Abstract
Differential organ growth during gravitropic response is caused by differential accumulation of auxin, that is, relative higher auxin concentration in lower flanks than in upper flanks of responding organs. Auxin responsive reporter systems such as DR5::GUS and DR5::GFP have usually been used as indicators of gravitropic response in roots and hypocotyls of Arabidopsis. However, in the inflorescence stems, the reporter systems don’t work well to monitor gravitropic response. Here, we aim to certify appropriate gravitropic response indicators (GRIs) in inflorescence stems. We performed microarray analysis comparing gene expression profiles between upper and lower flanks of Arabidopsis inflorescence stems after gravistimulation. Thirty genes showed > 2-fold differentially increased expression in lower flanks at 30 min, of which 19 were auxin response genes. We focused on IAA5 and IAA2 and verified whether they are appropriate GRIs by real-time qRT-PCR analyses. Transcript levels of IAA5 and IAA2 were remarkably higher in lower flanks than in upper flanks after gravistimulation. The biased IAA5 or IAA2 expression is disappeared in sgr2–1 mutant which is defective in gravity perception, indicating that gravity perception process is essential for formation of the biased gene expression during gravitropism. IAA5 expression was remarkably increased in lower flanks at 30 min after gravistimulation, whereas IAA2 expression was gradually decreased in upper flanks in a time-dependent manner. Therefore, we conclude that IAA5 is a sensitive GRI to monitor asymmetric auxin signaling caused by gravistimulation in Arabidopsis inflorescence stems.
Keywords: Arabidopsis thaliana, shoot gravitropism, inflorescence stem, IAA5, gene expression, gravitropic response indicator
Introduction
Plants perceive gravity and change their growth direction as a survival strategy for adaptation to environment. The shoot shows negative gravitropism, and the root shows positive gravitropism. Gravitropism is thought to consist of 4 sequential processes: gravity perception, signal conversion, signal transduction, and asymmetric organ growth. Gravity is perceived in statocytes (gravity sensing cells), which are endodermal and columella cells in shoots and in roots, respectively.1,2 The statocytes contain amyloplasts, which are starch accumulating plastids to work as statolith. When plants are reoriented, amyloplasts in statocytes sediment to new cell bottom according to the gravity vector. Amyloplast sedimentation, which is a physical signal, should be converted into biochemical signals in the statocytes. There are several candidates of the biochemical signals. Cytosolic alkalinization observed in columella cells in Arabidopsis roots is a possible signal triggered by amyloplast sedimentation.3 In addition, transient changes in concentration of inositol 1,4,5-triphosphate4 and/or Ca2+5,6 observed in the responding organs are also suggested as the signals, although it is still unclear whether such changes occur in the statocytes and how they are involved in gravitropic response. In any case, the signal is finally transmitted to the elongating organ to trigger asymmetric organ growth.
Cholodny-Went hypothesis suggests that auxin asymmetrically distributes in lower side of gravistimulated organ, and then higher auxin concentration, which promotes or inhibit cell elongation in shoots or in roots, respectively, results in organ curvature.7 Indeed, it has been shown that indol-3-acetic acid, a major endogenous auxin, was asymmetrically distributed after gravistimulation in rice and corn coleoptile.8-10 Furthermore, our knowledge about the molecular mechanism to cause asymmetric auxin distribution is growing. Intracellular polar localization of PIN3, an auxin efflux carrier protein mainly expressed in statocytes, is involved in generation of asymmetric auxin distribution in Arabidopsis roots.11
Transcriptome analyses during early gravitropic response in Arabidopsis roots have shown that numerous genes including auxin response genes such as AUX/IAA and SAUR family genes are upregulated.12-15 Comparison of gene expression profiles between lower and upper flanks of Brassica oleracea hypocotyls or rice shoot base has demonstrated that auxin response genes are asymmetrically induced in the lower flanks of organs after gravistimulation, probably reflecting that auxin distribution.14,15
DR5 is a synthetic auxin responsive promoter which has multiple copies of auxin responsive elements (AuxRE).16 DR5::GUS and DR5::GFP reporters produce β-glucuronidase (GUS) and green fluorescent protein (GFP), respectively, from the DR5 promoter in response to auxin. Thus, histochemical staining and fluorescence of their transgenic plants can indicate local auxin signal intensity.17,18 Asymmetric auxin signaling after gravistimulation in primary roots and hypocotyls is also detected with DR5::GUS and DR5::GFP reporters in Arabidopsis.11,19-21 In contrast, DR5::GUS does not work in inflorescence stems,22 and there are no reports on the availability of DR5::GFP to monitor auxin signaling in gravitropic response of inflorescence stems. In addition, DII-VENUS, a fluorescent reporter, was recently developed as an auxin sensor.23 Since DII-VENUS is degraded in response to auxin, its abundance inversely correlates with auxin activity. Thus, DII-VENUS allows rapid detection of dynamic changes in auxin distribution at the cellular level. Actually, DII-VENUS can reflect asymmetric auxin distribution after gravistimulation,24 although it has not been known whether it is suitable for monitoring auxin distribution during gravitropic response in inflorescence stems. Therefore, an appropriate reporter system is essential to study the details of gravitropic signaling processes in inflorescence stems.
Here, we focused to find appropriate gene(s) as gravitropic response indicators (GRIs) to monitor gravitropic signaling in inflorescence stems of Arabidopsis. We expected that genes differentially induced in lower flanks of inflorescence stems in response to gravistimulation can be used as GRIs. In this study, we identified 2 GRIs among auxin response genes found by differential transcriptome analysis between upper and lower flanks of the stems. We also demonstrated that the asymmetric expression of the GRIs is largely dependent on the gravity perception.
Results
Screening of GRI candidates in inflorescence stems of Arabidopsis
Inflorescence stems of Arabidopsis thaliana show the first visible bending at 20 min after gravistimulation by placing horizontally, and then the stems reached the vertical position in about 90 min.25 To find genes, gravitropic response indicators (GRIs) which exhibit increased expression in the lower flanks relative to the upper flanks in inflorescence stems after gravistimulation, we performed comparative transcriptomic analyses between each flank at 10 min and 30 min with microarray. As a result, none of the characterized genes showed >2-fold differentially increased expression in lower flanks at 10 min (data not shown). In contrast, 30 genes were showed >2-fold differentially increased expression in the lower flanks relative to the upper flanks at 30 min (Table 1), indicating that differential gene expression between each flank becomes detectable from 10 to 30 min in our experimental condition. The 30 genes, which can be considered as GRI candidates, included 5 Aux/IAA family genes and 14 SAUR family genes, which are auxin response genes (Table 1).26 The result implies that auxin asymmetric distribution may precede the differential expression of auxin response genes in inflorescence stems.
Table 1. The list of genes whose expressions were increased > 2-fold in lower flanks relative to the upper flanks of inflorescence stems at 30 min after gravistimulation.
| AGI | Annotation | Fold change |
|---|---|---|
| At2g22810 | Aminocyclopropane-1-carboxylate synthase 4 (ACS4) | 8.63 |
| At1g15580 | Indole-3-acetic acid inducible 5 (IAA5) | 6.60 |
| At5g66580 | Unknown protein | 4.70 |
| At3g57260 | Beta-1,3-glucanase 2 (BGL2) | 4.58 |
| At4g13790 | SAUR-like auxin responsive protein family | 3.83 |
| At5g18010 | Small auxin up RNA 19 (SAUR19) | 3.71 |
| At5g18080 | Small auxin up RNA 24 (SAUR24) | 3.57 |
| At5g18030 | SAUR-like auxin responsive protein family | 3.44 |
| At5g18020 | Small auxin up RNA 20 (SAUR20) | 3.35 |
| At4g38850 | Small auxin upregulated 15 (SAUR15) | 3.29 |
| At5g18050 | Small auxin up RNA 22 (SAUR22) | 3.24 |
| At5g18060 | Small auxin up RNA 23 (SAUR23) | 3.22 |
| At1g29500 | SAUR-like auxin responsive protein family | 3.10 |
| At1g29440 | Small auxin up RNA 63 (SAUR63) | 3.03 |
| At1g29460 | SAUR-like auxin responsive protein family | 2.96 |
| At5g02760 | Arabidopsis PP2C clade D 7 (APD7) | 2.89 |
| At1g29450 | SAUR-like auxin responsive protein family | 2.84 |
| At4g14560 | Indole-3-acetic acid inducible 1 (IAA1) | 2.83 |
| At5g52900 | Membrane-associated kinase regulator 6 (MAKR6) | 2.82 |
| At5g47370 | Homeobox-leucine zipper protein 2 (HAT2) | 2.62 |
| At4g25420 | Gibberellin 20-oxidase 1 (GA20OX1) | 2.37 |
| At1g78100 | Auxin upregulated F-box protein 1 (AUF1) | 2.31 |
| At5g39860 | Paclobutrazol resistance 1 (PRE1) | 2.26 |
| At1g04240 | Short hypocotyl 2 (SHY2/IAA3) | 2.25 |
| At1g29510 | Small auxin upregulated 68 (SAUR68) | 2.22 |
| At5g10760 | Eukaryotic aspartyl protease family protein | 2.22 |
| At1g15550 | Gibberellin 3-oxidase 1 (GA3OX) | 2.10 |
| At3g03820 | Small auxin up RNA 29 (SAUR29) | 2.10 |
| At3g23030 | Indole-3-acetic acid inducible 2 (IAA2) | 2.06 |
| At1g52830 | Short hypocotyl 1 (SHY1/IAA6) | 2.02 |
AGI: gene identification number by the Arabidopsis Genome Initiative. The gene annotation was retrieved from TAIR (http://arabidopsis.org/index.jsp). The genes in bold are AUX/IAA or SAUR family genes.
Expression level of IAA5 was increased in lower flanks of inflorescence stems after gravistimulation
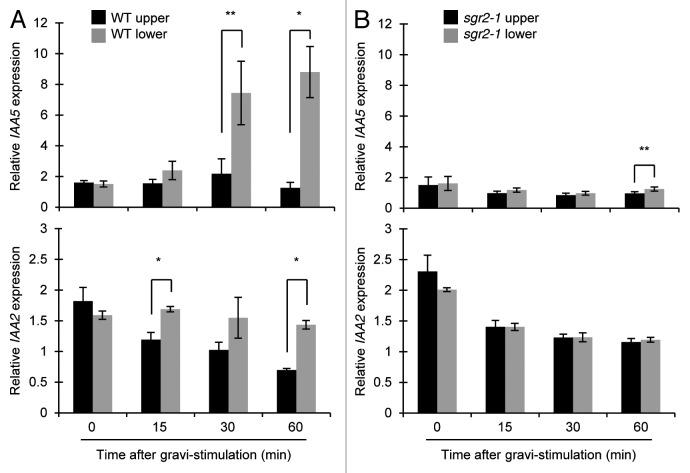
Among 19 auxin response genes of GRI candidates, IAA5 gene exhibits a 6.60-fold increased expression in the lower flanks relative to the upper flanks (Table 1). To validate this differential expression of IAA5, real-time qRT-PCR analysis was performed. Total RNA was extracted from upper and lower flanks of inflorescence stems after 0, 15, 30, and 60 min gravistimulation. Expression of IAA5 was significantly increased in lower flanks of inflorescence stems 30 and 60 min after gravistimulation (Fig. 1A). On the other hand, the level of IAA5 was stable in upper flanks from 0 min to 60 min (Fig. 1A).
Figure 1. Relative expressions of IAA5 and IAA2 after gravistimulation in upper and lower flanks of inflorescence stems. (A) wild type (WT), (B) sgr2–1. Relative expression level of IAA5 and IAA2 were normalized to expression of an actin gene (ACT8) in arbitrary units. Bars represent ± SD of 3 biological replicates. The statistical significance was tested using a Student t-test between upper and lower flanks at same time (*P < 0.01 and **P < 0.05).
It has been reported that a reporter gene using the promoter of IAA2 fused with uidA gene encoding GUS (β- glucuronidase) exhibits deeper staining in lower flanks than upper flanks of inflorescence stems 60 min after gravistimulation.22 Our microarray analysis showed that IAA2 exhibits a 2.06-fold increased expression in the lower flanks relative to the upper flanks (Table 1). Real-time qRT-PCR analysis showed that expression level of IAA2 in upper flanks decreased at 15 min and 60 min although that in lower flanks was stable at all time-points (Fig. 1A). The results indicate that the differential expression of IAA2 between upper and lower halves of stems was generated by a different manner from that of IAA5.
We also determined the change in expression levels of IAA5 and IAA2 genes in whole stems before and after gravistimulation. The expressions of both genes were remarkably changed at 30 and 60 min compared with those at 0 min (Fig. 2). However the patterns of those expressions were totally different, i.e., expression level of IAA5 was increased, whereas that of IAA2 was decreased in a time-dependent manner (Fig. 2). These results suggest that not all GRI candidate genes, which exhibit relatively increased expression in lower flanks, are upregulated after gravistimulation in the whole stems.
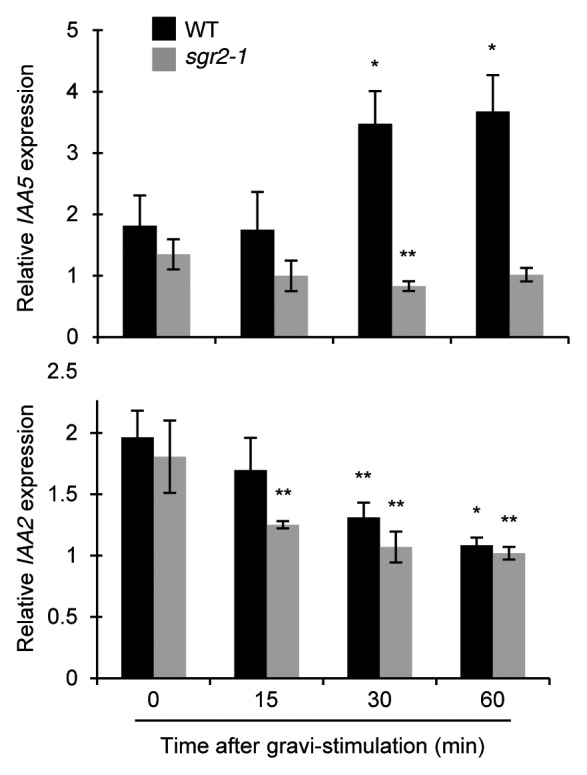
Figure 2. Relative expressions of IAA5 and IAA2 after gravistimulation in whole inflorescence stems. Relative expression level of IAA5 and IAA2 were normalized to expression of an actin gene (ACT8) in arbitrary units. Bars represent ± SD of 3 biological replicates. The statistical significance was tested using a Student t-test against relative expression at 0 min of WT (*P < 0.05) or sgr2–1 (**P < 0.05).
Gravity perception was essential for differential expression of IAA5 and IAA2
Since our gravistimutation, reorientation of plants, may contain mechanical stimulations, it is important to test whether the response of GRI gene expression is dependent on the gravity perception that happens in the statocytes. The sgr2–1 mutant has a defect in gravity perception due to the abnormal amyloplast sedimentation in the shoot statocytes, resulting in little gravitropic response in inflorescence stems.27-29 To examine whether expression changes in IAA5 and IAA2 upon gravistimulation are dependent on gravity perception, real-time qRT-PCR analyses was performed with sgr2–1 mutant stems. Neither increase of IAA5 expression in the lower flanks nor decrease of IAA2 expression in the upper flanks was detected in the sgr2–1 inflorescence stems upon gravistimulation (Fig. 1B). In addition, there is no significant difference in the expression levels of IAA5 and IAA2 between upper and lower flanks at any time-points except for small difference for IAA5 at 60 min (Fig. 1B). In whole stems, expression level of IAA5 or IAA2 is almost constant or gradually decreased, respectively, after gravistimulation in sgr2–1 (Fig. 2). The decreased expression of IAA2 after gravistimulation was observed both in wild type and in sgr2–1, though the mechanism of the phenomena is unclear. These results indicated that intact gravity perception process was necessary for the differential expression of IAA5 and IAA2, and also suggested that the differential expression of IAA5 and IAA2 might mainly reflect asymmetric auxin distribution during gravitropism
Discussion
We performed comprehensive screening of differentially expressed genes in upper and lower flanks of inflorescence stems after gravistimulation (Table 1). The majority of 30 GRI candidate genes found in microarray screening is auxin response gene, which show expression biased to lower flanks, implying formation of differential distribution of auxin in inflorescence stems. We identified 2 auxin response genes, IAA5 and IAA2, as GRIs to monitor gravitropic response 30 min after gravistimulation. However, the mechanisms producing the differentially increased expression in lower flanks seem to differ from one another (Fig. 1, 2, discussed below). Furthermore, intact gravitropic perception process was essential for their expression changes, because the changes in expression of both genes disappeared in sgr2–1.
The expression of IAA5 was remarkably induced in lower flanks at 30 min after gravistimulation. In contrast, the expression of IAA2 was reduced in upper flanks from 15 to 60 min after gravistimulation. In the previous report by Nadella et al., histochemical analysis of GUS reporter activity driven by IAA2 promoter after gravistimulation showed that GUS staining seemed to be deeper in lower side than in upper side of WT inflorescence stems from 20 to 60 min.22 Assuming that the IAA2 promoter activity is decreased in the upper flanks, the result of staining pattern could not be formed because GUS protein is hardly degraded in such short time-span due to its high stability. One possible explanation is that the IAA2 promoter activity could be upregulated by gravistimulation but it cannot operate the level of IAA2 transcripts (Fig. 2) due to a post-transcriptional regulation, while the IAA2 mRNA might be more severely degraded in upper flanks than lower flanks in a post-transcriptional manner (Fig. 1A).
The difference in expression level between upper and lower flanks after gravistimulation of IAA5 is remarkably larger than that of IAA2. In addition, real-time qRT-PCR analyses using split or whole stems demonstrate that IAA5 transcript is increased specifically in lower flanks at 30 min after gravistimulation without decrease of the IAA5 transcript in upper flanks. The promoter of IAA5 contains AuxRE and IAA5 has been recognized as one of early/primary auxin-responsive gene.30,31 The characteristics of IAA5 expression makes it easier to understand the relationship between gene expression behavior and gravitropic response than that of IAA2. These facts indicate that IAA5 is a rapid and sensitive GRI to monitor asymmetric auxin signaling caused by gravistimulation in Arabidopsis inflorescence stems. The measurement of IAA5 asymmetric expression would be a new available method to quantify gravitropic response of florescence stems in addition to measuring the organ curvature. IAA5 as a GRI will contribute to research on molecular mechanism of signaling process during gravitropic response in inflorescence stems.
Materials and Methods
Plant materials and growth conditions
The Columbia accession of Arabidopsis thaliana was used as the wild type (WT). The sgr2–1 mutant was described previously.27,28 Seeds were sterilized and plated on Murashige and Skoog (MS) plates followed by treatment at 4 °C for 3 days. Seedlings were germinated under constant light at 23 °C for 10 d. Plants were transplanted and grown on soil under constant white light at 23 °C.
Total RNA extraction from gravistimulated inflorescence stem
To gravistimulate the inflorescence stems, plants harboring primary stems 4–8 cm in length were placed horizontally under non-directional dim light at 23 °C. Gravistimulated stems were decapitated 5 mm from the top of inflorescence, and segments 3 cm and 3.5 cm in length from the cutting position were used for whole stem analysis of real-time qRT-PCR and microarray analysis, respectively. For upper and lower flank analysis, these segments were sectioned longitudinally and manually. The segments were rapidly frozen by liquid nitrogen, and then total RNA was extracted with RNeasy Plant Mini kit (Qiagen, Venlo, Netherland). Six whole stem segments or 10 upper and lower half segments were used for RNA isolation as one sample of real-time qRT-PCR analyses.
Microarray analysis
Prior to experiment, total RNA was examined for purity using an Agilent 2100 Bioanalyzer (Agilent, California USA). 0.5 μg of total RNA was used for cDNA synthesis and cRNA labeling reaction with Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent, California USA). In this reaction, each cRNA was labeled both cyanine-3 and cyanine-5 in separate reactions. These labeled RNA were hybridized to Agilent Arabidopsis 2 Oligo Microarray (Agilent, California USA) and were washed. Signals were detected with Agilent technologies Microarray Scanner (Agilent, California USA). Two hybridization including dye swap was performed for each experiments.
Real-time qRT-PCR
cDNA was synthesized with ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, Tokyo, Japan) from 0.5 μg of total RNA. Real-time qPCR was performed using the LightCycler® 96 Real-Time PCR System (Roch Applied Science, Upper Bavaria, Germany) and KAPA SYBER® FAST qPCR Kit (KAPABIOSYSTEMS, Massachusetts USA) was used for preparation of real-time qPCR reaction mix. Reactions were run in triplicate in 3 independent experiments. The results of real-time qPCR were analyzed with LightCycler® 480 Software (Roch Applied Science, Upper Bavaria, Germany), and mRNA relative expression levels (arbitrary units) were determined using the relative standard curve for IAA5, IAA2, and ACT8, which were generated by serial dilutions of cDNA.32 ACT8 was used as an internal control for normalization. The following primers used for amplification were designed at a specific region for each genes: At1g15580-IAA5-qPCR-F1 (5′-TGAAGGAAAG TGAATGTGTA CCAA-3′), AT1g15580-IAA5-qPCR-R1 (5′-GCACGATCCA AGGAACATTT-3′), At3g23030-IAA2-qPCR-F1 (5′-GAAGAATCTA CACCTCCTAC CAAAA-3′), At3g23030-IAA2-qPCR-R1 (5′-CACGTAGCTC ACACTGTTGT TG-3′), At1g49240_qPCR-F (5′-TCAGCACTTT CCAGCAGATG-3′) and At1g49240_qPCR-R (5′-CTGTGGACAA TGCCTGGAC-3′).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Keitaro Sakaguchi and Eiki Yagi for their contribution to sample preparation. We also thank Ms Nauko Inui and Ms Kaori Kaminoyama for technical assistance. Financial support was provided by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (16085205 to M.T.M.) and a grant from PRESTO project by Japan Science and Technology Agency (to M.T.M.).
Glossary
Abbreviations:
- AuxRE
auxin responsive element
- GUS
β-glucuronidase
- GFP
green fluorescent protein
- GRI
gravitropic response indicator
- WT
wild type
References
- 1.Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M. Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 1998;14:425–30. doi: 10.1046/j.1365-313X.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- 2.Blancaflor EB, Fasano JM, Gilroy S. Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 1998;116:213–22. doi: 10.1104/pp.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fasano JM, Swanson SJ, Blancaflor EB, Dowd PE, Kao TH, Gilroy S. Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell. 2001;13:907–21. doi: 10.1105/tpc.13.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perera IY, Heilmann I, Chang SC, Boss WF, Kaufman PB. A role for inositol 1,4,5-trisphosphate in gravitropic signaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiol. 2001;125:1499–507. doi: 10.1104/pp.125.3.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plieth C, Trewavas AJ. Reorientation of seedlings in the earth’s gravitational field induces cytosolic calcium transients. Plant Physiol. 2002;129:786–96. doi: 10.1104/pp.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toyota M, Furuichi T, Tatsumi H, Sokabe M. Cytoplasmic calcium increases in response to changes in the gravity vector in hypocotyls and petioles of Arabidopsis seedlings. Plant Physiol. 2008;146:505–14. doi: 10.1104/pp.107.106450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans ML. Gravitropism: interaction of sensitivity modulation and effector redistribution. Plant Physiol. 1991;95:1–5. doi: 10.1104/pp.95.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodbolé R, Michalke W, Nick P, Hertel R. Cytoskeletal drugs and gravity-induced lateral auxin transport in rice coleoptiles. Plant Biol. 2000;2:176–81. doi: 10.1055/s-2000-9154. [DOI] [Google Scholar]
- 9.Parker KE, Briggs WR. Transport of Indole-3-Acetic Acid during Gravitropism in Intact Maize Coleoptiles. Plant Physiol. 1990;94:1763–9. doi: 10.1104/pp.94.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iino M. Mediation of tropisms by lateral translocation of endogenous indole-3-acetic acid in maize coleoptiles. Plant Cell Environ. 1991;14:279–86. doi: 10.1111/j.1365-3040.1991.tb01502.x. [DOI] [Google Scholar]
- 11.Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415:806–9. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- 12.Moseyko N, Zhu T, Chang HS, Wang X, Feldman LJ. Transcription profiling of the early gravitropic response in Arabidopsis using high-density oligonucleotide probe microarrays. Plant Physiol. 2002;130:720–8. doi: 10.1104/pp.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimbrough JM, Salinas-Mondragon R, Boss WF, Brown CS, Sederoff HW. The fast and transient transcriptional network of gravity and mechanical stimulation in the Arabidopsis root apex. Plant Physiol. 2004;136:2790–805. doi: 10.1104/pp.104.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu L, Mei Z, Zang A, Chen H, Dou X, Jin J, Cai W. Microarray analyses and comparisons of upper or lower flanks of rice shoot base preceding gravitropic bending. PLoS One. 2013;8:e74646. doi: 10.1371/journal.pone.0074646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci U S A. 2006;103:236–41. doi: 10.1073/pnas.0507127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–71. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–72. doi: 10.1016/S0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- 18.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–53. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 19.Rashotte AM, DeLong A, Muday GK. Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell. 2001;13:1683–97. doi: 10.1105/tpc.13.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci U S A. 2003;100:2987–91. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakusová H, Gallego-Bartolomé J, Vanstraelen M, Robert HS, Alabadí D, Blázquez MA, Benková E, Friml J. Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 2011;67:817–26. doi: 10.1111/j.1365-313X.2011.04636.x. [DOI] [PubMed] [Google Scholar]
- 22.Nadella V, Shipp MJ, Muday GK, Wyatt SE. Evidence for altered polar and lateral auxin transport in the gravity persistent signal (gps) mutants of Arabidopsis. Plant Cell Environ. 2006;29:682–90. doi: 10.1111/j.1365-3040.2005.01451.x. [DOI] [PubMed] [Google Scholar]
- 23.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–6. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 24.Band LR, Wells DM, Larrieu A, Sun J, Middleton AM, French AP, Brunoud G, Sato EM, Wilson MH, Péret B, et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc Natl Acad Sci U S A. 2012;109:4668–73. doi: 10.1073/pnas.1201498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukaki H, Fujisawa H, Tasaka M. Gravitropic response of inflorescence stems in Arabidopsis thaliana. Plant Physiol. 1996;110:933–43. doi: 10.1104/pp.110.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–85. doi: 10.1023/A:1015207114117. [DOI] [PubMed] [Google Scholar]
- 27.Kato T, Morita MT, Fukaki H, Yamauchi Y, Uehara M, Niihama M, Tasaka M. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell. 2002;14:33–46. doi: 10.1105/tpc.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morita MT, Kato T, Nagafusa K, Saito C, Ueda T, Nakano A, Tasaka M. Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell. 2002;14:47–56. doi: 10.1105/tpc.010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toyota M, Ikeda N, Sawai-Toyota S, Kato T, Gilroy S, Tasaka M, Morita MT. Amyloplast displacement is necessary for gravisensing in Arabidopsis shoots as revealed by a centrifuge microscope. Plant J. 2013;76:648–60. doi: 10.1111/tpj.12324. [DOI] [PubMed] [Google Scholar]
- 30.Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–49. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 2003;133:1843–53. doi: 10.1104/pp.103.030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]