Abstract
We have searched for sequence motifs that contribute to the recognition of human pre-mRNA splice sites by comparing the frequency of 8-mers in internal noncoding exons versus unspliced pseudo exons and 5′ untranslated regions (5′ untranslated regions [UTRs]) of transcripts of intronless genes. This type of comparison avoids the isolation of sequences that are distinguished by their protein-coding information. We classified sequence families comprising 2069 putative exonic enhancers and 974 putative exonic silencers. Representatives of each class functioned as enhancers or silencers when inserted into a test exon and assayed in transfected mammalian cells. As a class, the enhancer sequencers were more prevalent and the silencer elements less prevalent in all exons compared with introns. A survey of 58 reported exonic splicing mutations showed good agreement between the splicing phenotype and the effect of the mutation on the motifs defined here. The large number of effective sequences implied by these results suggests that sequences that influence splicing may be very abundant in pre-mRNA.
Keywords: splicing, pre-mRNA, motifs, exon, enhancers, silencers
A fundamental step in the transfer of genetic information from DNA to protein is the splicing of RNA transcripts. In this process, relatively small exons (∼100 nt) are selected from among generally much larger introns (thousands of nucleotides) and are joined to form mature mRNA. Pre-mRNA splicing is accomplished by two sequential transesterification reactions catalyzed by a very large ribonucleoprotein complex known as the spliceosome (Burge et al. 1999). A spliceosome is either recruited or assembled at the correct 5′ splice site (donor) and 3′ splice site (acceptor) in part through recognition of conserved sequences spanning the intron-exon junctions (Burge et al. 1999). However, the sequence conservation at the splice sites is incomplete, such that there are many false sites that match the consensus sequence as well or better than the true sites (Senapathy et al. 1990). For most long transcripts, it is clear that the exon represents the unit of initial recognition (Robberson et al. 1990; Berget 1995). However, incorporation of a requirement for a pair of splice sites defining an exon of limited length does not alleviate the problem. Pseudo exons, so defined, outnumber real exons by an order of magnitude (Sun and Chasin 2000). The additional information needed to distinguish true from false exons is thought to reside in splicing enhancer and splicing silencer sequence elements (Blencowe 2000; Wagner and Garcia-Blanco 2001; Cartegni et al. 2002; Ladd and Cooper 2002). Exonic splicing enhancers (ESEs) have been extensively studied in the context of alternative splicing (Black 2003). ESEs have also been implicated in at least some constitutive splicing (Mayeda et al. 1999; Schaal and Maniatis 1999a). Exonic or intronic splicing silencers have been less extensively studied (Wagner and Garcia-Blanco 2001; Ladd and Cooper 2002).
Many ESEs have been found by selecting RNA sequences from among large numbers of random oligomers whose insertion stimulates the use of a weak splice site (Liu et al. 1998, 2000; Schaal and Maniatis 1999b) or a poorly included chimeric exon (Tian and Kole 1995) in a cell-free splicing system or in transfected cells (Coulter et al. 1997). In many of these cases the ESEs have been shown to function in concert with specific splicing factor proteins. However, despite their protein specificity, these sequences are highly degenerate and their abundance in introns is ∼80% of their frequency in exons (Liu et al. 1998). As a result, they are not very effective in distinguishing true exons from pseudo exons (our unpublished observations).
Computational approaches have also been used to find ESEs. There is a sharp transition in sequence composition between introns and exons because of the fact that most exons code for protein, whereas introns do not. Most exons can be readily distinguished by this difference (Zhang 1998; Zhang et al. 2003), and gene finding programs that exploit it can be highly accurate (Burge and Karlin 1997). It is not clear what proportion, if any, of this salient information is used as an ESE in addition to protein coding. Some researchers have succeeded in getting around this problem by comparing two classes of exons, each of which codes for protein (Fedorov et al. 2001; Fairbrother et al. 2002). Fairbrother et al. (2002), reasoning that exons with weak splice sites were more likely to require ESEs for recognition than were exons with strong sites, identified exonic hexamers that were both more prevalent in the former and more prevalent than those found in the intronic flanks. When tested, these hexamers indeed promoted splicing. Alternatively spliced exons are usually associated with weak splice sites (Black 2003), so it is possible that their selection method was biased toward this class of exons.
Exonic splicing silencers (ESSs) are a second class of sequence elements that are known to regulate alternative splicing. The high proportion of human genomic sequences that can act to inhibit splicing when inserted into an exon suggests that ESSs may also play a role in splice site selection (Fairbrother and Chasin 2000). Sironi et al. (2004) searched for potential ESSs among hexamers that are underrepresented in exons compared with pseudo exons and exon flanks; one of three such hexamers tested had silencing activity.
We have circumvented the noise represented by exonic protein coding sequences by examining exons that do not code for proteins. We compared the frequencies of 8-mers in constitutively spliced noncoding exons with those in pseudo exons and the 5′ untranslated regions (UTRs) of intronless genes. Sequences overrepresented in the noncoding exons were designated as putative ESEs, and underrepresented sequences were designated as putative ESSs. Over 3000 8-mers were identified by these criteria. On testing, almost all of 20 sequences assayed conferred the predicted phenotype. The large number of effective oligomers implied by these results suggests that sequences that positively and negatively influence splicing may be very abundant in pre-mRNA.
Results
Computational strategy
We have used a computational approach to identify ESEs and ESSs associated with constitutively spliced exons. We focused on constitutive splicing as opposed to alternative splicing so as to tackle the more fundamental problem represented by the former and to avoid what are likely more complex mechanisms in the latter. The strategy we used to overcome the confounding presence of protein coding information was to restrict our search to non-protein-coding exons. In more than 40% of all human genes, protein synthesis is initiated in an exon other than the first (Davuluri et al. 2001). On examination of a database of unrelated human genes, 9% were found in which protein synthesis is initiated in exon 3 or higher. We focused on 502 internal non-protein-coding exons of these genes as examples of exons that should have a relatively high content of ESEs and a relatively low content of ESSs, but with no protein-coding information. We compared the sequence composition of these noncoding exons with that of two dissimilar types of sequences. The first was pseudo exons: intronic regions that have the appearance of exons in that they are bounded by sequences similar to acceptor and donor splice sites and are of typical exon size, but are not in fact spliced (Sun and Chasin 2000; Zhang et al. 2003). These sequences are expected to be low in ESEs and perhaps high in ESSs. We expected this comparison alone to also yield sequences that specify other kinds of information inherent in exons: sequences for nuclear transport, mRNA stability, and mRNA localization, for example. To circumvent this problem, we also compared the non-coding exons with a second class of sequences: the 5′ UTRs of intronless genes. These regions could contain the same nonsplicing information as the noncoding exons, but they should lack ESEs.
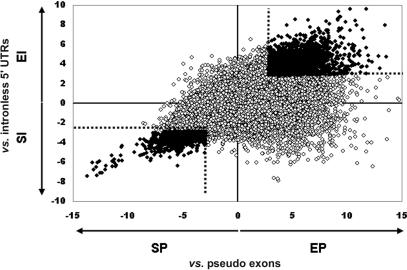
We compared the frequency of 8-mers (allowing one mismatch) in noncoding exons to these two counterparts. We chose 8-mers so as to include binding sites that could contain more information than the 5- or 6-mers that are commonly used in bioinformatics searches, and to facilitate the formation of stable synthetic heteroduplexes for later experimental testing. However, because the frequency of individual 8-mers was too low to gather sufficient data from our set of 502 noncoding exons, we allowed a single mismatch for each 8-mer considered. Pseudo exons were chosen from the introns adjacent to each noncoding exon in our database. We calculated z-scores as an index of the significance of a deviation between the frequency of each possible 8-mer in the non-coding exons versus the pseudo exons and in the non-coding exons versus the 5′ UTRs of intronless genes. This metric allows a determination of the statistical significance of a frequency difference between two populations without any knowledge of the underlying distribution. A p value can be assigned to any z-score (see Supplemental Material for the exact formula used). Because we allowed a single mismatch, the z-score for each 8-mer actually represents the average of 25 sequences, 24 of which differ from the nominal sequence by a single base substitution. The results are shown in Figure 1, in which the z-scores of noncoding exons versus pseudo exons are plotted on the abscissa, and the z-scores of noncoding exon versus 5′ UTRs of intronless genes are plotted on the ordinate. Although most of the 65,536 8-mers lie near the center of this two-dimensional scatter plot, many are either overrepresented or underrepresented in the noncoding exons. Choosing those 8-mers whose frequency difference corresponds to a p value of <0.002 for each comparison, we collected 2069 putative ESEs (PESEs; upper right quadrant of Fig. 1 beyond the dotted line) and 974 putative ESSs (PESSs, lower left quadrant beyond the dotted line). The thresholds of p < .002 predicts a false discovery rate (Storey and Tibshirani 2003) of 0.3% for PESEs and 0.7% for PESSs, on the basis of a bivariate normal distribution. These sequences were grouped into 69 PESS families and 80 PESE families by hierarchical clustering. The sequence logos for 10 PESS and 8 PESE clusters are shown in Figure 2, along with their information content and the size of the cluster.
Figure 1.
A scatter plot showing the scores of all possible 65,536 8-mers with respect to their relative abundance in three sequence classes. The axis numbers represent z-scores. Z-scores on the X-axis are from a comparison of the relative abundance of each 8-mer in noncoding internal exons versus pseudo exons; this number is called an EP index when it is >0 (for enhancer compared with pseudo exons) and an SP index when it is <0 (for silencer compared with pseudo exons). The z-scores on the Y-axis are from a comparison of the relative abundance of each 8-mer in noncoding internal exons versus the 5′ UTR of intronless genes; this number is called an EI index when it is >0 (for enhancer compared with intronless genes) and an SI index when it is <0 (for silencer compared with intronless genes). In all further discussion, the silencer indices SP and SI are expressed as their absolute values. The dotted line marks a z-score of 2.88, chosen as a threshold for z-scores considered to be of significance. A z-score >2.88 has a probability of <0.002 of occurring by chance. Points lying beyond this threshold in both comparisons are black and represent the set of putative exonic splicing enhancers or silencers characterized further. If all 8-mers were distributed equally in all data sets, then the probability that a point will lie outside the dashed lines (i.e., in both dimensions) by chance is <10-4.
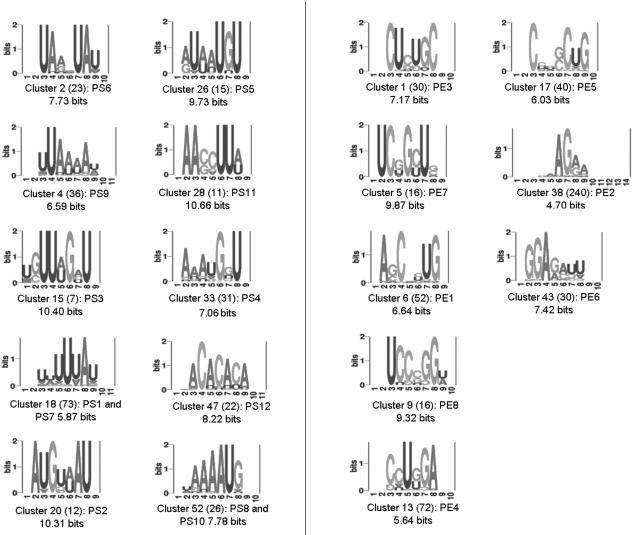
Figure 2.
Examples of putative exonic splicing silencer (PESS; left) and putative exonic splicing enhancer (PESE; right) sequence families. The 974 PESS and 2069 PESE 8-mer sequences were aligned and then clustered using ClustalW. Pictograms for 10 PESSs and 8 PESEs on the basis of the positional sequence scoring matrix underlying each cluster are shown. The number of sequences in the cluster is shown in parentheses and the name of an exact exemplar used for testing is given, as is the information content in bits.
Testing putative exonic splicing silencers
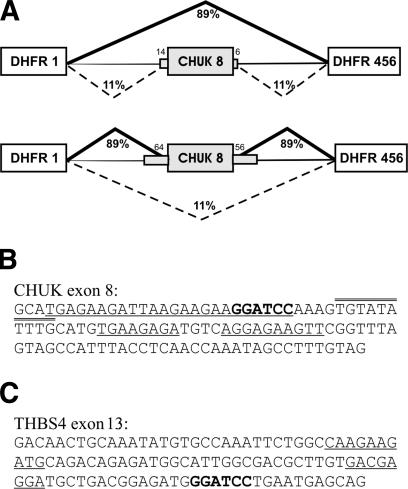
To determine whether these sequences could function as splicing regulatory elements, we tested several for their effect on the splicing of the central exon of a chimeric three-exon minigene. Two versions of the test minigene were used. The terminal exons of this construct were from the hamster dihydrofolate reductase (dhfr) gene separated by dhfr intron sequences. Exon 8 of the human conserved helix-loop-helix ubiquitous kinase (CHUK) gene was inserted into this minigene, either with or without ∼55 nt of flank beyond the splice site sequences. With the flanking sequences present, the central exon is included ∼90% of the time; without the flanking sequence, the central exon is skipped 90% of the time (Fig. 3A). This flanking sequence effect will be the subject of another report (X.H-F. Zhang, C. Leslie, L.A. Chasin, in prep.). The 108-nt CHUK exon 8 itself contains three clusters of PESEs and one cluster of PESSs (Fig. 3B). After insertion of various 8-mers at a position 22 nt from the 5′ end of this 108-nt exon, the effect on splicing was evaluated by transient transfection of human 293 cells and by quantifying the spliced transcripts that had either included or skipped the central exon.
Figure 3.
Minigenes used for testing effects on splicing. (A) Two versions of the exon 8 region of the human conserved helix-loop-helix ubiquitous kinase (CHUK) gene were inserted into a chimeric intron separating exon 1 and the combined exons 4-6 of the hamster dihydrofolate reductase (dhfr) gene. Large boxes depict exons, stubby gray boxes show flanking regions of the CHUK exon 8, and thin horizontal lines represent hamster intron sequence. In the upper figure, the CHUK exon 8 flanks are limited to the splice site consensus region from -14 upstream and to +7 downstream of the exon-intron junction; CHUK exon 8 is spliced poorly, as indicated. In the lower figure, additional CHUK exon 8 flanking sequences have been added from -62 upstream and to +75 downstream. The addition of this flanking sequence greatly improves exon inclusion, as shown. (B) Sequence of the 108-nt CHUK exon 8. PESE sequences are underlined and PESS sequences are double overlined. The BamHI site used for the insertion of tested 8-mers is in bold. (C) Sequence of the 108-nt thrombospon4 (Tbsh4) exon 13, annotated as in B.
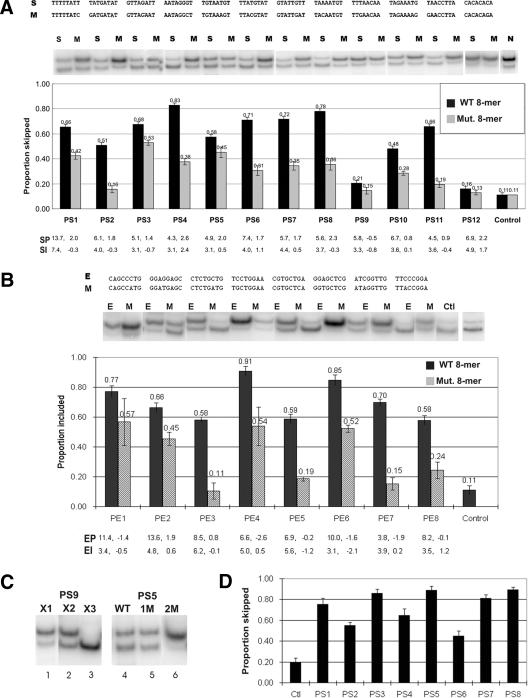
We first tested PESS sequences, as there is little information concerning the role of silencing in constitutive splicing. Twelve PESSs were chosen to represent families of diverse sizes and sequences. For 11 of the 12 cases, the possible functional significance of the sequence was not considered. The exception, PS4, was chosen because it constituted a core binding site for hnRNP A1 (AUAGGGU), which can act as an ESS (Del Gatto-Konczak et al. 1999). As can be seen by the black bars in Figure 4A, 10 of 12 putative silencers significantly increased exon skipping when inserted into the exon embedded in its flanks; the average increase in skipping was sixfold over the control, such that the central exon was now skipped 50%-80% of the time instead of 10%. Interestingly, whereas PESS PS9 had almost no effect on splicing, insertion of two tandem 8-mers of this sequence was effective, and three 8-mers virtually abolished central exon splicing (Fig. 4C, columns 1-3). Because two new PESSs were created at the joints of these tandem repeats, it is possible that one of these secondary 8-mers (UUAACAAU, ACAAUUUA) was responsible for the heightened silencing. However, inasmuch as three copies were much more effective than two, the conclusion that two or more PESSs can act synergistically can still be drawn.
Figure 4.
The effect of 8-mer insertions on splicing. (A) Testing PESSs for splicing inhibition. The indicated 8-mer PESS sequences were inserted into a BamHI site at position +22 in CHUK exon 8 using the lower construct shown in Figure 3A. Plasmids were transfected into human 293 cells by lipofection, and RNA was extracted after 24 h and assayed for splicing by RT-PCR using radioactive dATP as a precursor. Band intensity was quantified with a PhosphorImager; proportion skipped indicates skipped band/(skipped band + included band). The bands correspond to the column below them and show the results of one transfection experiment; the graph shows the average of two transfections, and the error bars indicate the range. Black bars represent insertion of the PESS shown at the top (S), gray bars represent insertion of a single base substitution mutant sequence also shown at the top (M); the SP and SI scoring indices (defined in the legend for Fig. 1) of each PESS and each mutant sequence are shown at the bottom. (B) Testing PESEs for splicing enhancement. The indicated PESE 8-mers were inserted into CHUK exon 8 using the upper construct shown in A. Splicing was assayed exactly as in B. (E) PESE; (M) single base substitution mutant sequence. Proportion included indicates included band/(skipped band + included band). Two transfections were carried out for each construct; the error bars indicate the range of the two measurements. (C) The effect of insert sequence variations on splicing silencing. (Left) Multiple copies of a PESS can act synergistically to inhibit splicing: one, two, and three copies of the 8-mer PS9 (see A) were inserted into CHUK exon 8 and assayed for splicing as described in A. (Right) A double base substitution is more effective than a single base substitution in destroying silencing activity: The original PESS P5 and mutants harboring one or two single base substitutions were inserted into CHUK exon 8 and assayed for splicing as in A. The 8-mers were UGUAAUGU, UGUAAAGU, and UGGAAAGU, respectively; the SP indices were 4.92, 1.95, and 1.74, respectively; and the SIs were 3.14, 0.50, and -4.07, respectively. (D) Testing PESSs for silencing in a second exon. A minigene analogous to that shown in Figure 3A was constructed using human thrombospondin 4 exon 13 as the central exon. Eight PESS sequences were inserted into a BamHI site at a position 16 nt upstream of the 3′ end of the exon (Fig. 3C) and tested for silencing as described in A.
Next we tested the specificity of this effect by introducing a single base substitution (SBS) in each putative silencer sequence; the changes were designed to reduce the z-score index to as close to zero as possible. None of the changes created a PESE. The silencing effect was significantly reduced in each case (Fig. 4A, gray bars), and the average decrease was over threefold (range = 1.4-8.4). In one case in which the SBS was ineffective, we introduced a second SBS; the double change completely reversed the splicing inhibition (Fig. 4C, columns 4-6). We also tested six 8-mers with low silencer scores (ACCCU AUC, AUACAUAA, AACAAUAC, CAUUUCUA, CCA UGACC, and CCAUAUAC): None produced significant silencing (data not shown). Thus, the induction of exon skipping was not caused by the mere introduction of a foreign sequence. We have previously shown that the insertion of much larger sequences (>100 nt) does not usually compromise splicing (Chen and Chasin 1994; Fairbrother and Chasin 2000). Interestingly, one of the two PESSs that did not inhibit splicing as a single copy, PS12, constitutes a CACACACA repeat—a sequence that has been characterized as an intronic enhancer (Hui et al. 2003).
We considered the possibility that the decreased proportion of exon-included species affected by an insertion was actually the result of the introduction of an in-frame nonsense codon in the central exon, leading to nonsense-mediated decay (NMD) of this species. In fact, although CHUK exon 8 contains no in-frame nonsense codons, the introduction of any 8-mer at the BamHI site used produces a frame shift and the generation of several in-frame stop codons, including one that is 60 nt upstream of the 3′ end of the exon. This position is outside the region of immunity from NMD, which is usually estimated as 50-55 nt from the 3′ end of the penultimate exon (Maquat 2004; Neu-Yilik et al. 2004). Nevertheless, NMD does not appear to be operating in this system, as 12 of the insertions (two PESSs, four mutated versions, and six arbitrary sequences) caused little or no increase in the proportion of skipped species despite the generation of the same nonsense codons. Exceptions to NMD have been previously noted (Enssle et al. 1993; Danckwardt et al. 2002; Maquat 2004; Neu-Yilik et al. 2004, and references therein). Moreover, we previously found that a nonsense mutation that caused NMD when expressed from the endogenous dhfr gene (used as the host minigene here) did not exhibit this phenotype upon transfection (Urlaub et al. 1989).
As a further test of an NMD effect, we repeated the PESS assay using a different target exon in our test minigene, the 108-nt exon 13 of the human Thbs4 (thrombospondin 4) gene. In this case the original central exon contains an in-frame nonsense codon at a position 11 bases from the 3′ end of the exon, from which it is not expected to elicit NMD. Now the frame shift induced by the insertion of an 8-mer removes this nonsense codon. We tested eight of the PESSs that exhibited strong silencer activity in the CHUK exon 8. As can be seen in Figure 4D, each of the eight PESSs again induced silencing, increasing exon skipping fourfold, from 19% to an average of 73%. We conclude that our results reflect splicing differences and not NMD. In addition to addressing the NMD issue, this experiment shows that these eight PESSs work similarly in two completely different exons and at two different relative locations (22 bases downstream of the 5′ end in CHUK exon 8, and 16 bases upstream of 3′ end of the exon in Thbs4 exon 13).
Insertion of 8-mers into the test exon can create PESS and PESE elements in the new joint sequences. In particular, the BamHI site used for the insertion lies at one end of a resident PESE in CHUK exon 8 (AAGGAUCC), and the insertions usually created an overlapping PESE at this location, extended by one base. In three cases, the insertion of the test PESSs created additional overlapping PESSs; in these instances we cannot be sure which 8-mer is responsible for the silencing. However, because the choice of an 8-mer to represent a regulatory sequence was arbitrary, these overlapping sequences can be just as well viewed as a single, somewhat longer, element
Testing putative splicing enhancers
Enhancers were tested next. Eight predicted PESEs were inserted into the central exon lacking its flanks. No PESSs were created at the joints by the insertion of these PESEs. As can be seen in Figure 4B (black bars), the eight PESEs increased inclusion of the central exon from the baseline of 11% to between 58% and 91% (average 6.4-fold). Once again, SBS mutations that reduced the z-score index to near zero decreased the enhancer effect significantly in seven of eight cases and dramatically (more than threefold) in four cases (Fig. 4B, gray bars). None of the mutations created a PESS. Two of the eight PESEs tested were chosen because they resembled known ESEs: PE1, the SC35 consensus sequence (Liu et al. 2000), and PE2, a purine-rich element (Schaal and Maniatis 1999b). The remaining six PESEs represented novel candidates in that they are not represented in the PESEs predicted by Fairbrother et al. (Fairbrother et al. 2002), nor are they found by ESEfinder (Cartegni et al. 2003). About half of our PESEs are novel by these criteria, implying that many such new ESEs remain to be discovered.
Global distribution of PESEs and PESSs
These PESEs and PESSs were predicted on the basis of relative abundance or scarcity, respectively, in noncoding exons compared with nonsplicing sequences. We asked whether these oligomers showed a similar differential distribution in protein-coding exons—the major class of exons. We collected data from 78,000 internal protein-coding exons of lengths >50 nt and calculated the frequencies of PESEs and PESSs occurring at each position, starting 100 nt upstream and ending 100 nt downstream of the exon-intron borders. Because the exons were of different lengths, we combined 25 nt from each end with 50 nt from the center to create a 100-nt version; if the exon was <100 nt, we just used the ends. For comparison, we repeated the same process for 20,580 repeat-free pseudo exons. We calculated the same frequencies for 148,000 regions of 100 nt located at the centers of introns.
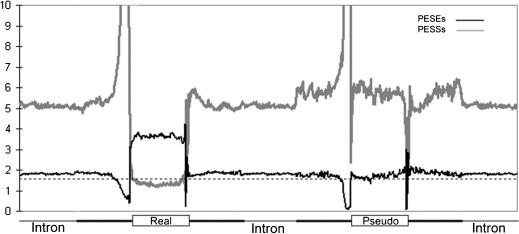
The results for the real exons and introns are shown on the left in Figure 5. It can be seen that the frequency of PESSs (gray line) dropped dramatically at the transition between intronic flanks and the (composite) exon. Spikes are seen as expected at the very edge of the exons because of the conservation of the splice site consensus sequence. There is a peak of PESS frequency in the region of the upstream flank harboring the polypyrimidine tract, again as expected because runs of U are common in these PESSs. Less expected is a smaller peak just beyond the donor site consensus in the downstream flank: We speculate that this peak in negative signals may contribute to a locking in on the positive signal represented by the splice sites. The frequencies of PESEs (black line) behaved in exactly the opposite manner, rising precipitously within the composite exon. Within the exon the average frequency of PESEs was approximately constant; that is, the frequencies were not very different within the 25-nt ends and the 50-nt center. Beyond ∼50 nt from the exon, the frequency of both PESSs and PESEs reached a level characteristic of deep intron sequences. Sharp transitions in sequence composition between exons and introns have previously been noted (Burge and Karlin 1997; Zhang 1998). It should be remembered that the PESEs and PESSs analyzed here were chosen on the basis of noncoding exons; that is, the transitions seen here were not produced by selection for protein coding potential.
Figure 5.
Statistical analysis of PESSs and PESEs in coding exons and introns. The frequencies of each of the 974 PESSs and 2069 PESEs were determined for each position in 78,000 human coding exons (50-250 nt long) and in 100 nt of their immediate flanks and in 100 nt regions from the center of 148,000 introns. Numbers on the ordinate indicate the average frequency of a PESS or PESE per nucleotide position multiplied by 100,000. The heavy gray curve represents the PESSs, and the black curve the PESEs. Indications below the curve: The box marked Real represents a composite exon standardized to 100 nt as described in the text and in Supplemen tal Material; their intronic flanks are indicated by heavy lines. The thin lines refer to intronic sequences of 100 nt extracted from the center of each intron; this same central intron data is presented three times for easy reference. The box marked Pseudo shows the same analysis performed on 20,580 pseudo exons drawn from repeat-free regions of introns; this set of pseudo introns did not overlap with the pseudo exon set used to derive the z-scores in Figure 1. The broken horizontal line depicts the average frequency of any given 8-mer in a random sequence (1/65,536).
In contrast to the real exons, the pseudo exons lacked an elevated PESE content (Fig. 5, right, black line). Also in contrast to the real exons, the PESS frequency in pseudo exons did not sharply drop compared with the pseudo exon's flanks. PESS frequency was, in fact, as much as 20% higher in both the pseudo exon body and flanks compared with deep introns (Fig. 5, right, gray line), suggesting a possible role of PESSs in silencing false splice sites. It should be noted that the pseudo exons analyzed here did not overlap the pseudo exon set used for prediction. It is interesting to compare the frequencies of these putative regulatory sequences with that predicted by chance, using the average probability of all possible 8-mers (1/65,536 = 0.0000153). The latter frequency is shown by the dashed horizontal line in Figure 5. The average PESE 8-mer frequency in introns is close to that value, but the PESS frequency in introns is several times greater, again supporting the idea (Fairbrother and Chasin 2000) that intronic sequences have evolved to create a generally inhospitable environment for splicing. Looking at the total number of PESEs and PESSs, the average real 140-nt internal exon would contain 10.5 PESEs and 1.9 PESSs, whereas the typical pseudo exon counterpart would contain 4.4 PESEs and 6.9 PESSs. These average PESE/PESS ratios differ by a factor of 8.6; splicing decisions may be made on the basis of these ratios.
Numerous mutagenesis studies have shown that ESS sequences are often juxtaposed with ESE sequences in alternatively spliced exons. Thus, one might expect to see a significantly higher level of PESS elements in alternatively spliced exons compared with constitutive exons. We analyzed a data set of 281 alternatively spliced exons and found that they contained 8.0 PESEs and 2.2 PESSs per 140 nt, for a PESE/PESS ratio of 3.6 compared with 5.5 for constitutive exons (and 0.64 for pseudo exons). This lower PESE/PESS ratio may play a role in allowing alternatively spliced exons to be skipped
Discussion
It is interesting to compare the 8-mer PESEs found here with the 6-mer PESEs found by Fairbrother et al. (2002), using a different computational strategy. One of their two criteria was to compare exons with exons: those with strong splice sites to those with weak splice sites. In this way they also avoided the isolation of sequences on the basis of protein coding potential. Over 80% of their 237 hexamers can be found in our PESE collection, with 1351 hits. In contrast, 10 random sets of 237 hexamers produced an average of only 308 hits. The overlap between these two PESE sets, isolated by different criteria, supports the validity of each set.
Several laboratories have used iterative selections starting with random oligomers to define sequences that bind splicing factors or that promote splicing in vitro or in vivo (Tacke and Manley 1995; Tian and Kole 1995; Coulter et al. 1997; Liu et al. 1998, 2000; Schaal and Maniatis 1999b). The full PESE 8-mers found here overlap with ∼40% of these ESE sequences; the overlap after scrambling the ESE sequences was only one-third of this value. That the overlap is not 100% may be related to the fact that the CpG content of these ESEs is unusually high in every case cited above; the average is more than 10% of all dinucleotides. In contrast, the CpG content of our PESEs is 4.2%, closer to the value for exons in general (2.8%). We may have selected against such CpG-rich sequences because we demanded that our PESEs stand out compared with 5′ UTRs, which are themselves rich in CpG (Davuluri et al. 2001). In addition, all but one of the studies cited above assayed the splicing of terminal exons, whereas only internal exons were considered in our experiment
To gauge the physiological relevance of the sequences identified here, we surveyed exonic mutations that result in a splicing deficiency, but that are not located within the consensus splice site sequence. If the PESEs and PESSs found here are important governors of splicing in vivo, then we would expect many of these mutations to have either destroyed a PESE or created a PESS. Of 58 splicing mutations examined (33 in the hprt gene, see Supplementary Table S1), more than half (55%) fulfilled this expectation: 19 represent disruptions in PESEs, and 16 created PESSs. In contrast, only 11% of missense mutations with no splicing phenotype affected PESS or PESE sequences (see Supplementary Table S2). It is interesting to note that the creation of PESSs was nearly as common as the disruption of PESEs, as predicted by Kashima and Manley (2003).
The threshold values we used to define a PESS or PESE were necessarily arbitrary at this stage. We can use the mutational data described above to direct us to a less conservative definition: If the threshold is reduced to include sequences with z-score indices down to 2.0 (p < 0.03), rather than 2.88 (p < 0.002), for each dimension, then we capture 81% of the splicing mutations referred to above as either disrupting a PESE or creating a PESS (compared with 20% for missense mutations; for details see Supplementary Table S3). This relaxation generates 4984 PESEs and 3579 PESSs with predicted false discovery rates of 5% and 7%, respectively. This new total of over 8500 putative regulatory elements represents one in eight of all possible 8-mers, leading to the conclusion that sequences that can regulate splicing are highly abundant. Further support of this idea lies in the effect of the single base mutations in PESSs shown in Figure 4. Although in every case the mutation decreased the silencing phenotype, substantial silencing activity remained in most cases. Of nine informative cases, eight mutant 8-mers still increased skipping at least 2.5-fold (Fig. 4A); most of these 8-mers had silencing indices between 0 and 2 (compared with the threshold of 2.88 for both indices to be predicted as a PESS). A similar situation was obtained considering PESEs. Here again, four of eight mutant sequences still enhanced splicing by more than fourfold, and the mutant 8-mers exhibited low statistical indices (Fig. 4B). Thus, although our combined statistical index performed well in predicting PESSs and PESEs, it is apparently not a reliable predictor of the lack of PESS or PESE activity.
Previous experiments from other laboratories also point to a plethora of PESEs. From the computational study of Fairbrother et al. (2002), 6% of all hexamers are predicted to have ESE activity, or approximately eight hexamers per exon of average size 140 nt. ESEs selected from random sequences and shown to enhance splicing in response to specific SR proteins are highly degenerate, with the same 5-8-nt consensus-defining region rarely being found twice (Liu et al. 1998, 2000). The combined prevalence of these sequences in exons is at least four per 140 nt, which can be extrapolated to at least eight if the full complement of SR proteins is considered. Both of these frequencies are similar to the average of 10.5 found for the PESEs defined here. Because all three sets do not overlap completely, the combined total number of PESEs per exon must be even larger. Even allowing for clustering and overlap of individual PESEs, the picture that emerges is one in which more than half of an exon is made up of ESEs and in which much of the remainder is composed of ESSs. It follows that general RNP structure (e.g., an H-complex) may reflect more information than has been hitherto acknowledged.
Compared with ESEs, there have been far fewer systematic searches for ESSs. In our own previous work we found that 7 of 19 sequences (∼100-mers) randomly chosen from the human genome inhibited splicing when inserted into an exon (Fairbrother and Chasin 2000). Reexamination of these seven inhibitory sequences showed that they contained at least one PESS, with the average content being 3.80 (over twice that expected by chance), whereas in 12 noninhibitory sequences, the average content was 1.25. That more than 90% of the tested PESSs inhibited splicing indicates that the great majority of the PESS sequences we have identified by computation could be playing a physiological role.
The large difference (8.6-fold) in PESE/PESS ratio between real exons and pseudo exons raises the question of whether this metric can be used to distinguish these two classes. Unfortunately, the distribution of ratio values is quite wide, such that if a threshold ratio value is chosen to capture 80% of real exons, it will also capture 20% of pseudo exons (this being the optimum condition for combined sensitivity and specificity). Thus, the final distinction of these two classes awaits further work. In the meantime, this ratio may be a useful adjunct to other criteria (Zhang et al. 2003).
Our lists of PESE and PESS 8-mers undoubtedly contain some ineffective sequences and omit other effective sequences. Nevertheless, we believe it is the most comprehensive list of splicing regulatory sequences yet collected and should serve as a basis for examining the biochemical mechanisms governing accurate splice site recognition.
Materials and methods
Additional details can be found in Supplemental Material.
Data sets
A nonredundant human transcript dataset was downloaded from ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot in September 2003. These transcript sequences were aligned to human genomic sequences obtained from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens using the Spidey program (http://www.ncbi.nlm.nih.gov/spidey/spideysource.html). We required that a valid alignment to have more than 98% identity, more than 95% mRNA coverage and that all identified exons be flanked by canonical splice sites. Under these restrictions, 16,930 transcripts yielded alignments, from which 166,538 exons and 149,608 introns were identified. Based on these sequences, three subsets were created as follows:.
1) Noncoding internal exons (NC). By comparing full-length genes with annotated mRNA sequences, 2495 NCs (∼1.5% of all exons) were extracted from 5′ UTRs. We discarded any exon if a single base substitution or a single base addition or deletion could generate an open reading frame, reasoning that these could be misannotated coding exons containing single sequencing errors. The majority of exons were eliminated by this filter. We eliminated exons that are mostly skipped in splicing, as deduced from the human dbEST database (ftp://ftp.ncbi.nih-.gov/blast/db/est_human.tar.gz). We discarded exons that have <70% inclusion. After these two filters, 502 noncoding exons remained for analysis. A list of the noncoding exons used can be found at http://www.columbia.edu/cu/biology/faculty/chasin/xz3/noncode.txt.
2) Pseudo exons adjacent to the noncoding exons (PE). We applied the same criteria as in our previous study (Zhang et al. 2003) to extract 2876 PEs: intronic sequences 50-250 nt long that are flanked by sequences resembling splice sites (acceptor consensus values of at least 75 and donor consensus values of at least 78). To further ensure that the pseudo exons set resembled the noncoding exon set in general base composition (e.g., from the same set of isochores), we collected pseudo exons from the introns adjacent to the 502 noncoding exons. After removal of any pseudo exons that were present as ESTs, we were left with 2309 pseudo exons. A list of the pseudo exons used for comparison can be found at http://www.columbia.edu/cu/biology/faculty/chasin/xz3/pseudos5.doc.
3) 5′ UTRs of intronless genes (IL). Among the 16,930 full-length genes, we extracted 1220 intronless genes and parsed their 5′-UTRs according to the annotation. We then applied the same criteria that we did for NCs to eliminate possible coding-exon contamination. The number of 5′ UTRs of intronless gene that made up this dataset was 864. A list of the intronless gene 5′ UTRs used can be found at http://www.columbia.edu/cu/biology/faculty/chasin/xz3/ilgenes.doc.
Calculations of scoring indices EP, EI, SP, and SI
EP represents the extent to which a given 8-mer is found in the noncoding exons as opposed to pseudo exons; this z-score was calculated as in Fairbrother et al. (2002). When this index is <0, the absolute value is taken as the silencer scoring index, SP. Similarly, EI and SI represent the scoring indices for noncoding exons compared with the 5′ UTRs of intronless genes. An index of 2.88 corresponds with p < .002, and an index of 2 corresponds with p < 0.03. The random chance for an 8-mer to pass both criteria at 0.002 is 10-4. A more detailed description of the calculations is provided in Supplemental Material. A list of all 8-mers and their corresponding z-scores is available as a 1.7 MB text file at http://www.columbia.edu/cu/biology/faculty/chasin/xz3/octamers.txt.
Clustering and sampling putative ESS/ESEs
We clustered the 974 PESSs and 2069 PESEs using a hierarchical clustering algorithm (Fairbrother et al. 2002). Using a dissimilarity cutoff of 3.2 in the dendrogram yielded 69 PESS clusters and 80 PESE clusters (see Supplemental Material).
Statistical analysis of the PESS/PESEs in coding exons
From among more than 120,000 internal coding exons, we chose to look at those 50-250 nt long and flanked by at least 100 nt of intron sequence on both sides. We extracted 100 nt of sequence from each of these 78,000 exons: 25 nt from each end and 50 from the center. If an exon was shorter than 100 nt, we only considered the two ends. For a composite intron, we collected all the corresponding introns that were at least 100 nt long. We also divided these into three parts: a 100-nt 5′ end, a 100-nt region at the center, and a 100-nt 3′ end. If an intron was shorter than 300 nt, we only considered its ends. We calculated the average frequency of all PESSs or PESEs at each position of these uniform exons and introns. Pseudo exons overlapping highly repeated sequences were excluded.
Constructs
A complete hamster dhfr minigene (pDCH1P12) was first constructed that contained exon1, intron1 (304 bp), exons 2 and 3 merged, an abbreviated intron 3 (900 bp), and exons 4-6 merged. This minigene was driven by the dhfr promoter and was terminated by the first dhfr polyA site. Exons 2 and 3 were then replaced with a unique NotI site to form pDCH1P12D. In the course of other studies, we have tested the splicing of several foreign exons inserted into this NotI site. When inserted into this site as a polymerase chain reaction product, the exon 8 of the human CHUK gene (Mock et al. 1995) is predominantly included when cloned with its flanking intron sequences (PDCHUK8F, 47 and 67 nt beyond the 3′ and 5′ splice sites, respectively) but is mainly skipped when cloned without these flanking sequences (pDCHUK8), making it a sensitive indicator for enhancement and for silencing. In the same way we constructed a minigene with exon 13 of the human thrombospondin4 gene inserted into the NotI site of pDCH1P12D without its flanks (pDTBSN413). The transcript of this minigene is spliced efficiently without its flanks.
We inserted PESS and PESE candidates into a unique BamHI site 22 nt downstream from the start of CHUK exon 8. We synthesized the two strands of the 8-mer sequence flanked by cohesive ends compatible with a BamHI site on each side. To facilitate future manipulations, the BamHI site was reconstructed on the upstream side of the insert and disrupted on the downstream side. For ligation of the annealed strands, we incubated 3 μL of double-strand insertions (0.6 μg) with 1 μL of BamHI-cut vectors (∼0.1 μg, without CIP treatment) in a 20-μL reaction at 16°C for 1-2 h; a 5-μL portion was used to transform DH5α competent cells. Recombinant plasmids were verified by sequencing. Tandem arrays of PS9 were constructed using synthetic oligomers that provided no space between repeats.
Splicing
Single representatives from eight clusters and two representatives from each of two additional clusters of PESSs were chosen for testing silencing. For testing PESEs, we focused on novel signals, choosing six not found by ESEfinder (Cartegni et al. 2003) or among the RESCUE 6-mers (Fairbrother et al. 2002). Two additional PESEs were chosen because they resemble known enhancers (see Results). Human 293 cells were transfected in 35-mm wells by the plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer. After 24 h, total RNA was isolated using RNAwiz (Ambion), treated with DNase I, and subjected to RT-PCR labeling with α-32P-dATP (Chen and Chasin 1993) under the following conditions: template, 3μL RT product; forward primer, CGCCAAACUUGGG GGAAGCA; reverse primer, CGGAACUGCCUCCAACUAUC; initial denaturation, 93°C for 5 min; denaturation, 93°C, 30 sec; annealing, 61°C, 30 sec; extension, 72°C, 1 min; 28 cycles; final extension, 72°C, 7 min. Results were quantified with a PhosphorImager.
Mutation analysis
Mutations in the hprt gene were collected from O'Neil et al. (1998) and Tu et al. (2000); mutations in other genes were taken from those collected by Cartegni et al. (2002). These mutations are listed in the Supplemental Material. A single point mutation always changes a set of eight overlapping 8-mers to a new set of eight sequences. If there were one or more putative enhancers in the original set but fewer or no enhancers in the new set, then this change was designated as an enhancer-disruption (ED) event. Conversely, if in the mutant set there were one or more putative silencers but none or fewer in the original set, then this change was designated as a silencer-creation (SC) event. Tabulated results can be found in Supplementary Tables S1 and S2.
Acknowledgments
We thank Will Fairbrother for providing an electronic version of the RESCUE-ESE hexamer sequences, Adrian Krainer for providing the list of sequences underlying the ESEfinder program, Harmen Bussemaker for a critical reading of the manuscript and helpful suggestions, Hongfei Zhang for help with the statistical analysis, and three anonymous reviewers for helpful criticisms. X.H-F. Zhang is a Columbia University Predoctoral Faculty Fellow. L.A.C. was supported by funds from Columbia University.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1195304.
Supplemental material is available at http://www.genesdev.org.
References
- Berget, S.M. 1995. Exon recognition in vertebrate splicing. J Biol Chem. 270: 2411-2414. [DOI] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 72: 291-336. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 25: 106-110. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world, 2nd ed. (ed. R.F. Gesteland, Cech, T. R. & Atkins, J. F.), pp. 525-560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 3: 285-298. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Wang, J., Zhu, Z., Zhang, M.Q., and Krainer, A.R. 2003. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31: 3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I.T. and Chasin, L.A. 1993. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol Cell Biol. 13: 289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1994. Large exon size does not limit splicing in vivo. Mol Cell Biol. 14: 2140-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter, L.R., Landree, M.A., and Cooper, T.A. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 17: 2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt, S., Neu-Yilik, G., Thermann, R., Frede, U., Hentze, M.W., and Kulozik, A.E. 2002. Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood. 99: 1811-1816. [DOI] [PubMed] [Google Scholar]
- Davuluri, R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat Genet. 29: 412-417. [DOI] [PubMed] [Google Scholar]
- Del Gatto-Konczak, F., Olive, M., Gesnel, M.C., and Breathnach, R. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 19: 251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enssle, J., Kugler, W., Hentze, M.W., and Kulozik, A.E. 1993. Determination of mRNA fate by different RNA polymerase II promoters. Proc Natl Acad Sci. 90: 10091-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother, W.G. and Chasin, L.A. 2000. Human genomic sequences that inhibit splicing. Mol Cell Biol. 20: 6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeh, R.F., Sharp, P.A., and Burge, C.B. 2002. Predictive identification of exonic splicing enhancers in human genes. Science. 297: 1007-1013. [DOI] [PubMed] [Google Scholar]
- Fedorov, A., Saxonov, S., Fedorova, L., and Daizadeh, I. 2001. Comparison of intron-containing and intron-lacking human genes elucidates putative exonic splicing enhancers. Nucleic Acids Res. 29: 1464-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, J., Stangl, K., Lane, W.S., and Bindereif, A. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 10: 33-37. [DOI] [PubMed] [Google Scholar]
- Kashima, T. and Manley, J.L. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 34: 460-463. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N., and Cooper, T.A. 2002. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 3: reviews0008. [DOI] [PMC free article] [PubMed]
- Liu, H.X., Zhang, M., and Krainer, A.R. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12: 1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., Chew, S.L., Cartegni, L., Zhang, M.Q., and Krainer, A.R. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 20: 1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 5: 89-99. [DOI] [PubMed] [Google Scholar]
- Mayeda, A., Screaton, G.R., Chandler, S.D., Fu, X.D., and Krainer, A.R. 1999. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 19: 1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock, B.A., Connelly, M.A., McBride, O.W., Kozak, C.A., and Marcu, K.B. 1995. CHUK, a conserved helix-loop-helix ubiquitous kinase, maps to human chromosome 10 and mouse chromosome 19. Genomics. 27: 348-351. [DOI] [PubMed] [Google Scholar]
- Neu-Yilik, G., Gehring, N.H., Hentze, M.W., and Kulozik, A.E. 2004. Nonsense-mediated mRNA decay: from vacuum cleaner to Swiss army knife. Genome Biol. 5: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, J.P., Rogan, P.K., Cariello, N., and Nicklas, J.A. 1998. Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat Res. 411: 179-214. [DOI] [PubMed] [Google Scholar]
- Robberson, B.L., Cote, G.J., and Berget, S.M. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 10: 84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal, T.D. and Maniatis, T. 1999a. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 19: 261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1999b. Selection and characterization of pre-mRNA splicing enhancers: Identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 19: 1705-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy, P., Shapiro, M.B., and Harris, N.L. 1990. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 183: 252-278. [DOI] [PubMed] [Google Scholar]
- Sironi, M., Menozzi, G., Riva, L., Cagliani, R., Comi, G.P., Bresolin, N., Giorda, R., and Pozzoli, U. 2004. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 32: 1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J.D. and Tibshirani, R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci. 100: 9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. and Chasin, L.A. 2000. Multiple splicing defects in an intronic false exon. Mol Cell Biol. 20: 6414-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke, R. and Manley, J.L. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14: 3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H. and Kole, R. 1995. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 15: 6291-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, M., Tong, W., Perkins, R., and Valentine, C.R. 2000. Predicted changes in pre-mRNA secondary structure vary in their association with exon skipping for mutations in exons 2, 4, and 8 of the Hprt gene and exon 51 of the fibrillin gene. Mutat Res. 432: 15-32. [DOI] [PubMed] [Google Scholar]
- Urlaub, G., Mitchell, P.J., Ciudad, C.J., and Chasin, L.A. 1989. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 9: 2868-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 21: 3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.Q. 1998. Statistical features of human exons and their flanking regions. Hum Mol Genet. 7: 919-932. [DOI] [PubMed] [Google Scholar]
- Zhang, X.H., Heller, K.A., Hefter, I., Leslie, C.S., and Chasin, L.A. 2003. Sequence information for the splicing of human pre-mRNA identified by support vector machine classification. Genome Res. 13: 2637-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]