Abstract
Mitochondrial quality control has an impact on many diseases, but intense research has focused on the action of 2 genes linked to heritable forms of Parkinson disease (PD), PINK1 and PARK2/parkin, which act in a common pathway to promote mitophagy. However, criticism has been raised that little evidence links this mechanism to sporadic PD. To gain a greater insight into the mechanisms of PINK1-PARK2 mediated mitophagy, we undertook a genome-wide RNAi screen in Drosophila and human cell models. Strikingly, we discovered several components of the lipogenesis pathway, including SREBF1, playing a conserved role in mitophagy. Our results suggest that lipids influence the stabilization of PINK1 during the initiation of mitophagy. Importantly, SREBF1 has previously been identified as a risk locus for sporadic PD, and thus implicates aberrant mitophagy as contributing to sporadic PD. Our findings suggest a role for lipid synthesis in PINK1-PARK2 mediated mitophagy, and propose a mechanistic link between familial and sporadic PD, supporting a common etiology.
Keywords: SREBF1, Parkin, FBXW7, Parkinson disease, lipids, Drosophila
Parkinson disease is a neurodegenerative movement disorder in which abnormal mitochondrial homeostasis is a common and consistent pathological feature. Over the past 20 y, genetic analyses of patients with PD have identified several genes whose mutation leads to the development of familial parkinsonism. A number of these genes play a role in maintaining a healthy mitochondrial network, underscoring the importance of mitochondrial homeostasis in PD. While these discoveries have greatly strengthened our understanding of the causes of familial PD, there has been much debate in the field over its relevance to the more common sporadic forms of PD.
Compelling insights have come from studying 2 PD-causing genes with an undisputed role in mitochondrial homeostasis, PINK1 and PARK2, which encode a mitochondrially-targeted serine-threonine kinase and a cytoplasmic E3-ubiquitin ligase, respectively. These genes act in a common pathway, coordinating the removal of failing mitochondria via mitophagy. Substantial evidence supports the idea that following mitochondrial injury PINK1 is stabilized on the outer mitochondrial membrane (OMM) and signals the recruitment of PARK2, whereupon it promiscuously ubiquitinates many mitochondrial proteins, labeling the organelle for destruction. Since its inception this mechanism has proven to be a polarizing idea in the field. Proponents find it an attractive mechanism that can explain several features of the disease, such as accumulation of oxidative damage, elevated mitochondrial DNA mutations, and cell-type selectivity in neurons with extreme demands on energy and calcium regulation. However, critics often retort that there is currently little direct evidence for this mechanism in vivo, and even less linking it to sporadic PD, holding this absence of evidence to be sufficient evidence of absence.
To gain a greater understanding of the mechanism of mitophagy, we performed a genome-wide RNAi screen for modulators of toxin-induced mitochondrial translocation of PARK2. The initial screen was performed in Drosophila S2R+ cells to take advantage of reduced genetic redundancy in this model system and to define conserved mediators of mitophagy. A primary hit group of Drosophila genes were identified that prevent park translocation and other aspects of Pink1-related mitochondrial homeostasis, including alterations in mitochondrial morphology and perinuclear aggregation of mitochondria. To define functionally conserved factors, the homologous human genes were tested for their affects on PARK2 translocation and mitophagy in HeLa cells. This approach produced a hit group of 20 conserved genes that promote mitophagy. Of these, 4 genes are known to form part of the major sterol regulatory element binding protein (SREBP) lipogenesis pathway; SREBF1 and SREBF2, master transcription factors, and FBXW7 and GSK3A, pathway regulators. This strongly suggested a mechanistic link between the synthesis of lipids and mitophagy.
We next performed a series of tests to verify that the observed effects were due to the interference of the lipogenesis pathway. First, we found that a chemical inhibitor of SREBP activation, genistein, reproduces the block in PARK2 translocation seen with SREBF1 siRNA. We also showed that the SREBF1-dependent block in PARK2 translocation can be partially rescued by the addition of exogenous lipids, both fatty acids and cholesterol. These results support a role for the canonical SREBP pathway in PARK2 translocation and suggest that the synthesis of lipids by SREBP target genes is necessary for effective PINK1-PARK2 mediated mitophagy.
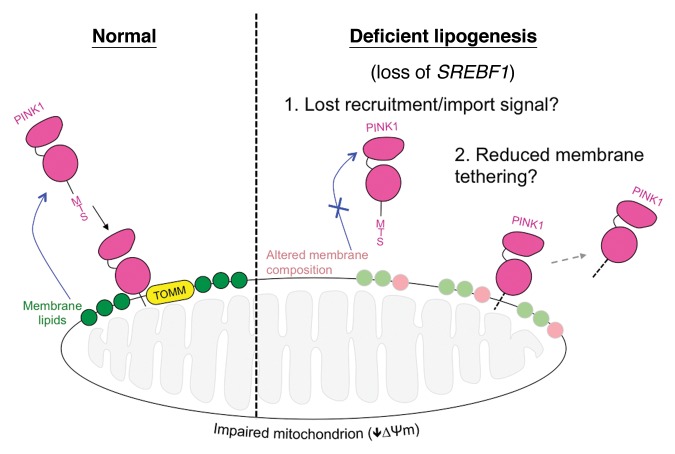
To gain insight into potential mechanisms by which lipids could be affecting mitophagy, we next looked upstream of PARK2 translocation. Following mitochondrial injury full-length PINK1 becomes stabilized on the OMM. Both SREBF1 silencing and genistein treatment lead to a decrease in the stabilization of PINK1. These results suggest that SREBF1 loss may block mitophagy by reducing the degree to which full-length PINK1 is stabilized on the OMM of injured mitochondria. Intriguingly, the exogenous addition of lipids produces a clear trend toward rescued PINK1 stabilization, though none of the conditions reached significance in this assay. Nevertheless, overall our findings suggest that SREBF1-dependent lipid synthesis may be a key factor in PINK1 OMM stabilization (Fig. 1). How alterations in the lipidome cause a change in PINK1 dynamics is not yet understood. It could be that alterations in the mitochondrial membrane lipid composition lead to a weakened association of PINK1 with the OMM. Alternatively, reduced lipid synthesis could deprive mitochondria of signaling events upstream or independent of PINK1 stabilization, such as the externalization of cardiolipin. Clearly, further work is needed clarify these mechanisms.
While this study provides novel insight into how the PINK1-PARK2 pathway coordinates the elimination of dysfunctional mitochondria, the broader significance of this work comes from the finding that SREBF1 has been identified as a risk locus for idiopathic PD in genome-wide association studies. A parsimonious implication of this is that genetic variation at the SREBF1 locus alters the expression of SREBF1, thereby promoting pathogenesis. We hypothesize that this is due to reduced SREBF1 expression inhibiting mitophagy. While our findings are in no way definitive, they provide further circumstantial evidence that defective mitophagy may play a role in the pathology of sporadic PD. Such findings also form a mechanistic bridge between the etiology of familial and sporadic PD, suggesting a common cause.
Figure 1. Proposed model for lipid influence on PINK1 stabilization on dysfunctional mitochondria. Under normal conditions, PINK1 is targeted to the mitochondrion via its mitochondrial targeting sequence (MTS) where it is imported by the translocase of outer membrane (TOMM) complex. Upon reduction in mitochondrial membrane potential (∆Ψm) full-length PINK1’s import is abolished and it becomes stabilized on the outer membrane, whereupon it stimulates mitophagy via the recruitment of PARK2 (not shown). Our data show that loss of the SREBF1 pathway and deficient lipogenesis significantly reduces full-length PINK1 stabilization, inhibiting mitophagy. We hypothesize that the dysregulation of certain lipids or membrane-lipid composition leads to reduced PINK1 stabilization by reducing recruitment or import, or interfering with membrane tethering. Other engulfment signals such as cardiolipin externalization may also be affected.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The original work was funded by a Wellcome/MRC Parkinson’s Disease Consortium grant to UCL/IoN, the University of Sheffield, and the MRC Protein Phosphorylation Unit at the University of Dundee (WT089698), the MEFOPA project funded through the European Union FP7 research program, and an ERC Starting Grant (no. 309742).