Abstract
Autophagy is reported to suppress tumor proliferation, whereas deficiency of autophagy is associated with tumorigenesis. ATG4B is a deubiquitin-like protease that plays dual roles in the core machinery of autophagy; however, little is known about the role of ATG4B on autophagy and proliferation in tumor cells. In this study, we found that ATG4B knockdown induced autophagic flux and reduced CCND1 expression to inhibit G1/S phase transition of cell cycle in colorectal cancer cell lines, indicating functional dominance of ATG4B on autophagy inhibition and tumor proliferation in cancer cells. Interestingly, based on the genetic and pharmacological ablation of autophagy, the growth arrest induced by silencing ATG4B was independent of autophagic flux. Moreover, dephosphorylation of MTOR was involved in reduced CCND1 expression and G1/S phase transition in both cells and xenograft tumors with depletion of ATG4B. Furthermore, ATG4B expression was significantly increased in tumor cells of colorectal cancer patients compared with adjacent normal cells. The elevated expression of ATG4B was highly correlated with CCND1 expression, consistently supporting the notion that ATG4B might contribute to MTOR-CCND1 signaling for G1/S phase transition in colorectal cancer cells. Thus, we report that ATG4B independently plays a role as a positive regulator on tumor proliferation and a negative regulator on autophagy in colorectal cancer cells. These results suggest that ATG4B is a potential biomarker and drug target for cancer therapy.
Keywords: ATG4B, autophagy, CCND1, colorectal cancer, MTOR, tumor proliferation
Introduction
Autophagy is a self-digesting process through which cells recruit damaged proteins and organelles to autophagosomes, which then fuse with lysosome for their degradation and recycling during nutrient deprivation.1 This evolutionarily conserved process plays an important role in many diseases, particularly in cancer development.1 Several tumor-suppressor genes have been shown to promote autophagy, including STK11/LKB1 (serine/threonine kinase 11)2 and TSC1/2 (tuberous sclerosis 1/2).3 In contrast, oncogenes, such as AKT/protein kinase B (v-akt murine thymoma viral oncogene homolog)4 and MTOR (mechanistic target of rapamycin),5 inhibit the initiation of autophagy. Moreover, autophagy functions as a tumor suppressor by reducing SQSTM1/p62-mediated tumor proliferation pathways.6,7 The liver-specific knockout of Atg5 or Atg7, essential autophagy-related (ATG) genes, accumulates SQSTM1 and increases spontaneous tumorigenesis, with the development of liver adenomas.8 BECN1 (mammalian ortholog of yeast Vps30/Atg6)9 and UVRAG (a potential activator of BECN1-dependent autophagy) induce autophagy and inhibit tumor cell growth and tumor formation in vivo.10 These results indicate that autophagy plays a suppressive role on cell proliferation to inhibit tumorigenesis.
At least 36 ATG genes are primarily involved in the progression of autophagy from phagophore initiation to autophagosome formation in mammalian cells.11 Among them, 2 ubiquitin-like (Ubl) proteins are required for the autophagy core machinery, including MAP1LC3 (microtubule-associated protein 1 light chain 3, a mammalian ortholog of yeast Atg8) and ATG12, which require ATG3 and ATG10 as the E2-like enzymes, respectively.12 ATG7 is a common E1-like enzyme for both Ubl proteins,13 and the ATG12–ATG5 complex serves as an E3-like enzyme that covalently attaches MAP1LC3 to phosphatidylethanolamine (termed MAP1LC3-II),14 which plays a crucial role in phagophore elongation.15 Moreover, ATG4 is the cysteine protease required for the activation of the MAP1LC3 precursor (proMAP1LC3)16,17 and the delipidation of MAP1LC3-II from autophagosomes for its recycling to facilitate autophagy.18 Autophagic activity is also diminished in both ATG4-deficient yeast and Atg4b-knockout mice.19-21 Nevertheless, inhibition of ATG4 activity by reactive oxygen species (ROS) is essential for starvation-induced autophagy,22 indicating that ATG4 activity is dynamically switched during autophagy conditions and the tight regulation of ATG4 is crucial for autophagy quality control.
There are 4 independent genes encoding ATG4 (4A, 4B, 4C, and 4D) in the human genome.23 ATG4B shows the most active and broadest proteolysis activity on human Atg8 orthologs,16,17,24,25 including MAP1LC3 and GABARAP/GABARAPL2 subfamilies, both of which are required for autophagosome formation.15 In addition, Atg4b-knockout mice exhibit decreased autophagic flux,19 whereas Atg4c-knockout mice displayed minimal effect on autophagy,26 indicating that ATG4B is functionally dominant in autophagic homeostasis compared with the other isoforms of ATG4. Although numerous studies have demonstrated the correlation between autophagy and tumor growth, the role of ATG4B on autophagy and the dependence of ATG4B-mediated autophagy on proliferation in cancer cells remain largely unknown. In this study, ATG4B knockdown blocked G1/S phase transition in colorectal cancer cells independent of autophagic flux. We further found that the role of ATG4B on tumorigenesis is prominent in xenograft tumors and colorectal cancer patients. Thus, our data show that ATG4B serves as an oncogene to promote tumorigenesis in colorectal cancer cells, which might be independent of autophagy.
Results
Knockdown of ATG4B induces autophagic flux in colorectal cancer cells
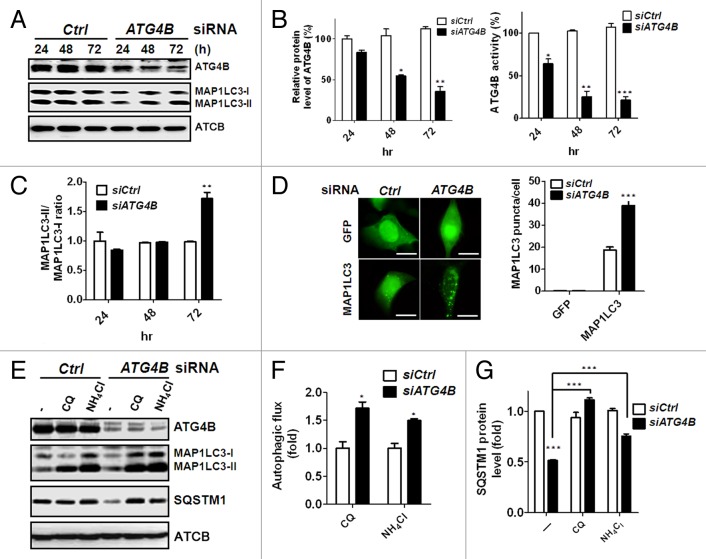
On the basis of reports of the dual roles of ATG4 on autophagy, we initially corroborated a role for ATG4B on autophagy in human colorectal cancer cells. Knockdown of ATG4B with 3 individual siRNA against ATG4B attenuated ATG4B expression and increased the ratio between the lipidated (MAP1LC3-II) and nonlipidated (MAP1LC3-I) forms of MAP1LC3 in colorectal cancer cells (Fig. S1). To minimize off-target effects of siRNA, we used a siRNA pool to silence ATG4B for subsequent experiments. The ATG4B protein level was gradually decreased in the presence of siRNA against ATG4B from 24 h to 72 h (Fig. 1A), and both the protein expression and proteolysis activity of ATG4B were attenuated in the ATG4B-silenced cells (Fig. 1B). The ratio of MAP1LC3-II/LC3-I was increased at 72 h after ATG4B knockdown (Fig. 1C). Consistent with the immunoblotting results, ATG4B knockdown resulted in a large number of GFP-MAP1LC3 puncta (Fig. 1D). Because both induction of autophagy and a block in downstream steps increase the ratio of MAP1LC3-II/LC3-I and GFP-MAP1LC3 puncta, autophagic flux assay was used to distinguish between these 2 possibilities.27 The autophagy inhibitor chloroquine (CQ) or ammonium chloride (NH4Cl) was employed to determine autophagic flux in human colorectal cancer cells silenced with siRNA against ATG4B (Fig. 1E–G), with both inhibitors increasing MAP1LC3-II accumulation in the ATG4B-silenced HCT116 cells (Fig. 1E and F). Furthermore, SQSTM1, an autophagy adaptor,28,29 was degraded in the ATG4B-knockdown cells, whereas degradation was recovered in the presence of autophagy inhibitors (Fig. 1E and G). Similar results were obtained using human colorectal cancer Caco2 cells (Fig. S2), indicating that ATG4B is functionally dominant on the negative regulation of autophagy in human colorectal cancer cells.
Figure 1. Silencing ATG4B induced autophagic flux in human colorectal cancer cells. (A) Scrambled siRNA (5 nM, siCtrl) or siRNA against ATG4B (5 nM, siATG4B) was transfected into human colorectal cancer Caco2 cells, and the cells were harvested every 24 h for 72 h. The cells were lysed for immunoblotting to determine the protein levels of ATG4B, MAP1LC3-I, MAP1LC3-II, and ACTB. (B) The knockdown efficiency of ATG4B was quantitated for each time point using ACTB as the normalization control (left panel). The proteolysis activity of ATG4B in the silenced cells was measured with the MAP1LC3-PLA2 reporter assay (right panel). (C) The ratio of MAP1LC3-II/MAP1LC3-I as an autophagy marker was employed to determine the effect of silencing ATG4B on autophagy. (D) Human colorectal cancer HCT-116 cells harboring GFP or GFP-MAP1LC3 were transfected with 5 nM siRNA for 72 h, and GFP-MAP1LC3 puncta were observed under fluorescence microscopy (left panel). Scale bar: 10 μm. The number of GFP-MAP1LC3 puncta for each cell was quantified (right panel). (E) HCT116 cells were transfected with the siRNA for 72 h and treated with or without (-) 20 μM CQ or 2 mM NH4Cl for 2 h. MAP1LC3-II accumulation and SQSTM1 degradation were examined by immunoblotting to determine the autophagic flux. The levels of (F) MAP1LC3-II accumulation and (G) SQSTM1 degradation in the cells were quantitated using ACTB as the normalization control. The results are expressed as the mean ± SEM from 3 individual experiments.
ATG4B promotes tumor growth in colorectal cancer cells
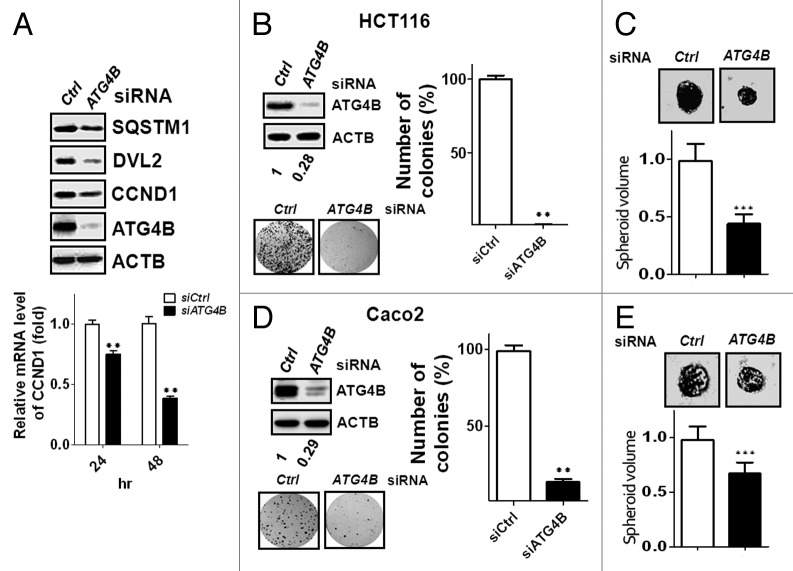
In terms of molecular mechanisms of autophagy on tumor growth, autophagy is able to trigger SQSTM1-mediated degradation of DVL (dishevelled segment polarity protein), a key mediator of WNT (wingless-type MMTV integration site family) for both the canonical and noncanonical pathways, to block WNT signaling during induction of autophagy, which in turn diminishes expression of its target genes, including CCND1 (cyclin D1).7 Aberrant expression of WNT activates its signaling through autocrine mechanisms in colorectal sarcomas and sarcoma cell lines.30-32 To examine the involvement of ATG4B-mediated autophagy on WNT signaling and tumor proliferation, we used human colorectal cancer HCT116 cells, a WNT3A expressed and a CTNNB1/β-catenin (catenin [cadherin-associated protein], β 1, 88 kDa)-mutated cell line that is highly responsive to WNT signaling.31,33 Besides degradation of SQSTM1, protein levels of both DVL2 and CCND1 were decreased in colorectal cancer HCT116 cells silenced with ATG4B siRNA, and mRNA level of CCND1 was reduced in the ATG4B-knockdown cells (Fig. 2A). The viability was also diminished in cells with siRNAs against ATG4B (Fig. S3). Moreover, silencing ATG4B significantly reduced the colony formation in HCT116 cells (Fig. 2B). To mimic the in vivo conditions, cell culture with Matrigel was employed to determine the effect of ATG4B on cancer growth in 3-dimensional spheroid formation (Fig. 2C). Spheroid volume was notably decreased in the ATG4B-silenced HCT116 cells (Fig. 2C). Similar results were observed in human colorectal cancer Caco2 cells (Fig. 2D and E), revealing that ATG4B is required for the tumor growth of colorectal cancer cells.
Figure 2. Knockdown of ATG4B reduced CCND1 expression and inhibited tumor growth in colorectal cancer cells. (A) Human colorectal cancer HCT116 cells were transfected with 5 nM siRNA for 72 h, and the protein levels of SQSTM1, DVL2, and CCND1 were detected by immunoblotting (upper panel). The mRNA level of CCND1 was measured by real-time PCR (lower panel). (B) HCT116 cells were transfected with 5 nM scrambled or ATG4B siRNA for the clonogenic assay. Representative data and quantitative results are shown in the left and right panels, respectively. (C) The siRNA-transfected cells were seeded on Matrigel-coated wells for spheroid formation. The cells were fixed to observe spheroid formation (upper panel), and the spheroid sizes were measured with ImageJ using the scrambled siRNA-transfected cells as the normalization control (n = 50, lower panel). Human colorectal cancer Caco2 cells were transfected with siRNA for (D) clonogenic assay or (E) spheroid formation. The results are expressed as the mean ± SEM from 3 individual experiments.
Silencing ATG4B inhibits G1/S phase transition in colorectal cancer cells
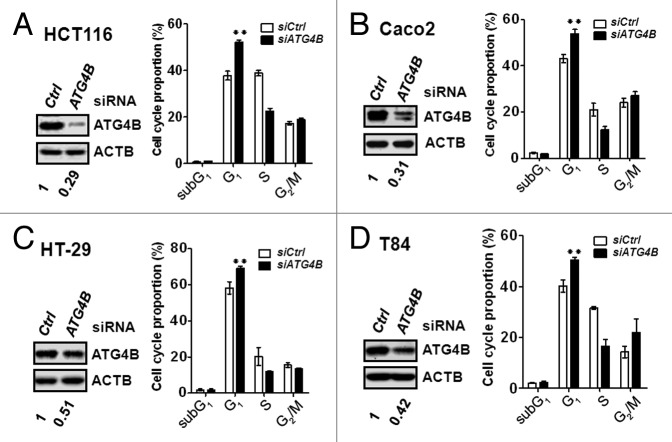
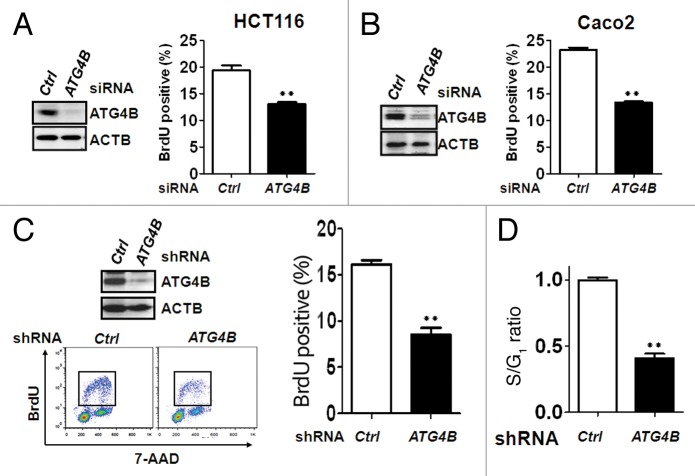
CCND1 forms a complex with cyclin-dependent kinase CDK4 or CDK6 and serves as a sensor for cell cycle transition from the G1 to the S phase. To examine whether ATG4B is involved in the control of cell cycle and contributes to the tumor growth of colorectal cancer cells, the cell cycle profiles of ATG4B-knockdown cells were measured using flow cytometry (Fig. S4). The percentage of cells in G1 phase was increased, whereas the percentage of cells in S phase was accordingly attenuated in the HCT116 cells transfected with the siRNA against ATG4B, indicating that silencing ATG4B arrested cell cycle at the G1 phase (Fig. 3A). Similar effects were observed in 3 other colorectal cancer cell lines, Caco2, HT-29, and lung-metastatic colorectal cancer T84 cells (Fig. 3B–D), suggesting that ATG4B may promote the transition from G1 to S phase in various stages of colorectal cancer cells. To more precisely measure the percentage of the cells in S phase, HCT116 or Caco2 cells were transfected with the siRNA against ATG4B and analyzed using the BrdU incorporation assay (Fig. S5). ATG4B knockdown reduced the number of BrdU-positive cells, which represents the percentage of cells in S phase (Fig. 4A and B). Similarly, HCT116 cells stably harboring shRNA against ATG4B were used for BrdU-7AAD staining and showed a significant reduction in the percentage of cells in S phase (Fig. 4C). The ratio of S/G1 phase was markedly decreased in the ATG4B stable-knockdown cells (Fig. 4D). These results indicate that ATG4B promotes cell cycle transition from the G1 to S phase.
Figure 3. Silencing ATG4B arrested the cell cycle at the G1 phase in human colorectal cancer cell lines. Human colorectal cancer (A) HCT116, (B) Caco2, (C) HT-29, and (D) T84 cells were transfected with 5 nM scrambled siRNA (siCtrl) or siRNA against ATG4B (siATG4B) for 72 h. The ATG4B protein level in the silenced cells was validated by immunoblotting (left panel). The knockdown cells were fixed and stained with propidium iodide to examine proportions of the cell cycle using flow cytometry. The flow cytometry data were analyzed and quantitated with FlowJo (right panel). The results are expressed as the mean ± SEM from 3 individual experiments.
Figure 4. Silencing ATG4B inhibited G1/S phase transition in human colorectal cancer cells. (A) Human colorectal cancer HCT116 (left panel) or (B) Caco2 (right panel) cells were transiently transfected with 5 nM scrambled or ATG4B siRNA for 72 h for subsequent immunoblotting (left panel) and BrdU incorporation assays (right panel). The BrdU-positive cells, representing proliferating cells in the S phase of the cell cycle, were quantified. (C) Human colorectal cancer HCT16 cells stably expressing scrambled or ATG4B shRNA were used for BrdU/7-AAD staining, followed by flow cytometry analysis. The BrdU-positive cells (tangle) were quantitated with FlowJo. (D) The ratio of S/G1 phase for the cells as (C) was further quantified. The results are expressed as the mean ± SEM from 3 individual experiments.
Autophagic flux is not required for ATG4B knockdown-induced G1 arrest
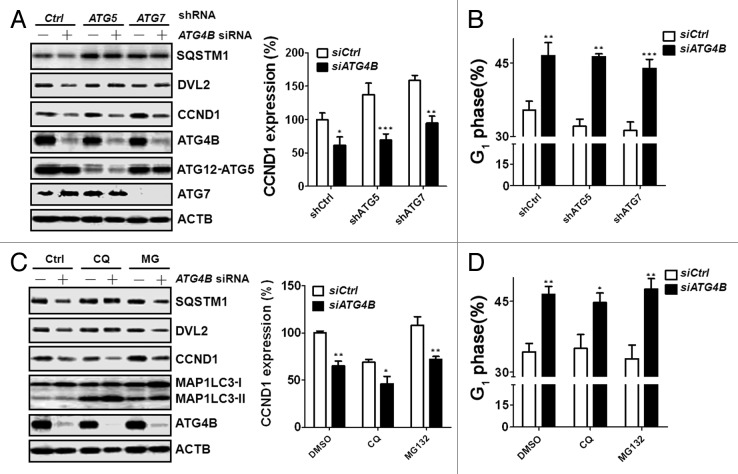
To inspect whether autophagic flux is involved in the reduction of CCND1 expression and G1 arrest of the cell cycle in ATG4B-knockdown cells, the siRNA against ATG4B was introduced into autophagy-deficient HCT116 cells stably expressing shRNA against ATG5 or ATG7 (Fig. 5A). CCND1 expression was slightly increased in the autophagy-deficient cells, which is consistent with the results from Atg5- or Atg7-knockout liver cells.34 Surprisingly, although silencing ATG4B-induced degradation of SQSTM1 and DVL2 was restored in the autophagy-deficient cells, the expression of CCND1 was not restored (Fig. 5A). The arrested G1 phase caused by silencing ATG4B in autophagy-deficient cells also remained unchanged (Fig. 5B). Moreover, because ATG5- and ATG7-independent macroautophagy35 may occur to affect CCND1 expression, we further treated the ATG4B-knockdown cells with chloroquine (CQ) and found that decreased CCND1 expression resulting from ATG4B knockdown was not recovered in the CQ-treated cells (Fig. 5C). Although the basal level of CCND1 expression was slightly increased in the cells treated with MG132 due to the inhibition of protein degradation, the expression of CCND1 was relatively low in the HCT116 cells transfected with siRNA against ATG4B (Fig. 5C). Accordingly, both CQ and MG132 treatments did not recover the G1 phase arrest in the ATG4B-silenced cells (Fig. 5D), suggesting that reduction in CCND1 expression and G1 phase arrest caused by ATG4B knockdown is independent of autophagic flux.
Figure 5. Autophagic flux was not required for G1 phase arrest in ATG4B-silenced cells. (A) Scrambled (-) or ATG4B (+) siRNA (5 nM) was introduced into autophagy-deficient (shATG5 or shATG7) human colorectal cancer HCT116 cells for 66 h. The protein levels of SQSTM1, DVL2, and CCND1 were determined by immunoblotting, and the knockdown efficiency of ATG4B, ATG5, or ATG7 was also examined. (B) The proportion of cells in G1 phase was analyzed by flow cytometry. (C) Scrambled (-) or ATG4B (+) siRNA was transfected into human colorectal cancer HCT116 cells for 56 h, followed by treatment with CQ (10 μM) or MG132 (1μM) for 16 h prior to harvesting. The protein levels of SQSTM1, DVL2, and CCND1 were determined by immunoblotting. (D) The proportion of the cells as (C) in G1 phase was analyzed by flow cytometry. The image of the immunoblotting results was quantitated using ACTB as the normalization control. The results are expressed as the mean ± SEM from 3 individual experiments.
Dephosphorylation of MTOR is involved in G1 arrest induced by depletion of ATG4B
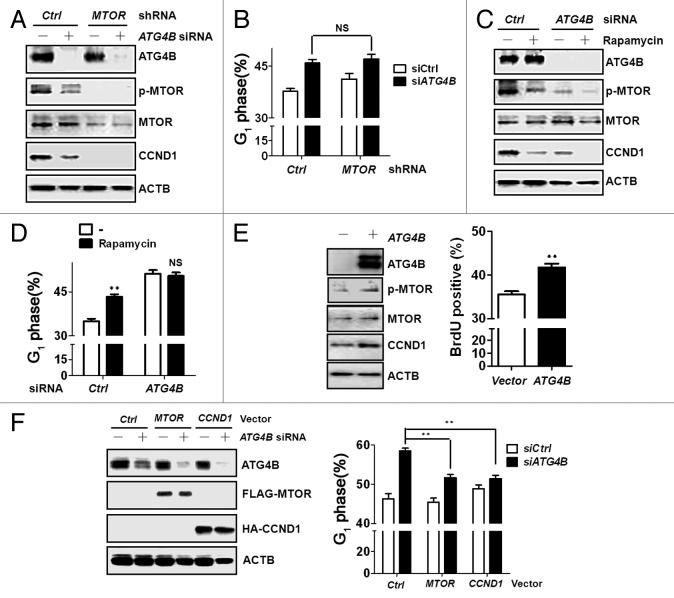
MTOR, a member of the PtdIns 3-kinase-related kinase (PIKK) family, masters signaling in G1/S phase progression of cell cycle and inhibits autophagy,5,36 which shows similar functions with ATG4B observed in this study. In terms of MTOR in cell cycle control, MTOR is able to elevate CCND1 transcription and translation to promote G1/S phase progression.36 To evaluate the involvement of MTOR in ATG4B-mediated G1/S phase transition, MTOR phosphorylation was examined in the ATG4B-knockdown cells. Both MTOR phosphorylation and CCND1 expression were decreased in ATG4B-knockdown cells, while the diminished level of MTOR and CCND1 expression was observed in the control cells with shRNA against MTOR (Fig. 6A). Transfection of siRNA against ATG4B into the cells stably harboring shRNA against MTOR did not synergize G1 phase arrest compared with the cells with scrambled shRNA (Fig. 6B). Similar effects on MTOR phosphorylation, CCND1 expression, and G1 phase proportion were also observed in ATG4B-silenced cells in the presence or absence of rapamycin, an inhibitor of MTOR (Fig. 6C and D). Moreover, overexpression of ATG4B increased MTOR phosphorylation, CCND1 expression, and number of BrdU-positive cells (Fig. 6E). Ectopic expression of MTOR and CCND1 diminished G1 arrest caused by silencing ATG4B in human colorectal cancer HCT116 cells (Fig. 6F). These results indicate that MTOR-CCND1 signaling may contribute to ATG4B-mediated cell proliferation.
Figure 6. MTOR dephosphorylation was involved in G1 phase arrest in ATG4B-silenced cells. (A) Scrambled (-) or ATG4B (+) siRNA (5 nM) was introduced into human colorectal cancer HCT116 cells stably harboring scrambled shRNA (Ctrl) or shRNA against MTOR for 72 h. MTOR phosphorylation and protein level of MTOR, CCND1, and ATG4B were subsequently determined by immunoblotting. (B) The proportion of G1 phase for cells as (A) was analyzed by flow cytometry. (C) Scrambled (-) or ATG4B (+) siRNA was transfected into human colorectal cancer HCT116 cells for 56 h, followed by treatment with rapamycin (0.2 μM) for 16 h prior to harvesting. The cells were harvested for immunoblotting or (D) assessment of the proportion in the G1 phase. (E) HCT116 cells were transfected with a vector expressing cherry or cherry tagged ATG4B for immunoblotting (left panel) or BrdU staining (right panel). The percentage of BrdU-positive cells in cherry-expressing cells was quantitated with FlowJo. (F) The cells were transfected with scrambled or siRNA against ATG4B for 48 h, followed by transfection of MTOR or CCND1 expression vectors for 24 h. The cells were harvested for immunoblotting (left panel) or cell cycle analysis. The results are expressed as the mean ± SEM from 3 individual experiments. NS: not significant.
ATG4B promotes G1/S phase transition in xenograft tumors
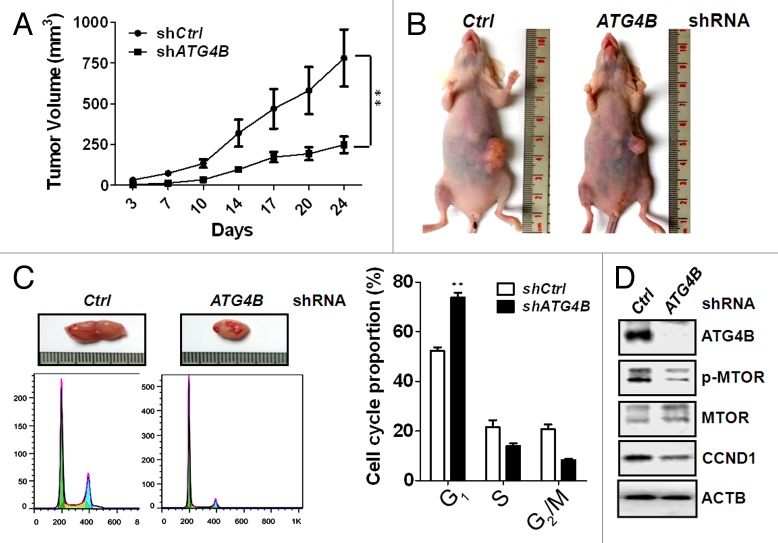
To determine the role of ATG4B on tumor growth in vivo, HCT116 cells stably expressing scrambled shRNA or ATG4B shRNA were xenografted into immunodeficient mice. The growth of xenograft tumors derived from ATG4B stable-knockdown cells was significantly slower than the cells with scrambled shRNA (Fig. 7A and B). The xenograft tumors were further excised for dissociation to obtain single cell population. The dissociated cells were harvested to examine cell cycle profiles and molecular mechanisms in vivo (Fig. 7C and D). The percentage of ATG4B-knockdown cells in G1 phase was significantly increased compared with the cells with scrambled shRNA (Fig. 7C). MTOR phosphorylation and CCND1 expression were also diminished in the cells with shRNA against ATG4B (Fig. 7D), supporting the notion that ATG4B may facilitate MTOR phosphorylation to increase CCND1 expression for G1/S phase transition.
Figure 7. Silencing ATG4B reduced G1/S phase transition and MTOR phosphorylation in vivo. (A) Mice were injected with 2 x 106 of human colorectal cancer HCT116 cells harboring scrambled shRNA or shRNA against ATG4B. The tumor volume in each mouse was measured every 3 to 4 d (5 for each group). (B) The representative mice with xenograft tumors at d 24 postinjection are shown. (C) The xenografted tumor was cut from sacrificed mice and dissociated to single cell population for cell cycle analysis (left panel). The cell cycle proportion was analyzed and quantitated with FlowJo (right panel) (n = 3). (D) The cell lysates as (C) were also harvested for immunoblotting to determine MTOR phosphorylation and protein level of MTOR, CCND1, and ATG4B (n = 3).
Elevated ATG4B expression is correlated with CCND1 expression in colorectal cancer patients
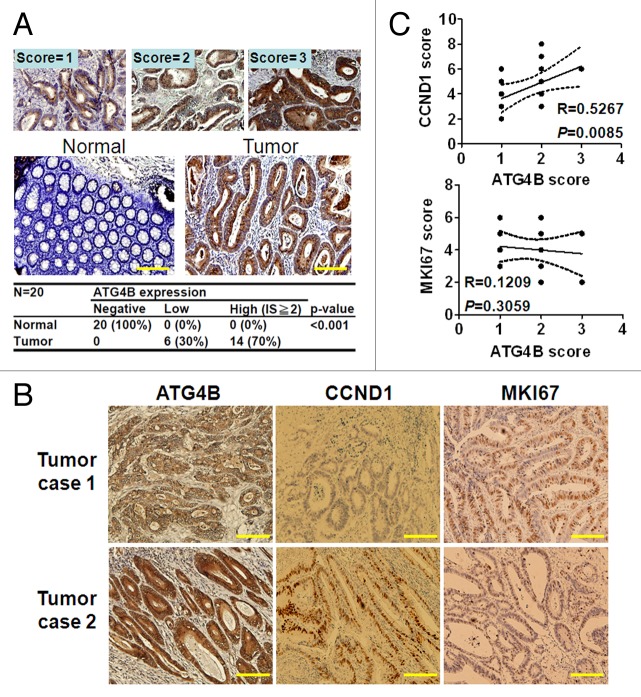
In the cell culture and mouse models, ATG4B was found to serve as an oncogene and facilitate G1/S phase progression of the cell cycle via CCND1 expression for tumor growth in colorectal cancer cells. To determine whether ATG4B expression level is correlated with tumor proliferation in colorectal cancer patients, 20 colorectal cancer patients (5 for each tumor stage) were recruited to examine ATG4B and CCND1 expression with immunohistochemical staining using MKI67 (a typical proliferation marker) as a control. Significantly (P < 0.001), the expression level of ATG4B, CCND1, and MKI67 was increased in the colorectal cancer cells compared with the stroma and goblet cells (Fig. 8A; Fig. S6), which were considered adjacent normal cells. For the correlation between ATG4B and CCND1 or MKI67, representative sections for both low and high ATG4B-expressing tumors from 2 colorectal cancer patients are shown in Figure 8B. There was a strong correlation between ATG4B and CCND1 expression in tumor tissues of colorectal cancer patients (R = 0.5267, P = 0.0085), whereas there was no correlation between ATG4B and MKI67 (Fig. 8C). This is likely because maximal expression of both ATG4B and CCND1 occurs in the G1 phase,37 but maximal expression of MKI67 occurs during mitosis.38 We also analyzed ATG4B expression in tumor cells at different stages, and found that the intensity of high ATG4B expression was nearly correlated with a late stage (II–IV) of tumors (73%) (Fig. S7). These results further support oncogenesis role of ATG4B in colorectal cancer.
Figure 8. Elevated ATG4B expression was correlated with CCND1 expression in tumor specimens of colorectal cancer patients. (A) The ATG4B expression level in colorectal cancer was determined by immunohistochemistry, and the intensity score (IS) was categorized into 3 grades based on the ATG4B protein level (upper panel). ATG4B was only expressed in the cytoplasm of the colorectal tumor cells and not in the adjacent normal tissues (stroma and goblet cells, scored as 0). The ATG4B protein level in the tumor and adjacent normal cells is quantified in the lower panel (P < 0.001). (B) Representative tumor sections stained for ATG4B, CCND1, and MKI67 from 2 colorectal cancer patients were shown. (C) The Allred scoring system was employed to quantitate CCND1 and MKI67 protein level. The correlation between ATG4B and CCND1 or MKI67 expression was calculated through the Pearson χ2 test. Twenty tissue samples from colorectal cancer patients were used to determine the expression and correlation. Scale bar: 200 μm.
Discussion
Dysregulation of autophagy is highly associated with cancer, particularly in tumorigenesis and chemotherapy resistance.6,8,39,40 ATG4 is required for basal homeostasis of autophagy in eukaryotic cells,16,19-21 whereas spatiotemporal inactivation of ATG4 ensures conjugation of MAP1LC3 to autophagosome membrane and facilitates autophagy in cells during starvation.22 The role of ATG4 on autophagy and tumor growth in cancer cells remains unclear. In this study, we found that silencing ATG4B, a dominant member of ATG4 in human cells, induced autophagic flux and arrested the cell cycle at G1 phase in colorectal cancer cells. Interestingly, the depletion of ATG4B-induced autophagic flux was not required for growth suppression in human colorectal cancer cells. Moreover, MTOR dephosphorylation was involved in reduced CCND1 expression and inhibited G1/S phase transition in ATG4B-knockdown cells, which was also proved in a mouse xenograft model for human colorectal cancer HCT116 cells. Furthermore, elevated ATG4B expression was strongly correlated with CCND1 expression in colorectal cancer patients. These results suggested that ATG4B functions as an oncogene and modulates MTOR-CCND1 signaling to facilitate G1/S phase transition in colorectal cancer cells independent of autophagic flux.
ATG4 is functionally redundant and plays dual roles on autophagy in mammalian cells. Human ATG4B cleaves all Atg8 orthologs at a considerably low concentration (1 nM) in vitro.17 ATG4A16,17,22,24 and activated ATG4D25 demonstrate preferential proteolysis activity on the GABARAP/GABARAPL2 subfamily of Atg8 orthologs. Using the highly sensitive MAP1LC3-PLA2 reporter assay, ATG4A and activated ATG4D were able to cleave MAP1LC3 in vitro (Fig. S8). The MAP1LC3 and GABARAP/GABARAPL2 subfamilies are required for phagophore elongation and autophagosome sealing, respectively, which are crucial for autophagosome biogenesis.15 These results suggested the functional redundancy of ATG4 on autophagy machinery in mammalian cells. In addition, ATG4B might locally interfere with the conjugation of MAP1LC3-II to autophagosome membrane in mammalian cells.22 Overexpression of ATG4B reveals an inhibitory effect on MAP1LC3-II conjugation because it is likely that ATG4B competes with conjugating enzymes on MAP1LC3 binding.41 Our present study found that ATG4B inhibited autophagic flux in several colorectal cancer cell lines. This finding indicated that a small amount of ATG4B or other isoforms might be sufficient for the homeostasis of autophagy in normal cells, whereas excess ATG4B leads to a reduction in conjugation of MAP1LC3-I to PE, or deconjugation of the MAP1LC3-II from premature phagophore, which inhibits autophagosome formation, thereby decreasing the autophagic flux in cancer cells. Alternatively, post-translational modification is required for ATG4D activation,25 while phosphorylation of ATG4B at Ser383 site has been detected in human cells.42,43 It raises a possibility that post-translational modification of ATG4B alters its function on autophagy in colorectal cancer cells.
Numerous studies have revealed that autophagy induction plays a suppressive role in tumor growth.6,7 Defects in autophagy augment SQSTM1-mediated WNT signaling7 and NFKB activation,6 both of which induce CCND1 expression to promote G1/S transition during the cell cycle. However, based on genetic and pharmacological approaches, our data show that ATG4B might play an autophagy-independent role on cell proliferation in colorectal cancer. Additional functions of several ATG genes have been shown in cancer cells but not with regard to autophagy. ATG5 is induced in cancer cells under genotoxic stress and contributes to mitotic catastrophe in an autophagy-independent manner.44 BECN1 functions as a tumor suppressor, and a monoallelic deletion in the gene has been identified in several types of cancer.45 However, cells with monoallelic-deleted BECN1 exhibit the promotion of cell proliferation46 yet are still able to mount autophagic flux under stress condition,47,48 suggesting an autophagy-independent role of BECN1 on cell proliferation.48 Our results show that ATG4B promotes G1/S phase transition for colorectal cancer proliferation in an autophagy-independent manner, which presented a new function of ATG4B on tumorigenesis.
Several oncogenes or tumor suppressors independently act as upstream regulators on tumorigenesis and autophagy. For example, DAPK1 (death-associated protein kinase 1) not only phosphorylates BECN1 to promote autophagy49 but also induces CDKN2A/p19 and TP53/p53 (TRP53 in mice)-mediated apoptosis to suppress tumor transformation.50 MTOR phosphorylates and inactivates Unc-51 kinase (ULK1) to inhibit autophagy,5 whereas MTOR increases CCND1 expression to promote cell proliferation through the inhibition of EIF4EBP1/4EBP1 (eukaryotic translation initiation factor 4E binding protein 1) or activation of AKT.36,51 In this study, ATG4B might facilitate MTOR phosphorylation to independently inhibit autophagy and promote cell proliferation in tumor cells. The effect was not caused by direct binding between ATG4B and MTOR complexes, because no interaction between ATG4B and MTOR was observed in co-immunoprecipitation experiments (data not shown). Therefore, ATG4B may confer tumor proliferation through indirect regulation on MTOR phosphorylation in colorectal cancer cells. Regardless, further studies are required to clarify the detailed mechanism of ATG4B on tumor proliferation.
To date, there is no report demonstrating the role of ATG4B on tumor proliferation in cancer cells. According to our present data, ATG4B knockdown reduced MTOR phosphorylation and CCND1 expression to attenuate the G1/S transition in both colorectal cancer cells and xenograft tumors. Consistently, ATG4B expression was strongly correlated with CCND1 expression in tumor cells from colorectal cancer patients. Overexpression of CCND1 has been found to initiate neoplasm in colorectal cancer patients,52,53 supporting the notion that ATG4B might modulate CCND1 expression through MTOR phosphorylation for tumor proliferation in colorectal cancers. More interestingly, atg4b−/− mice only show a slight defect on otoconial development of the inner ear, without growth inhibition or other clear abnormalities.19,54 Our data also showed that the growth rate of atg4b−/− 3T3 cells was similar to that of Atg4b+/+ 3T3 cells (Fig. S9), revealing that the regulation of ATG4B on cell proliferation in colorectal cancer cells might be different from normal cells due to the complicated nature of cancer. Taken together, our data indicated that ATG4B increases MTOR phosphorylation and CCND1 expression, promoting the transition of G1/S phase for tumor proliferation in colorectal cancer cells. Our findings show that ATG4B is the first ATG gene that serves as an oncogene in colorectal cancer cells, potentially providing a novel cancer marker and drug target for colorectal cancer therapy.
Materials and Methods
Plasmids and siRNA transfection
Human colorectal cancer cells, including HCT116, Caco2, T84, and HT-29, from ATCC were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Invitrogen, 12100-046) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 mg/ml). The cells were seeded at 20% to 30% confluence and reversely transfected with RNAiMAX (Life Technologies, 13778-150) in the presence of 5 nM scrambled siRNA (Life Technologies, 12935-112) or siRNA against ATG4B (Life Technologies, 20218, s23245, s23246) for 72 h. For plasmid transfection, the cells were transfected with 2 μg FLAG-MTOR (26603) or HA-CCND1 (8948) expression plasmids (Addgene) or cherry-tagged ATG4B (subcloned from FLAG-ATG4B reported previously17) using 2 μl Lipofectamine 2000 (Invitrogen, 11668-027) for 24 h. The cells were harvested for either protein expression analysis by immunoblotting or cell cycle analysis by flow cytometry.
Generation of shRNA stable cell line
shRNAs against ATG4B (TRCN0000073801), ATG5 (TRCN0000151963), ATG7 (TRCN0000007584), and MTOR (TRCN0000199323) were obtained from The RNAi Consortium (TRC, Taiwan). pLKO.1 Scrambled shRNA was purchased from Addgene (1864). The plasmids (2 μg) were transfected into 293FT cells (1 × 106 cells) using 2 μl of Lipofectamine 2000 (Life Technologies, 11668-027). The supernatant was harvested after 2 d, and the cell debris was removed for the infection of human colorectal cancer HCT116 cells. The infected HCT116 cells were selected with puromycin (1 μg/ml) for 10 ds to obtain cells stably harboring shRNA. The knockdown efficiency was verified by immunoblotting.
Autophagic flux measurement and immunoblotting
For more precise monitoring of autophagy activity, siRNA-transfected cells were treated with or without 20 μM CQ (Sigma-Aldrich, C6628) or 2 mM NH4Cl (Sigma-Aldrich, A9434) for 2 h prior to harvesting. The cell lysate was used to detect MAP1LC3-II accumulation by immunoblotting to verify autophagic flux.27 For immunoblotting, the cells were briefly rinsed in PBS (Biological Industries, 02-023-1) and lysed with RIPA buffer (1% NP40 [MDBio, 101-9016-45-9], 50 mM TRIS-HCl, pH 7.5, 150 mM NaCl, 0.25% sodium deoxycholate [Sigma-Aldrich, D6750], 0.1% sodium dodecyl sulfate [SDS; Calbiochem, 428015], and protease inhibitor cocktail [Roche, 11873580001]). The cell lysates were resolved by SDS-PAGE and transferred electrophoretically onto nitrocellulose membranes. The membrane was blocked with 5% skim milk and then incubated with primary antibodies against ATG4B (A2981), MAP1LC3 (L7543), Flag (F1804) and ACTB (β-actin, A5441) (all purchased from Sigma-Aldrich), SQSTM1 (BD PharMingen, 610832), HA (Roche, 12CA5) and CCND1 (cyclin D1, 2926), DVL2 (3224), ATG5 (8540), ATG7 (8558), p-MTOR (5536), and MTOR (2983) (all purchased from Cell Signaling Technology) for overnight at 4 °C. The proteins were probed with an HRP-labeled secondary antibody (Santa Cruz, sc-2004 or sc-2005) and detected with an ECL reagent. The membrane was scanned and analyzed for the protein expression level with the ChemiDoc XRS Imaging System (Bio-Rad).
ATG4B activity measured using MAP1LC3B-PLA2 substrate
The ATG4B activity in cells was assayed according to previous reports using the reporter substrate MAP1LC3B-PLA2 and fluorescent dye 2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino) hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine (NBD-C6-HPC) (Invitrogen, N-3786).17,55 Briefly, ATG4B-silenced cells were harvested and lysed in lysis buffer containing 25 mM TRIS-HCl, pH 8.0, 100 mM NaCl, 1 mM CaCl2, 5 mM MgCl2, 5% glycerol, 0.1% NP-40, 1 mM PMSF, and 1 mM DTT on ice followed by sonication for 2 s to break cells. The proteins in the supernatant fraction were diluted and mixed with assay buffer containing 20 mM TRIS-HCl, pH 8.0, 2 mM CaCl, 1 mM DTT, 20 μM NBD-C6-HPC, and 100 nM MAP1LC3B-PLA2. The fluorescence intensity was kinetically measured within 1 h using the Fluoroskan Ascent FL reader (Thermo Fisher Scientific, Waltham, MA, USA) at room temperature with excitation and emission wavelengths of 485 and 530 nm, respectively. The ATG4B activity in the silenced cells was normalized to control cells and quantified using Prism 5 software (GraphPad).
Real-time PCR
The cells transfected with siRNA were used for extraction total RNA with TRIzol Reagent (Invitrogen, 15596-018). The 1 μg of RNA was reverse-transcribed with SuperScript II RNase H-Reverse Transcriptase (Invitrogen, 18064-014) for cDNA synthesis. The amount of CCND1 mRNA relative to GAPDH was analyzed by real-time PCR performed in StepOnePlusTM system (Applied Biosystems) with SYBR Green Master Mix (Applied Biosystems, 4385612). The primers for the genes are as follows: CCND1 forward 5′-CGCCCCACCC CTCCAG-3′ and reverse 5′-CCGCCCAGAC CCTCAGACT-3′, GAPDH forward 5′-TGCACCACCA ACTGCTTAGC -3′ and Reverse 5′- GGCATGGACT GTGGTCATGA G -3′.
Clonogenic assay
The cells were seeded in 6-well plates at a density of 500 or 1000 cells/well in the presence of scrambled siRNA or siRNA against ATG4B. The cells were then cultured in complete media, which was refreshed every 3 d for 2 wk. The cell colonies were fixed with 2% paraformaldehyde and stained with 20% ethanol containing 0.25% crystal violet at room temperature for 30 min. The stained cells were washed with PBS for 3 times to observe colonies; colonies over 1 mm were counted in at least 3 independent experiments.
Three-dimensional spheroid culture
The 3-dimensional cell culture was performed as previously described, with minor modifications.56 Briefly, 50 µl of collagen- and laminin-enriched Matrigel (BD Biosciences, 356237) was coated in each well of a 96-well plate at 37 °C for 30 min. Human colorectal cancer cells were trypsinized to obtain a single-cell suspension, and the cells were centrifuged at 1000 × g for 5 min. The cells were resuspended in DMEM medium containing 2.5% Matrigel at a density of 2 × 104 cells / ml, and 100 µl of the cells was seeded into each well. The medium was refreshed every 3 d for 14 d until the cells form a spheroid scaffold. The cells were further fixed with 2% paraformaldehyde and visualized under a microscope. The volume of spheroids (n = 50) was measured using ImageJ software.
Flow cytometry for cell cycle and proliferation analysis
For the cell cycle analysis, the knockdown or treated cells were washed with PBS containing 1 mM EDTA and then fixed with 75% ethanol at -20 °C overnight. The fixed cells were then washed with PBS containing 1% fetal bovine serum and further stained with propidium iodide (50 µg/ml; Sigma-Aldrich, P4170) and RNase A (25 µg/ml; Sigma-Aldrich, R6513) on ice for 30 min to profile the cell cycle. To precisely determine the G1/S phase transition of the cells, BrdU/7AAD staining was performed according to the manufacturer’s instructions (BD PharMingen, 559619). Briefly, the cells were treated with 10 µM of BrdU for 1 h prior to fixation, and the cells were further probed with a fluorescein isothiocyanate (FITC)-conjugated anti-BrdU antibody and 7-aminoactinomycin D (7AAD). The stained cells were analyzed for the proportion of cells in different cell cycle phases using the FACScan (Becton Dickinson) and FlowJo (Tree Star).
Tumor xenograft
Five-wk-old immunodeficient mice (nu/nu, female) were purchased and acclimated for 5 d prior to tumor implantation. Human colorectal cancer HCT116 cells (2 × 106) stably harboring scrambled shRNA or shRNA targeting ATG4B were mixed with Matrigel (1:1) and subcutaneously implanted into mice. Tumors were measured every 3 to 4 d with vernier calipers, and tumor volumes were calculated by the formula (larger diameter) × (smaller diameter)2 × π/6. All animal experiments were approved by Institutional Animal Care and Use Committee at Kaohsiung Veterans General Hospital. Tumors were further cut from sacrificed mice and dissociated mechanically and enzymatically with a gentleMACs dissociator according to the manufacturer’s instructions (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). The dissociated cells were seeded on culture dish at 37 °C for 2 h to eliminate the cell debris and contamination from mouse blood cells. The adherent cells were then harvested for cell cycle analysis and immunoblotting.
Immunohistochemistry and scoring
As the retrospective cohort for this study, 20 surgically resected colorectal cancers (5 for each tumor stage) were collected between 2004 and 2005; no chemotherapy was administered prior to surgery, and the tissue was pathologically assessed at Kaohsiung Veterans General Hospital. Informed consent was provided by all the patients in this study, and the study was approved by IRB of Kaohsiung Veterans General Hospital (No. VGHKS11-CT12-08). The colorectal tissues were analyzed by immunohistochemistry staining: the tissue sections were stained with an anti-ATG4B antibody (Sigma-Aldrich, A2981) or CCND1 (Abcam, SP4) or MKI67 (Genemed, GM001), followed by a BondTM Polymer Refine Detection kit (Leica Biosystems, DS9800) to observe protein expression in both the tumor and adjacent normal cells. All the tumor cells within each microscopic field were counted, and then the positive rates of ATG4B expression in the cells were calculated. The ATG4B immunoreactivity of the tumor cells varied considerably; the estimated average staining intensity was scored as 0 (all cells negative), 1+ (weak expression), 2+ (moderate expression), and 3+ (strong expression). In the subsequent data analysis, an expression level of ATG4B scored 0 and 1+ was considered “low,” whereas 2+ and 3+ were categorized as “high.” Moreover, since the staining pattern of CCND1 and MKI67 on tumors was heterogeneous, the expression level was scored using the Allred scoring system based on the sum of a proportion score and intensity score, ranging from 2 to 8. Expression level of CCND1 and MKI67 scored ≥ 5 was considered as “high” expression. The expression level of ATG4B, CCND1, and MKI67 in the tissues was scored by Dr Chih-Wen Shu and pathologist Dr Hung-Wei Pan at least twice.
Statistical analysis
All the data are expressed as the mean ± SEM from at least 3 individual experiments. The statistical analysis was performed using a nonparametric 2-tailed Student t test with Prism 5.0 (Graph-Pad). For correlation between ATG4B and CCND1 expression in immunohistochemical results, the P value was calculated through the Pearson χ2 test. P values lower than 0.05 were considered significant (*P < 0.05, **P < 0.01, ***P < 0.001).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr López-Otín for kindly providing Atg4b+/+ and atg4b−/− mouse embryonic fibroblast (MEF) cells from which we generated immortalized Atg4b+/+ and atg4b−/− 3T3 cells. We also thank Dr Jie Chen and Dr Philip Hinds for sharing plasmids FLAG-MTOR and HA-CCND1, respectively. This work was supported by National Science Council (NSC 101 and 102-2311-B-075B-001 to C-WS) and Kaohsiung Veterans General Hospital (VGHKS102-007 to C-WS, VGHKS103-G01-1 to C-JH, and VGHKS100-058 to C-ML).
Glossary
Abbreviations:
- ATG
autophagy related
- ATG4B
autophagy-related 4B, cysteine peptidase
- CCND1
cyclin D1
- CQ
chloroquine
- DVL
disheveled segment polarity protein
- WNT
wingless-type MMTV integration site family
- MAP1LC3
microtubule-associated protein 1 light chain 3
- MTOR
mechanistic target of rapamycin
- ROS
reactive oxygen species
- SQSTM1
sequestosome 1
- Ubl
ubiquitin-like
- MKI67
marker of proliferation Ki-67
- BECN1
Beclin 1, autophagy related
- GABARAP
GABA(A) receptor-associated protein
- GABARAPL2
GABA(A) receptor-associated protein-like 2
- BrdU
bromodeoxyuridine
- 7AAD
7-aminoactinomycin D
- ACTB
actin, beta
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–24. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 3.Papadakis M, Hadley G, Xilouri M, Hoyte LC, Nagel S, McMenamin MM, Tsaknakis G, Watt SM, Drakesmith CW, Chen R, et al. Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat Med. 2013;19:351–7. doi: 10.1038/nm.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–9. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 8.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu CW, Liu PF, Huang CM. High throughput screening for drug discovery of autophagy modulators. Comb Chem High Throughput Screen. 2012;15:721–9. doi: 10.2174/138620712803519734. [DOI] [PubMed] [Google Scholar]
- 12.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–64. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser SE, Qiu Y, Coats JE, Mao K, Klionsky DJ, Schulman BA. Structures of Atg7-Atg3 and Atg7-Atg10 reveal noncanonical mechanisms of E2 recruitment by the autophagy E1. Autophagy. 2013;9:778–80. doi: 10.4161/auto.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 15.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 17.Shu CW, Drag M, Bekes M, Zhai D, Salvesen GS, Reed JC. Synthetic substrates for measuring activity of autophagy proteases: autophagins (Atg4) Autophagy. 2010;6:936–47. doi: 10.4161/auto.6.7.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–76. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 19.Mariño G, Fernández AF, Cabrera S, Lundberg YW, Cabanillas R, Rodríguez F, Salvador-Montoliu N, Vega JA, Germanà A, Fueyo A, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatogawa H, Ishii J, Asai E, Ohsumi Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 2012;8:177–86. doi: 10.4161/auto.8.2.18373. [DOI] [PubMed] [Google Scholar]
- 21.Yu ZQ, Ni T, Hong B, Wang HY, Jiang FJ, Zou S, Chen Y, Zheng XL, Klionsky DJ, Liang Y, et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8:883–92. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–60. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariño G, Uría JA, Puente XS, Quesada V, Bordallo J, López-Otín C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–38. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–66. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 29.Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–95. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 30.Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, Hundsrucker C, Kerr G, Sandmann T, Anchang B, et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun. 2013;4:2610. doi: 10.1038/ncomms3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holloway KR, Calhoun TN, Saxena M, Metoyer CF, Kandler EF, Rivera CA, Pruitt K. SIRT1 regulates Dishevelled proteins and promotes transient and constitutive Wnt signaling. Proc Natl Acad Sci U S A. 2010;107:9216–21. doi: 10.1073/pnas.0911325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 34.Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, Jaeschke H, Ding WX. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–50. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 36.Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest. 2011;121:1231–41. doi: 10.1172/JCI44145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welsh CF, Roovers K, Villanueva J, Liu Y, Schwartz MA, Assoian RK. Timing of cyclin D1 expression within G1 phase is controlled by Rho. Nat Cell Biol. 2001;3:950–7. doi: 10.1038/ncb1101-950. [DOI] [PubMed] [Google Scholar]
- 38.Urruticoechea A, Smith IE, Dowsett M. Proliferation marker Ki-67 in early breast cancer. J Clin Oncol. 2005;23:7212–20. doi: 10.1200/JCO.2005.07.501. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Lotze MT, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 40.Ma XH, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, Li LZ, Amaravadi RK. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17:3478–89. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–9. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Kéri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–64. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 44.Maskey D, Yousefi S, Schmid I, Zlobec I, Perren A, Friis R, Simon HU. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. doi: 10.1038/ncomms3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 46.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leone RD, Amaravadi RK. Autophagy: a targetable linchpin of cancer cell metabolism. Trends Endocrinol Metab. 2013;24:209–17. doi: 10.1016/j.tem.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–9. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5:720–2. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- 50.Raveh T, Droguett G, Horwitz MS, DePinho RA, Kimchi A. DAP kinase activates a p19ARF/p53-mediated apoptotic checkpoint to suppress oncogenic transformation. Nat Cell Biol. 2001;3:1–7. doi: 10.1038/35050500. [DOI] [PubMed] [Google Scholar]
- 51.Averous J, Fonseca BD, Proud CG. Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene. 2008;27:1106–13. doi: 10.1038/sj.onc.1210715. [DOI] [PubMed] [Google Scholar]
- 52.Nosho K, Kawasaki T, Chan AT, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS, Ogino S. Cyclin D1 is frequently overexpressed in microsatellite unstable colorectal cancer, independent of CpG island methylator phenotype. Histopathology. 2008;53:588–98. doi: 10.1111/j.1365-2559.2008.03161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 54.Read R, Savelieva K, Baker K, Hansen G, Vogel P. Histopathological and neurological features of Atg4b knockout mice. Vet Pathol. 2011;48:486–94. doi: 10.1177/0300985810375810. [DOI] [PubMed] [Google Scholar]
- 55.Shu CW, Madiraju C, Zhai D, Welsh K, Diaz P, Sergienko E, Sano R, Reed JC. High-throughput fluorescence assay for small-molecule inhibitors of autophagins/Atg4. J Biomol Screen. 2011;16:174–82. doi: 10.1177/1087057110392996. [DOI] [PubMed] [Google Scholar]
- 56.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–65. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.