it was shown that the mutant precursor tRNATrp is adenylated in a poly(A) polymerase-dependent manner, and the presence of adenosines is required for most of the observed tRNATrp instability. It was thus concluded that a mechanism requiring adenylation and exonucleolytic cleavage was responsible for eliminating mutant precursor tRNATrp from E. coli. In this report, we provide strong evidence that a pathway similar to the one described in E. coli exists in S. cerevisiae. Our working model is that Trf4p marks pre-tRNAiMet lacking m1A58 by polyadenylation, which targets the tRNA for rapid degradation by the exosome. A tacit assumption of this model is that the polyadenylated pre-tRNAiMet is too short-lived to be detected by Northern blot analysis of total RNA in trm6-504 cells.
We gained considerable support for our model by identifying polyadenylated pre-tRNAiMet in a trm6-504 rrp6Δ mutant (Fig. 6). The fact that polyadenylated pre-tRNAiMet is found in trm6-504 rrp6Δ but not in the rrp6Δ or wild-type strain indicates that hypomodified, but not wild-type pre-tRNAiMet, is subject to polyadenylation. The increased abundance and shift to longer poly(A) tail lengths we observed in the trm6-504 rrp6Δ+hcTRF4 mutant compared to trm6-504 rrp6Δ further supports the proposed role for Trf4p as the poly(A) polymerase responsible for adenylation of hypomodified pre-tRNAiMet. However, we cannot rule out the possibility that Pap1p, the major poly(A) polymerase in yeast, is responsible for some or all of the pre-tRNAiMet adenylation. In this regard, a Pap1p-dependent polyadenylation of wild-type pre-snoRNAs U14, U18, snR73, and snR72 in a rrp6Δ mutant has been described (Allmang et al. 1999a; van Hoof et al. 2000a). Furthermore, Van Hoof et al. (2000a) proposed that polyadenylation represents a normal but nonessential step in the 3′-end processing of wild-type pre-snoRNAs. This is in contrast to the polyadenylation of pre-tRNAiMet described here, which is limited to the hypomodified molecules in trm6-504 cells and seems more consistent with an RNA surveillance pathway that eliminates abnormal tRNAs. Moreover, the poly(A) tail lengths on hypomodified pre-tRNAiMet are ∼10-50 nucleotides in length (Fig. 6), in contrast to the 40-100 nucleotide lengths of poly(A) tails found on pre-sno and snRNAs (Allmang et al. 1999a; van Hoof et al. 2000a). This distinction may reflect differences in the biochemical activities of yeast Pap1p and Trf4p, or it could underlie the differences between a pathway of normal pre-snRNA and snoRNA processing versus degradation of hypomodified pre-tRNAiMet and possibly other abnormally processed stable RNAs. In stark contrast to the polyadenylation of pre-tRNAiMet in yeast, degradation of mutant pre-tRNATrp in E. coli is preceded by adenylation of the tRNA precursor with only 1-3 adenosines (Li et al. 2002).
We propose that the degradation of mutant tRNAs in yeast and bacteria represent similar mechanisms that exist to eliminate aberrant tRNAs from cells that might otherwise negatively affect their growth. Elucidating the exact role of Trf4p in the degradation of hypomodified tRNAiMet and demonstration that adenylation is a prerequisite for degradation will allow us to refine our model and deepen our understanding of the role played by the exosome in nuclear RNA surveillance and degradation.
Materials and methods
Plasmid constructions
Plasmids used in this study are described in detail in Table 1. B269 and CB432 were generous gifts from Dr. S. Butler (University of Rochester, Rochester, NY) and Dr. M. Christman (Boston University, Boston, MA). B227, B228, B229, and pSup1.1 and 1.2 were identified as complementing plasmids from a genomic library during suppressor screening. The entire RRP44 ORF plus 600 base pairs upstream and 175 base pairs downstream were amplified by PCR using primers JA126 and JA127 and B228 DNA as a template. The PCR product was digested with XhoI and XmaI and cloned into SalI/XmaI-digested YCplac33 (B234) or YEpLac195 (B251). B285 was created by cloning a 1.1-kb EcoRI fragment isolated from EcoRI-digested B227 into EcoRI-digested YIpLac211. B254 was created by cloning an ∼3.0 kb SspI/PmeI fragment from pSup1.1 into SmaI-digested YCplac33. Cloned DNAs were propagated through transformation of E. coli DH10B cells (Invitrogen) by electroporation. Yeast transformations were done as described (Ito et al. 1983).
Yeast strains and media
Standard genetic techniques and culture media were employed as described (Sherman et al. 1974b). Growth analysis using plates containing 3-AT was done as described (Hinnebusch and Fink 1983). Yeast strains used in this study are described in Table 3. Standard genetic techniques and culture media were employed as described (Sherman et al. 1974a). Y193 was created by self mating of H2457 after transformation with YCp50-HO plasmid, containing the HO endonuclease gene required for mating type switching (Herskowitz and Jensen 1991). Diploid cells were identified by their inability to mate with mating testers of Mat a or α genotype. Tetrad dissection and analysis of Y193 yielded four isogenic haploid spores (Y189, Y190, Y191, and Y192) from a single tetrad. Y190 and Y192 were used to create Y200 and Y201 by introducing ClaI-digested B145 (YIpTRM6) into Y190 (trm6-504) and selecting for Ura+ transformants on synthetic complete medium lacking uracil (SCUra). The transformants containing both TRM6 and trm6-504 genes were identified by growth on Sc-Ura at 36°C. Eviction of the integrated plasmid by homologous recombination was selected by replica plating transformants to SC + Ura containing 1.0 μg/mL 5-FOA (Boeke et al. 1987; Rothstein 1991). 5-FOA-resistant colonies retaining the TRM6 allele were identified by their growth on YPD at 36°C and failure to grow on SC plates containing (30mM) 3-AT (Hinnebusch and Fink 1983). Y298, Y299, and Y300 were created by mating F23, F24, and F22 to Y190 (trm6-504), respectively. The resulting diploids were subjected to sporulation, and tetrad analysis was conducted to identify haploids containing trm6-504 and the corresponding mutant. Y306 was created by transformation of sup2 trm6-504 with BglII-digested B285 to integrate the URA3 gene within the IFM1 locus upstream of RRP44. Ura+ transformants were selected on SC-Ura medium and tested for growth on YP glycerol medium (YPG). Genomic DNA was isolated from one positive respiratory-deficient transformant incapable of growth on YPG medium, and disruption of the IFM1 locus was verified by PCR analysis of genomic DNA with primers JA239, JA240, and JA241 (Table 4).
Table 3.
Genotypes of strains used
| Strain | Genotype | Reference |
|---|---|---|
| H2457 | MATα trm6-504, gcn2-101, his1-29, ura3-52, ino1 (HIS4-lacZ, ura3-52) | Anderson et al. 1998 |
| Y193 | MATa/MATα trm6-504/trm6-504, gcn2-101/gcn2-101, his1-29/his1-29 ura3-52/ura3-52, ino1/ino1 (HIS4-lacZ, ura3-52) | This study |
| Y190 | MATa trm6-504, gcn2-101, his1-29, ura3-52, ino1 (HIS4-lacZ, ura3-52) | This study |
| Y192 | MATα trm6-504, gcn2-101, his1-29, ura3-52, ino1 (HIS4-lacZ, ura3-52) | This study |
| Y200 | MATa TRM6, gcn2-101, his1-29, ura3-52, ino1, (HIS4-lacZ, ura3-52) | This study |
| Y201 | MATα TRM6, gcn2-101, his1-29, ura3-52, ino1, (HIS4-lacZ, ura3-52) | This study |
| Sup1 | MATa trf4-20, trm6-504, gcn2-101, his1-29, ura3-52, ino1, (HIS4-lacZ, ura3-52) | This study |
| Sup2 | MATα rrp44-20, trm6-504, gcn2-101, his1-29, ura3-52, ino1, (HIS4-lacZ, ura3-52) | This study |
| Sup3 | MATαsup3, trm6-504, gcn2-101, his1-29, ura3-52, ino1, (HIS4-lacZ, ura3-52) | This study |
| Y298 | MATa rrp6Δ:KanMX4 trm6-504, ura3 | This study |
| Y299 | MATα ski2Δ:KanMX4 trm6-504, ura3 | This study |
| Y300 | MATα trf4Δ:KanMX4 trm6-504, ura3 | This study |
| Y306 | MATα rrp44-20, trm6-504, gcn2-101, his1-29, ura3-52, ino1, ifml::URA3<B285> (HIS4-lacZ, ura3-52) | This study |
| F22 | MATα his3Δ, leu2Δ, lys2Δ, ura3Δ, trf4Δ:KanMX4 clone#11777 | Invitrogen |
| F23 | MATα his3Δ, leu2Δ, lys2Δ, ura3Δ, rrp6Δ:KanMX4 clone#16265 | Invitrogen |
| F24 | MATα his3Δ, leu2Δ, lys2Δ, ura3Δ, ski2Δ:KanMX4 clone#15307 | Invitrogen |
| F27 | MATα his3Δ, leu2Δ, lys2Δ, ura3Δ | Invitrogen |
Table 4.
Oligonucleotides used
| Oligo | Target | Sequence |
|---|---|---|
| JA126 | RRP44 | TCCCCCCGGGTTTGGGAAATGTCGTGCTCGACGT |
| JA127 | RRP44 | CCGCTCGAGGTCCACCACCAAAATGTCAA |
| JA11 | tRNAiMet | TCGGTTTCGATCCGAGGACATCAGGGTTATGA |
| JA99 | 5S rRNA | TCGCGTATGGTCACCCACTACA |
| JA239 | YIpLac211 | GTTCTGCTATGTGGCGCGGTATTATC |
| JA240 | YIpLac211 | CTCTCAAGGATCTTACCGCTGTTG |
| JA241 | IFM1 5′ | GATGCCCCAGACATTAGAAGCC |
| JA125 | 5.8S rRNA | GCGTTSTTCATCGATGC |
| JA151 | tRNACAALeu | TGGTTGCTAAGAGATTCGAAC |
Generation and identification of trm6-504 suppressors
Selection of suppressors was carried out using strains Y190 and Y192. One hundred individual colonies from each mating type were patched and grown at 26°C overnight on YPD medium. The plates containing the patches were replica-plated to YPD and grown at 36°C for 3-7 d. Individual papillae were picked and streaked for single colonies on YPD plates and grown at 26°C. Two individual colonies from each streak were patched onto YPD plates, grown at 26°C, and replica-plated to YPD and grown at 36°C, 16°C, or 30°C, and to SC-His supplemented with 30 mM 3-AT in order to identify Ts+ and 3-ATs revertants. We identified 150 such revertants, and 49 exhibited a cold-sensitive phenotype (Cs-). A revertant's dominance or recessiveness was determined by mating each revertant of one mating type to the corresponding opposite mating type trm6-504 Sup+ strain (Y190 or Y192) and testing for complementation of the revertant phenotype. Complementation of the revertant phenotypes Ts+ 3-ATs to Ts-, 3-ATr indicated that the revertant was recessive. Complementation groups were assigned by mating revertants from Y190 with revertants from Y192 and testing for complementation of Ts+, 3-ATs phenotypes. Complementation to Ts- and 3-ATr indicated that the revertants were in different complementation groups, and failure to complement placed the revertants in the same complementation group. Of the 49 Cs- revertants, we assigned three complementation groups, designating them sup1-3. The wild-type alleles of sup1 and sup2 were identified by transforming representative strains of each complementation group with a single-copy yeast genomic plasmid library (Rose et al. 1987) and plating on minimal media lacking uracil at 30°C overnight. After this preincubation period at a permissive temperature, transformation plates were incubated at 16°C for 3 d. Growth at 16°C was taken to indicate complementation of the Cs- phenotype linked to the suppressor gene. The Cs+ transformants were tested for growth on YPD plates at 36°C and 16°C, and on plates containing 3-AT as described above. Plasmids were rescued from Ts-, Cs+, and 3-ATr transformants in E. coli (Hoffman and Winston 1987), and the rescued plasmids were used to confirm complementation of the cognate mutants. Plasmids that passed this test were analyzed to determine the ends of the genomic DNA inserts by DNA sequencing using primers complementary to sequences flanking the BamHI site of YCp50. These sequences were compared to the yeast genome using the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces). sup1 was confirmed as TRF4 by transforming the sup1 strain with scTRF4 (B254) and also by creating a trm6-504 trf4Δ double mutant and showing that deletion of TRF4 suppressed the trm6-504 mutant phenotypes. To confirm the identity of sup1 as a mutant allele of TRF4, we conducted marker-rescue experiments using two integrative plasmids containing the 5′ or 3′ half of TRF4 derived from YIpLac211 (Gietz and Sugino 1988). Integration of plasmid trf4mp2 bearing the 5′ half of TRF4 (∼1.5-kb SspI-HindIII fragment) but not plasmid trf4mp3 bearing 3′ half of TRF4 (∼1.5-kb HindIII-PmeI fragment) complemented the sup1 phenotypes and restored trm6-504 mutant phenotypes. Eviction of the integrated plasmids was selected by replica-plating transformants to 5-FOA-containing medium, and the 5FOAr colonies were screened for trm6-504 phenotypes. Of the 20 5-FOAr colonies from the transformant containing the 5′ half of TRF4, 12 exhibited Sup+ and eight showed sup- phenotype.
sup2 was confirmed as RRP44 by linkage analysis. After mating Y190 ura3-52, gnc2-101, trm6-504, Sup+ to Y306 ura3-52, gcn2-101, trm6-504, sup2, ifm1::URA3, the resulting diploid was subjected to sporulation and tetrad analysis. In 18 out of 18 tetrads we observed cosegregation of the Ts+, Cs-, and 3-ATs phenotype of sup2, with the Ura+ phenotype of the disrupted ifm1::URA3 allele integrated near RRP44.
RNA isolation and blotting
Total RNA was isolated as described (Kohrer and Domdey 1991). For RNA blotting, total RNA was separated on a 6% polyacrylamide-bis-acrylamide (19:1) gel cast in 8 M ureaand 1× Tris-Borate-EDTA (TBE) buffer (Maniatis et al. 1982) by electrophoresis at 450 V for 1 h unless otherwise stated. The separated RNA was transferred to a Hybond N+ membrane (Amersham Pharmacia) at 12 V for 5 h in 0.5× TBE. The blot was probed using radiolabeled oligonucleotides (Table 4) for specific RNAs in 0.25 M Na2HPO4, 1 mM EDTA, 1% BSA, and 7% SDS buffer. The oligonucleotides were radiolabeled by incubating (γ-32P)ATP (Amersham Pharmacia) and T4 Polynucleotide Kinase (New England Biolabs) with 10 pmole of oligonucleotide at 37°C for 1 h.
tRNA isolation and modification studies
Total RNA was extracted from yeast grown in YPD (Kohrer and Domdey 1991). tRNA was isolated by DEAE-cellulose chromatography of total RNA (Hatfield et al. 1979). Three hundred μg of purified tRNA was lyophilized and analyzed by HPLC as described (Gehrke et al. 1982; Gehrke and Kuo 1990). The amount of m1A present in a tRNA sample was determined by taking the area under the m1A peak and dividing it by the area of the peak for m1G, m5C, or ψ from the same sample. This normalized m1A content was compared to the same value derived from tRNA isolated from the wild-type strain, Y200.
Oligo d(T) purification of PolyA+ RNAs
Yeast strains, wild type, wild-type +hcTRF4, rrp6Δ, trm6-504 rrp6Δ, and trm6-504 rrp6Δ +hcTRF4 were grown to an optical density at A600nm of 0.5-0.6 in 500 mL SC-Ura at 30°C. Cells were harvested at 6000 × g for 5 min at 4°C and washed with 1× phosphate buffered saline (PBS). The pellet was resuspended in 2.0 mL of lysis buffer (20 mM Tris at pH 7.4, 50 mM LiCl, 1% SDS, 1% BME, 1 mg/mL Heparin, 10 mM Vanadyl-ribonucleoside complex [New England Biolabs] 300 μg/mL Proteinase K [New England Biolabs]) and passed through a French pressure cell (20,000 psi) twice to effect cell lysis. The lysate was incubated at 42°C for 15 min, and then EDTA was added to a final concentration of 10 mM, and incubation at 42°C was continued for 15 min. The lysate was incubated at 65°C for 10 min and put on ice until cool. Lithium chloride was added to a final concentration of 0.5 M, and the lysate was centrifuged at 6000 × g for 5 min at 4°C. Fifty microliters of the cleared lysate was removed for total RNA extraction as described (Kohrer and Domdey 1991). The remaining lysate was incubated with 0.1 g of oligo d(T) Cellulose (New England Biolabs) pre-equilibrated with binding buffer (10 mM Tris at pH 7.4, 1 mM EDTA, 0.5% SDS and 0.5 M LiCl) at room temperature for 25 min. with constant mixing. The lysate-oligo-d(T) mixture was applied to an econocolumn (Bio-Rad) and washed with 10 mL of binding (20× bed volume). Poly(A)+ RNA was eluted using 5.0 mL of elution buffer (10 mM Tris, 1 mM EDTA and 0.05% SDS). The eluate was heated to 65°C, cooled on ice, and brought to 0.5 M LiCl before being incubated and chromatographed with oligo-d(T) a second time. The poly(A)+ RNA was eluted from oligo-d(T) with elution buffer by collection 0.5-mL fractions. The O.D.260 of each fraction was determined, and fractions having O.D.260 of 0.2 or greater were pooled, phenol chloroform-extracted, and precipitated with 0.3 M sodium acetate and three volumes of ethanol.
Oligo d(T) RNaseH treatment of PolyA+ RNAs
First, 1.0 μg of PolyA+ RNA from trm6-504/rrp6Δ +hcTRF4 was dried with 300 ng of oligo d(T18) and resuspended in 10 μL of hybridization buffer (25 mM Tris at pH 7.5, 1 mM EDTA, and 50 mM NaCl) and incubated at 68°C for 20 min, 42°C for 10 min, 30°C for 10 min, and then placed on ice for 5.0 min. The reaction was started by the addition of 10 μL of 2× RNaseH buffer containing 250 units RNaseH (40 mM Tris at pH 7.5, 20 mM MgCl2, 100 mM NaCl, 2 mM DTT, and 60 μg/mL of BSA) and incubated at 30°C for 1 h. A duplicate reaction was incubated at 30°C for 3 h with the addition of 0.5 μL (250 units) RNaseH (NEB) after 1 h. The reaction was stopped by the addition of 0.1 mL of stop buffer (4 μg/mL E. Coli tRNAPhe, 20 mM EDTA, and 300 mM NaoAc) The samples were phenol chloroform-extracted and precipitated with three volumes of ethanol.
Acknowledgments
We thank Drs. Michael F. Christman, J. Scott Butler, César Nombela, and Roy Parker for the generous gifts of reagents. We thank Glen R. Björk and Kerstin Jacobsson for conducting the HPLC analysis. G.R.B. and K.J. are supported by grants (BU-2930) and (proj 680) from the Swedish Natural Science Research Council and Swedish Cancer Society. We thank Dr. Scott Butler for critical reading of this manuscript. This work was supported by NSF grant D.B.I. 0100667.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1183804.
References
- Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., and Tollervey, D. 1999a. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18: 5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D., and Mitchell, P. 1999b. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes & Dev. 13: 2148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., Cuesta, R., Carlson, B.A., Pak, M., Asano, K., Bjork, G.R., Tamame, M., and Hinnebusch, A.G. 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 12: 3650-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, Y., Takahashi, S., Kobayashi, T., Kajiho, H., Hoshino, S., and Katada, T. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20: 4684-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L. and Koonin, E.V. 1999. DNA polymerase β-like nucleotidyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27: 1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa, R. and Sigler, P.B. 1991. The 3 Å crystal structure of yeast initiator tRNA: Functional implications in initiator/elongator discrimination. EMBO J. 10: 3105-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork, G.R. 1995. Biosynthesis and function of modified nucleosides. In tRNA: Structure, biosynthesis and function (eds. D. Söll and U. RajBhandary), pp. 165-205. ASM Press, Washington, DC.
- Boeke, J.D., Trueheart, J., Natsoulis, G., and Fink, G.R. 1987. 5-fluoroorotic acid as a selective agent in yeast molecular genes. Methods Enzymol. 154: 164-175. [DOI] [PubMed] [Google Scholar]
- Briggs, M.W., Burkard, K.T., and Butler, J.S. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273: 13255-13263. [DOI] [PubMed] [Google Scholar]
- Brown, J.T., Bai, X., and Johnson, A.W. 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard, K.T. and Butler, J.S. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell Biol. 20: 604-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, O., Cuesta, R., Anderson, J., Gutierrez, N., Garcia-Barrio, M.T., Hinnebusch, A.G., and Tamame, M. 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4167-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano, I.B., Brzoska, P.M., Sadoff, B.U., Chen, H., and Christman, M.F. 1996a. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes & Dev. 10: 2564-2576. [DOI] [PubMed] [Google Scholar]
- Castano, I.B., Heath-Pagliuso, S., Sadoff, B.U., Fitzhugh, D.J., and Christman, M.F. 1996b. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 24: 2404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby, D., Leboy, P.S., and Guthrie, C. 1981. Yeast tRNA precursor mutated at a splice junction is correctly processed in vivo. Proc. Natl. Acad. Sci. 78: 415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta, R., Hinnebusch, A.G., and Tamame, M. 1998. Identification of GCD14 and GCD15, novel genes required for translational repression ofGCN4 mRNA in Saccharomyces cerevisiae. Genetics 148: 1007-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, S., Li, C.M., Levy, D.L., Brown, J., Snow, P.M., and Campbell, J.L. 2003. Saccharomyces cerevisiae DNA polymerase epsilon and polymerase σ interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol. Cell Biol. 23: 2733-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, S.P. and Yarus, M. 1980. The structure and aminoacylation of a temperature-sensitive tRNATrp (Escherichia coli). J. Biol. Chem. 255: 1128-1137. [PubMed] [Google Scholar]
- Eschenlauer, J.B., Kaiser, M.W., Gerlach, V.L., and Brow, D.A. 1993. Architecture of a yeast U6 RNA gene promoter. Mol. Cell Biol. 13: 3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio, M.T., Naranda, T., Cuesta, R., Hinnebusch, A.G., Hershey, J.W.B., and Tamame, M. 1995. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes & Dev. 9: 1781-1796. [DOI] [PubMed] [Google Scholar]
- Gehrke, C.W. and Kuo, K.C. 1990. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography, p A3-A71. In Chromatography and modification of nucleosides (eds. C.W. Gehrke and K.C. Kuo). Elsevier, Amsterdam. [DOI] [PubMed]
- Gehrke, C.W., Kuo, K.C., McCune, R.A., Gerhardt, K.O., and Agris, P.F. 1982. Quantitative enzymatic hydrolysis of tRNA: Reversed-phase high-performance liquid chromatography of tRNA nucleoside. J. Chromatogr. 230: 297-308. [PubMed] [Google Scholar]
- Gietz, R.D. and Sugino, A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527-534. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., Hurt, E., and Simos, G. 2000. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes & Dev. 14: 830-840. [PMC free article] [PubMed] [Google Scholar]
- Harashima, S. and Hinnebusch, A.G. 1986. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell Biol. 6: 3990-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield, D., Matthews, C.R., and Rice, M. 1979. Aminoacyl-transfer RNA populations in mammalian cells chromatographic profiles and patterns of codon recognition. Biochim. Biophys. Acta 564: 414-423. [DOI] [PubMed] [Google Scholar]
- Herskowitz, I. and Jensen, R.E. 1991. Putting the HO gene to work: Practical uses for mating-type switching. In Methods in enzymology: Guide to yeast genetics and molecular biology (eds. C. Guthrie and G.R. Fink), pp. 132-146. Academic Press, San Diego. [DOI] [PubMed]
- Hilleren, P. and Parker, R. 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33: 229-260. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538-542. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. 1997. Translational regulation of yeast GCN4: A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272: 21661-21664. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. and Fink, G.R. 1983. Repeated DNA sequences upstream from HIS1 also occur at several other coregulated genes in Saccharomyces cerevisiae. J. Biol. Chem. 258: 5238-5247. [PubMed] [Google Scholar]
- Hoffman, C.S. and Winston, F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267-272. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17: 162-180. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukada, Y., Murata, K., and Kimura, A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J.S., Anderson, A.R., and Parker, R.P. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, M.J. and Bystrom, A.S. 2002. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA 8: 324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer, K. and Domdey, H. 1991. Preparation of high molecular weight RNA. In Methods in enzymology: Guide to yeast genetics and molecular biology (eds. C. Guthrie and G.R. Fink), pp. 398-405. Academic Press, San Diego. [DOI] [PubMed]
- Kushner, S.R. 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184: 4658-4665; discussion 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Reimers, S., Pandit, S., and Deutscher, M.P. 2002. RNA quality control: Degradation of defective transfer RNA. EMBO J. 21: 1132-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T.H. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell Biol. 22: 8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M. and McNally, M.T. 2003. mRNA decay: X (XRN1) marks the spot. Mol. Cell 11: 1126-1128. [DOI] [PubMed] [Google Scholar]
- Lund, E. and Dahlberg, J.E. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082-2085. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. 1982. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Maquat, L.E. 2002. Molecular biology. Skiing toward nonstop mRNA decay. Science 295: 2221-2222. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2001. mRNA turnover. Curr. Opin. Cell Biol. 13: 320-325. [DOI] [PubMed] [Google Scholar]
- ____. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosomemediated 3′ → 5′ degradation. Mol. Cell 11: 1405-1413. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457-466. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. 2002. Nuclear RNA turnover. Cell 108: 431-434. [DOI] [PubMed] [Google Scholar]
- Noguchi, E., Hayashi, N., Azuma, Y., Seki, T., Nakamura, M., Nakashima, N., Yanagida, M., He, X., Mueller, U., Sazer, S., et al. 1996. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 15: 5595-5605. [PMC free article] [PubMed] [Google Scholar]
- Rose, M., Novick, P., Thomas, J., Botstein, D., and Fink, G.R. 1987. A Saccharomyces cerevisiae plasmid bank based on a centromere-containing shuttle vector. Gene 60: 237-243. [DOI] [PubMed] [Google Scholar]
- Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. In Methods in enzymology: Guide to yeast genetics and molecular biology (eds. C. Guthrie and G.R. Fink), pp. 281-301. Academic Press, San Diego. [DOI] [PubMed]
- Sadoff, B.U., Heath-Pagliuso, S., Castano, I.B., Zhu, Y., Kieff, F.S., and Christman, M.F. 1995a. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 141: 465-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1995b. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 141: 465-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W.H., Thelander, L., Yates, J.R., and Russell, P. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109: 563-573. [DOI] [PubMed] [Google Scholar]
- Sarkar, S., Azad, A.K., and Hopper, A.K. 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 96: 14366-14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Lawrence, C.W. 1974a. Methods of yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- ____. 1974b. Methods of yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- van Hoof, A. and Parker, R. 1999. The exosome: A proteasome for RNA? Cell 99: 347-350. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Messenger RNA degradation: Beginning at the end. Curr. Biol. 12: R285-R287. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., Lennertz, P., and Parker, R. 2000a. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell Biol. 20: 441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Staples, R.R., Baker, R.E., and Parker, R. 2000b. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell Biol. 20: 8230-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Frischmeyer, P.A., Dietz, H.C., and Parker, R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262-2264. [DOI] [PubMed] [Google Scholar]
- Walowsky, C., Fitzhugh, D.J., Castano, I.B., Ju, J.Y., Levin, N.A., and Christman, M.F. 1999. The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J. Biol. Chem. 274: 7302-7308. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Castano, I.B., De Las Penas, A., Adams, C., and Christman, M.F. 2000. Pol κ: A DNA polymerase required for sister chromatid cohesion [see comments]. Science 289: 774-779. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Castano, I.B., Adams, C., Vu, C., Fitzhugh, D., and Christman, M.F. 2002. Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol σ DNA polymerase. Genetics 160: 381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wang, W., and Peltz, S.W. 2001. Curbing the nonsense: The activation and regulation of mRNA surveillance. Genes & Dev. 15: 2781-2785. [DOI] [PubMed] [Google Scholar]
- Wolfner, M., Yep, D., Messenguy, F., and Fink, G.R. 1975. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J. Mol. Biol. 96: 273-290. [DOI] [PubMed] [Google Scholar]
- Yuo, C.Y. and Weiner, A.M. 1989. Genetic analysis of the role of human U1 snRNA in mRNA splicing: I. Effect of mutations in the highly conserved stem-loop I of U1. Genes & Dev. 3: 697-707. [DOI] [PubMed] [Google Scholar]
- Zuo, Y. and Deutscher, M.P. 2001. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 29: 1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]