Abstract
Nonhomologous end joining (NHEJ) and homologous recombination (HR) represent the two major pathways of DNA double-strand break (DSB) repair in eukaryotic cells. NHEJ repairs DSBs by ligation of cognate broken ends irrespective of homologous flanking sequences, whereas HR repairs DSBs using an undamaged homologous template. Although both NHEJ and HR have been clearly implicated in the maintenance of genome stability, how these apparently independent and mechanistically distinct pathways are coordinated remains largely unexplored. To investigate the relationship between HR and NHEJ modes of DSB repair, we generated cells doubly deficient for the NHEJ factor DNA Ligase IV (Lig4) and the HR factor Rad54. We show that Lig4 and Rad54 cooperate to support cellular proliferation, repair spontaneous DSBs, and prevent chromosome and single chromatid aberrations. These findings demonstrate a role for NHEJ in the repair of DSBs that occur spontaneously during or after DNA replication, and reveal overlapping functions for NHEJ and Rad54-dependent HR in the repair of such DSBs.
Keywords: DNA break repair, nonhomologous end joining, homologous recombination, Lig4, Rad54
Repair of DNA double-strand breaks (DSBs) is a crucial event in the maintenance of genome stability. Unrepaired or misrepaired DSBs can lead to mutations or loss of genetic information and can have significant consequences, such as cellular transformation or cell death. In mammalian cells, DSB repair occurs by at least two distinct pathways: nonhomologous end joining (NHEJ), which can ligate broken DNA ends irrespective of sequence homology, and homologous recombination (HR), which uses homologous sequences on an undamaged template to promote repair. Although both pathways are required for the repair of induced or spontaneous DSBs and for the maintenance of genome stability, it is unclear how pathway choice occurs (Ferguson and Alt 2001; Levitt and Hickson 2002; Thompson and Schild 2002; van den Bosch et al. 2002; Mills et al. 2003). Differential usage through the cell cycle likely represents one level of pathway control, but additional levels of regulation may play a role. Furthermore, although NHEJ is generally thought to predominate in G1, NHEJ and HR activities may also overlap in certain cell cycle phases (Valerie and Povirk 2003). In the latter context, possible interplay between NHEJ and HR remains largely untested.
NHEJ is employed in both the repair of general DSBs and in the repair of site-specific DSBs generated by the lymphoid-specific RAG endonuclease at antigen receptor variable region gene segments during early B and T lymphocyte development (for a review, see Bassing et al. 2002b). There are six known NHEJ factors (for a review, see Lieber et al. 2003). These include the Ku70 and Ku80 DNA end-binding complex (Ku) and the DNA Ligase IV (Lig4) and XRCC4 end-ligation complex, all of which are evolutionarily conserved from yeast through mammals. The remaining two factors, more recently evolved, are the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and the Artemis endonuclease, which also may function in concert. The activities of these proteins have been best characterized in the context of V(D)J recombination. Such studies have indicated that Ku, Lig4, and XRCC4 are required for the basic end-ligation activity that is fundamental to all NHEJ reactions, whereas DNA-PKcs and Artemis show more restricted roles, including processing of DNA ends, such as the hairpin coding ends generated during V(D)J recombination, that cannot be directly ligated by the conserved factors (for reviews, see Jeggo and O'Neill 2002; Lieber et al. 2003).
Inactivating mutations of each of the NHEJ factors have been generated in mice. Murine cells deficient for DNA-PKcs or Artemis show genomic instability and variable ionizing radiation (IR) sensitivity, but no proliferation defects (Gao et al. 1998a; Rooney et al. 2003). Except for severe combined immunodeficiency (SCID), due to defects in V(D)J recombination DNA-PKcs and Artemis deficient mice show no other major developmental phenotypes (Gao et al. 1998a; Taccioli et al. 1998; Rooney et al. 2002, 2003). By contrast, deficiency for Ku, XRCC4, or Lig4 results in more severe phenotypes, including proliferation defects, substantial IR sensitivity, and genomic instability (Nussenzweig et al. 1996; Gu et al. 1997a,b; Barnes et al. 1998; Frank et al. 1998; Gao et al. 1998b). Moreover, mice deficient for these conserved NHEJ factors exhibit substantial apoptosis of newly generated neurons, in addition to a SCID phenotype (Barnes et al. 1998; Frank et al. 1998; Gao et al. 1998b; Gu et al. 2000). Although Ku-deficient mice are viable, XRCC4 and Lig4-deficient mice die before birth, likely as a result of very severe neuronal apoptosis (Gu et al. 1997b; Barnes et al. 1998; Frank et al. 1998; Gao et al. 1998b). In this regard, p53 deficiency alleviates XRCC4- or Lig-4-deficient neuronal apoptosis and embryonic lethality, indicating these phenotypes result from a p53-dependent response to unrepaired DSBs, but does not rescue lymphocyte development, because NHEJ per se is not restored (Frank et al. 2000; Gao et al. 2000). The phenotypic severity of XRCC4- and Lig4-deficient mice is thought to arise, at least in part, because these two factors are absolutely required for NHEJ. Finally, because Ku and DNA-PKcs appear to be involved in processes other than NHEJ, XRCC4 and Lig4 are the factors most specific for NHEJ (Gao et al. 1998a; Sekiguchi et al. 2001).
Like NHEJ, HR is also required for the maintenance of general genome stability, and different subsets of HR reactions probably require different classes of HR factors (for a review, see Valerie et al. 2003). The evolutionarily conserved chromatin remodeling factor, Rad54, is a member of the Rad52 family of HR factors (Ivanov and Haber 1997; Symington 2002; Tan et al. 2003). By contrast to other classes of HR factors, disruption of Rad54 results in relatively mild overall phenotypes. Rad54-deficient embryonic stem (ES) cells exhibit sensitivity to IR, and show a decrease in HR as measured by gene targeting efficiency or DSB-initiated sister chromatid gene conversion (Essers et al. 1997; Dronkert et al. 2000). Rad54-deficient cells also display a decrease in the mitomycin C (MMC)-stimulated sister chromatid exchange rate and a mild susceptibility to spontaneous genomic instability (Dronkert et al. 2000; Jaco et al. 2003). However, Rad54 is not required for cellular viability or proliferation, and Rad54 knockout mice are viable, fertile, not tumor prone, and are proficient V(D)J recombination. These findings indicate that the HR defects seen in Rad54-/- cells are not severe enough to exert significant developmental consequences (Essers et al. 1997). The mild phenotypes associated with Rad54 deficiency may result from functional redundancy with other HR factors or may reflect a role for Rad54 in only a minor subset of total DSB repair. In this context, a significant role for Rad54-mediated recombinational repair may be obscured by the activity of other pathways, such as NHEJ.
To test the hypothesis that a more critical role for mouse Rad54 would be revealed by the absence of NHEJ, Rad54-deficient mice were bred onto a Lig4-deficient background. The Lig4-deficient background was selected because this factor appears most specific (along with XRCC4) to NHEJ. Thus, this breeding allowed us to monitor the role of Rad54 in a background without residual NHEJ activity. We find that Rad54 deficiency, in this context, leads to a significant increase in the occurrence of chromosomal abnormalities, thus revealing a novel role for Rad54. Moreover, the current findings provide evidence for the participation of NHEJ in the repair of spontaneous DSBs incurred during or after DNA replication, and reveal an overlap between NHEJ and Rad54-mediated HR in repairing these breaks.
Results
Rad54 is not required for viability of Lig4 p53 doubly deficient mice
To determine if Rad54 deficiency would compromise the postnatal viability of p53-deficient Lig4-deficient (LP) mice, we generated Rad54 Lig4 p53 triply deficient (RLP) mice by breeding. Because Rad54 mice show no overt developmental defects and are fertile, Rad54-/- deficiency was bred to homozygosity in Lig4 p53 double heterozygotes. These were then intercrossed to generate Rad54 Lig4 p53 triple mutant mice. In total, five viable triple mutant mice were recovered from these crosses demonstrating conclusively that Rad54 is not required to support the pre- or postnatal survival of LP mice (Table 1A). Similar to LP mice, the RLP mice were all smaller than normal littermates and three RLP mice developed pro-B cell lymphomas within 12 wk. This is consistent with previous findings showing that Rad54-/- Ku80-/- p53-/- mice also rapidly develop pro-B cell lymphomas (Difilippantonio et al. 2002). Two RLP mice died prior to analysis, and therefore the presence of lymphoma and cause of death in these mice could not be determined. However, in the three RLP mice that were analyzed, the pro-B cell tumor latency was similar to that in LP mice (Zhu et al. 2002). The molecular characterization of these pro-B cell lymphomas revealed no significant differences between LP double mutant tumors and RLP triple mutant tumors (Zhu et al. 2002).
Table 1.
Genotype frequencies
| A) Liveborn mice | ||||||
|---|---|---|---|---|---|---|
| Genotype | Observed | Expected | Ratio | |||
| p53+/+ | 15 | 6 | 1/16 | |||
| Lig4+/+ | p53+/− | 18 | 12 | 1/8 | ||
| p53−/− | 5 | 6 | 1/16 | |||
| p53+/+ | 10 | 12 | 1/8 | |||
| Rad54−/− | Lig4+/− | p53+/− | 34 | 24 | 1/4 | |
| p53−/− | 8 | 12 | 1/8 | |||
| p53+/+ | 0 | 6 | 1/16 | |||
| Lig4−/− | p53+/− | 2 | 12 | 1/8 | ||
| p53−/− | 5 | 6 | 1/16 | |||
| TOTAL | 97 | |||||
| B) E13.5 embryos | ||||
|---|---|---|---|---|
| Genotype | Observed | Expected | Ratio | |
| Lig4+/+ | 10 | 6 | 1/4 | |
| Rad54−/− | Lig4+/− | 9 | 13 | 1/2 |
| Lig4−/− | 7 | 6 | 1/4 | |
| TOTAL | 26 |
Although loss of Rad54 resulted in no overt developmental consequences in LP mice, we wished to test if Rad54 deficiency would exacerbate the developmental defects associated with Lig4 mutation when p53 was intact. Rad54-/-, Lig4+/-, or Rad54+/- Lig4+/- animals were intercrossed to generate Rad54 Lig4 doubly deficient (RL) embryos, as well as all appropriate control genotypes. Additionally, Rad54+/-Lig4+/-p53+/- (triply heterozygous) mice were intercrossed to generate genotype-matched control embryos that were p53+/+, or p53+/-, or p53-/-. Fetuses were isolated at day 13.5 of embryonic development (E13.5) to determine if RL embryos could be recovered. From all fetuses, genomic DNA was prepared for determination of genotype, mouse embryonic fibroblasts (MEFs) were isolated and cultured for cellular studies (see below), and heads were fixed in Bouin's fixative and prepared for histological analysis. RL embryos (E13.5) were obtained at approximately expected Mendelian ratios, demonstrating that Rad54 deficiency did not grossly alter viability of E13.5 Lig4-/- embryos (Table 1B). One hallmark developmental defect associated with Lig4 deficiency is a high incidence of apoptosis in developing, newly postmitotic neurons as they migrate outward from the cerebral ventricular zone (VZ; Barnes et al. 1998; Frank et al. 1998, 2000). As expected, Lig4-deficient embryos exhibited frequent pycnotic nuclei distributed throughout the VZ and the intermediate zone (IZ; data not shown). With respect to neuronal apoptosis, Lig-/- and RL embryonic brains appeared qualitatively and quantitatively similar (data not shown), demonstrating that Rad54 deficiency does not grossly alter the neuronal defects observed in Lig4-/- embryos.
Rad54 Lig4 double deficiency results in a severe proliferation defect
RL MEFs (and littermate controls) were isolated and cellular proliferation was assayed in culture. Passage-matched, low passage (P0 or P1) cells were seeded at low density, in duplicate, and cell counts determined at 2, 4, 6, 8, 10, or 12 d after plating (Fig. 1A). As expected, wild-type cells exhibited variability in proliferation capability, with one line growing as poorly as Lig4-/- cells. However, all were growth proficient and, on average, more so than any of the mutant cells (Fig. 1A). Rad54-/- cells exhibited no appreciable proliferation defects, either in growth rate or saturation density (Fig. 1A). By contrast, and consistent with previously published results (Frank et al. 1998), Lig4-/- cells were deficient for proliferation and experienced growth cessation at lower cell density relative to wild-type controls (Fig. 1A). RL cells were even more severely compromised for growth, exhibiting minimal expansion over the 12-d culture period (Fig. 1A). This profound defect in proliferation is dependent on intact p53, as RLP MEFs exhibited proliferation potential that was significantly restored relative to RL cells (Fig. 1A).
Figure 1.
Proliferation analysis of Rad54 and Lig4 mutant MEFs. (A) Growth curves for duplicate cultures of MEFs of the genotypes indicated at the right. At day 0, 0.5 × 106 cells were seeded onto each of 12 plates. Cell counts were determined at subsequent 2-d intervals up to 12 d. (B) Cell cycle analysis of untreated MEFs of the indicated genotypes. The G1 and G2 DNA content peaks are indicated below. Percentage of cells in the sub-G1, G1, G2, and hyper-G2 gates are shown.
To determine if the severely growth deficient RL MEFs exhibited altered cell cycle distribution, wild-type, Rad54-/-, Lig4-/-, or RL cells were seeded at low density, cultured for 2 d, and then cells were analyzed by flow cytometry to determine cell cycle distribution (Fig. 1B). Wild-type and Rad54-/- MEFs, which proliferate well, exhibited a large percentage of cells in the G1 phase of the cell cycle (Fig. 1B, top panels). By contrast, Lig4-/- and RL cell populations showed a decrease in the percentage of cells in the G1 phase and a concomitant increase in the percentage of cells in the G2 phase (Fig. 1B, bottom panels). Additionally, Lig4-/- and RL cell populations showed a quantifiable increase in cells with a DNA content greater than G2 and an increase in the percentage of cells with a sub-G1 DNA content, indicative of increased cell death in these genotypes.
High level of unrepaired DSBs in Rad54-/- Lig4-/- cells
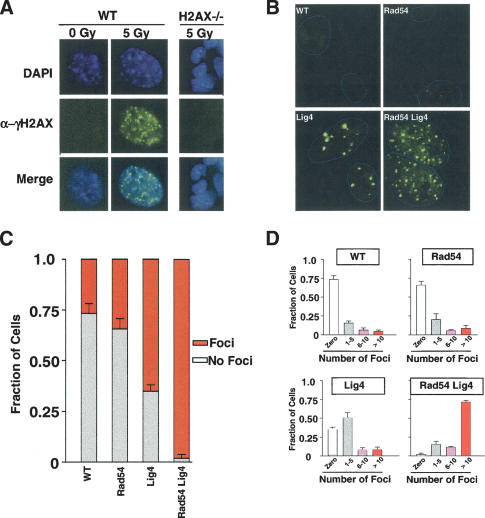
As both Lig4 and Rad54 have been previously characterized as DSB repair factors, one possible explanation for the severe proliferation defect in RL cells is growth arrest or cell cycle delay precipitated by a high level of DNA breakage. Therefore, we wished to assess the load of unrepaired, spontaneously occurring DSBs in RL versus control cells. The histone H2A variant H2AX becomes rapidly phosphorylated following the induction of DSBs and has been used as a marker for the presence of DSBs (Rogakou et al. 1998, 1999). Multiple, independently derived cell lines representing wild-type, Rad54-/-, Lig4-/-, and RL cells were subjected to indirect immunofluorescent detection of phospho-H2AX foci (γ-H2AX). As a control for the detection of DSBs, wild-type MEFs were cultured on cover slips and exposed to 5 Gy of γ irradiation 30 min prior to fixation and staining (Fig. 2A). Cells were counterstained with the DNA-binding dye DAPI, to detect nuclei.
Figure 2.
Immunofluorescent detection of γ-H2AX foci. (A) Wild-type MEFs stained for γ-H2AX foci following either mock treatment (0 Gy) or irradiation (5 Gy). (Upper panels) DAPI staining of nuclei. (Middle panels) γ-H2AX signal. (Bottom panels) Merged signals. Genotypes and irradiations conditions are as indicated. (B) Immunofluorescent detection of γ-H2AX foci in untreated MEFs of the indicated genotypes. Nuclear boundaries, as determined by DAPI staining (not shown), are indicated in blue and digitally overlaid on the FITC images. (C) Percentage of cells exhibiting γ-H2AX foci (red bars) versus no foci (white bars) is shown for each of the indicated genotypes. (D) Percentage of cells exhibiting zero (white bars), 1-5 (gray bars), 6-10 (pink bars), or more than 10 (red bars) foci is shown for each of the indicated deficiencies. All counts represent averages derived from three to five independent experiments.
A small percentage of unirradiated wild-type cells exhibited spontaneously occurring foci, consistent with previous observations (Rogakou et al. 1999; Paull et al. 2000; Ward and Chen 2001). However, the majority of unirradiated wild-type cells showed little detectable γ-H2AX staining (Fig. 2A). By contrast, following 5 Gy of γ irradiation, nearly 100% of wild-type cells exhibited high levels of γ-H2AX foci at 30 min postirradiation (Fig. 2A). As a control for the specificity of the α-γ-H2AX antibody used in this assay, cells deficient for H2AX (Bassing et al. 2002a) were exposed to 5 Gy of irradiation and treated identically to the wild-type cells as described above (Fig. 2A). No spontaneous or IR-induced foci were detected in H2AX-/- cells, thus confirming the antibody specificity (Fig. 2A; data not shown). Next wild-type, Rad54-/-, Lig4-/-, or RL MEFs were analyzed for the presence of spontaneously occurring γ-H2AX foci in unirradiated cells. Like wild-type cells, very few Rad54-/- cells exhibited spontaneous γ-H2AX focus formation (Fig. 2B-D). By contrast, greater than 50% of unirradiated Lig4-/- cells were positive for γ-H2AX foci, and nearly 100% of RL cells exhibited γ-H2AX foci (Fig. 2B,C). Notably, whereas γ-H2AX positive Lig4-/- cells exhibit a small number of foci per cell, RL cells contained numerous foci in every cell examined, indicating a dramatically elevated load of unrepaired, spontaneously occurring DSBs in RL cells, even relative to Lig4 deficient cells (Fig. 2D). Although the high level of spontaneously occurring γ-H2AX foci in RL cells was significantly reduced by additional deficiency for p53 (Supplementary Fig. S1), most RL cells did not express common apoptotic markers (Supplementary Fig. S2A-E) This demonstrates that, in most RL cells, the foci likely represent spontaneously occurring DSBs.
Chromatid instability in Rad54-/-Lig4-/- cells
Although RL cells exhibited high levels of spontaneous γ-H2AX foci, we sought an independent confirmation that RL cells were subject to frequent, spontaneous DNA damage. Therefore we determined if Rad54 deficiency would alter or exacerbate the genomic instability phenotype associated with Lig4 deficiency. As an assessment of genome instability, we scored chromosome abnormalities in DAPI-stained metaphase spreads derived from RL cells and appropriate controls. Metaphase spreads were readily obtained from wild-type, Rad54-/-, and Lig4-/- cells, but owing to the extreme proliferation defect of RL cells, few metaphase chromosomes were recovered, even following extended treatment with the microtubule inhibitor colcemid to enforce metaphase arrest. However, because p53 deficiency rescues proliferation of Lig4-/- or RL cells but does not affect DNA repair activity (Ferguson et al. 2000), we reasoned that actively cycling RLP cells would facilitate the recovery of metaphase chromosomes. Consistent with previously published data for Rad54 deficiency (Essers et al. 1997; Jaco et al. 2003), RP cells showed a low but detectable level of spontaneous instability of both chromosome and chromatid types (Fig. 3A). In contrast, LP cells exhibited a significantly higher rate of chromosome breakage, compared with RP cells (Fig. 3A). Notably, RLP cells displayed a substantial increase in the incidence of fragmented chromosomes relative to LP cells, and, in striking contrast to either RP or LP cells, also incurred a large number of events in which only a single sister chromatid was visibly broken or fragmented (Fig. 3A).
Figure 3.
Cytological examination of Rad54 and Lig4 mutant MEFs. (A) Indicated spontaneously occurring chromosomal anomalies were scored in DAPI-stained metaphase spreads. (P) p53-/-; (RP) Rad54-/- p53-/-; (LP) Lig4-/- p53-/-; (RLP) Rad54-/- Lig4-/- p53-/-. Error bars represent S.E.M. (B) Metaphase spreads from Rad54-/- Lig4-/- p53-/- (RLP). Numbered panels below correspond to magnified DAPI (first row) and SKY (bottom row) of the circled chromosomes above. (I) Broken single chromatid; (II) triradial; (III) complex translocation.
In addition to chromosomal fragmentation, Lig4 deficiency also results in an elevated incidence of spontaneous, random chromosomal translocations (Karanjawala et al. 1999; Ferguson et al. 2000). Because RLP cells exhibit dramatically increased rates of chromosome and chromatid breakage, relative to LP cells, we tested whether Rad54 deficiency would alter the pattern or incidence of chromosomal translocations in LP cells. Spectral karyotype (SKY) analysis was employed to visualize and identify the entire chromosomal complement in metaphase spreads prepared from RP, LP, or RLP cells, and chromosomal abnormalities were scored (Table 2). Numerous complex events, including triradial translocation products and dicentric chromosomes, were observed in metaphase spreads from RLP cells (Fig. 3B; Table 2). Other complex abnormalities included an apparent short arm fusion that produced a dicentric chromosome derived from two different original chromosomes, a complex fusion chromosome with three presumptive centromeres and a putative ring chromosome that may have resulted from a sister chromatid end-fusion event. In addition to fragmented chromosomes, SKY analyses revealed simple acrocentric translocation products as the most evident translocation species in Lig4-/- cells (Table 2).
Table 2.
Spectral karyotype analysis
| Rad54−/− p53−/− | Lig4−/− p53−/− | Rad54−/− Lig4−/− p53−/− | |
|---|---|---|---|
| Metaphases karyotyped | 33 | 32 | 21 |
| Fragmented chromatids and chromosomes | 6 | 10 | 14 |
| Translocations | 1 | 6 | 16 |
| SCa fusions and ring chromosomes | 2 | 1 | 3 |
| Dicentric | 0 | 1 | 8 |
| Tri-radial | 0 | 0 | 2 |
| Metaphases with structural abnormality (%) | 18 | 47 | 81 |
(SC) Single chromatid.
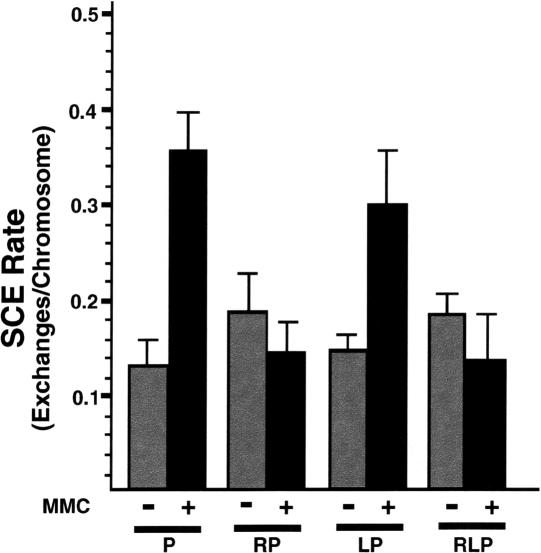
Because loss of Rad54 compromises DSB-stimulated gene conversion using a sister chromatid template and attenuates MMC-stimulated sister chromatid exchange (SCE; Dronkert et al. 2000), we considered the possibility that the elevated genome instability seen in RLP cells resulted from a defect in SCE. Therefore, the frequency of spontaneous and MMC-induced SCE was compared in RLP versus control cells (Fig. 4). MMC-treated cells lacking only p53 (P) showed an approximately threefold increase in the number of SCE events per chromosome scored. Cells lacking Lig4 and p53 (LP) showed an MMC-inducible increase in the SCE rate, whereas cells deficient for Rad54 (RP or RLP) did not exhibit an increase above the spontaneous SCE rate following MMC treatment (Fig. 4). These results are consistent with previously noted effects of MMC in ES cells (Dronkert et al. 2000), confirming that Rad54, but not Lig4, is important for DNA damage-stimulated SCE in MEFs. No differences in the rate of spontaneous SCE were observed between untreated RLP cells and controls (Fig. 4), indicating that the elevated genomic instability in these cells does not stem from an inability to effect SCE in response to spontaneously occurring DSBs.
Figure 4.
Sister chromatid exchange assay in mutant MEFs. SCE rate determined in BrdU-labeled, acridine-orange-stained MEF cell metaphase spreads of the indicated genotypes. (P) p53-/-; (RP) Rad54-/- p53-/-; (LP) Lig4-/- p53-/-; (RLP) Rad54-/- Lig4-/- p53-/-; (P + MMC) p53-/- cultured in the presence of MMC, as a positive control for induced SCE. Error bars represent S.E.M.
Discussion
An important role of Rad54 DSB repair is revealed by NHEJ deficiency
The findings described here uncover a critical role for the HR protein Rad54 in cellular proliferation and the maintenance of genome stability within a setting of deficient NHEJ. Cells simultaneously deficient for both Rad54 and Lig4 fail to proliferate in culture, show a multiplicity of spontaneously occurring DSBs as evidenced by γ-H2AX focus formation, and exhibit high levels of spontaneous cytogenetic abnormalities. Notably, the occurrence of unresolved DSBs and the spontaneous genomic instability in Lig4-/- cells are markedly less, and of a different type (see below), than in RL or RLP cells, indicating that simultaneous Rad54 and Lig4 deficiency produced a synthetic phenotype. Although a minor fraction of γ-H2AX-positive RL cells undergo apoptosis, most RL cells do not express common apoptotic makers, and thus appear to acquire γ-H2AX foci as a result of spontaneously occurring DSBs (Supplementary Fig. S2).
In the DT-40 transformed chicken B cell lymphoma line, which exhibits high rates of HR as measured by gene targeting efficiency, Rad54 disruption alone leads to significant defects in IR resistance, DSB repair, and maintenance of genome stability (Bezzubova et al. 1997; Takata et al. 1998). DT-40 cells doubly deficient for rad54 and ku70 show a relative increase in IR sensitivity but, unlike the RL cells described here, are not impaired for cellular proliferation, possibly owing to a preexisting lack of p53 expression in the DT-40 cell line (Takata et al. 1998; Fukushima et al. 2001). The differences in Rad54 phenotypes between DT-40 cells and mouse cells had previously been interpreted to reflect differences in the relative usage of HR-versus NHEJ-mediated repair. However, we now demonstrate a DSB repair role for murine Rad54 that can be revealed when NHEJ is impaired. Based on the relatively mild phenotypes conferred by Rad54 deficiency, this role was not predicted. Our findings help to reconcile the general DT-40 phenotypes with mouse cell phenotypes by demonstrating that Rad54 can also be important for genome stability in mammalian cells, but that its role is largely masked by NHEJ activities.
Rad54 and Lig4 cooperate to maintain chromatid stability
Cells deficient for both Rad54 and Lig4 incur a very high proportion of single-chromatid breaks and gaps, relative to either deficiency alone, thus demonstrating a defect in postreplication DSB repair. As DNA repair pathway usage varies with cell cycle progression, these defects may arise, in part, because cells in the G2 phase of the cell cycle exhibit a relatively greater dependence on Rad54 for maintenance of genome stability. Although an important role for Rad54 in postreplication DNA repair is not unexpected and may be unmasked by cell cycle delay in G2, an overlapping role for NHEJ in this phase of the cell cycle has been more speculative. One recent report of DNA-PKcs versus XRCC3 in the repair of IR-induced DSBs in CHO cells provided a suggestion that NHEJ can play a role in all phases of the cell cycle (Rothkamm et al. 2003). It has also been proposed that NHEJ can, in some cases, resolve DSBs arising from stalled replication forks, although this has remained controversial (Arnaudeau et al. 2001; Saintigny et al. 2001; Lundin et al. 2002; Rothkamm et al. 2003). We now demonstrate a role for Lig4, and by extension NHEJ, in the repair of DSBs that arise spontaneously during postreplication phases of the cell cycle, although the source of such breaks remains to be determined.
The findings described here also extend those described in a study of the genetic interaction between Lig4 and the DNA-damage-signaling kinase ATM (Sekiguchi et al. 2001). ATM is a mammalian DSB-responsive cell cycle checkpoint factor that participates in G1, S, and G2 checkpoints (Jeggo et al. 1998; Rotman and Shiloh 1999; Abraham 2001), and may have diverse functions in the activation of certain modes of DSB repair, such as HR (Jeggo et al. 1998; Morrison et al. 2000; Abraham 2001; Shiloh 2001; Yang et al. 2003). In the context of mammalian NHEJ, loss of ATM activity can abrogate the embryonic lethality and neuronal apoptosis associated with Lig4 deficiency, probably due to decreased activation of the p53 pathway. However, Lig4-/- ATM-/- MEFs exhibit a substantial proliferation defect relative to either single mutant (Lee et al. 2000; Sekiguchi et al. 2001). Lig4-/- ATM-/- MEFs also show increased rates of genomic instability, including elevated chromatid instability (Sekiguchi et al. 2001). Our findings now provide a mechanistic explanation for the proliferative failure and the genomic instability in Lig4-/- ATM-/- cells. The results described here indicate that the proliferation and genome stability defects in Lig4-/- ATM-/- cells likely result from combined loss of NHEJ and HR, and suggest that these phenotypes arise from loss of ATM functions that are distinct from those responsible for embryonic lethality and neuronal apoptosis of Lig4-/- mice. Consistent with this interpretation, ATM and Rad54 have been placed in the same genetic pathway with respect to repair of IR-induced DNA damage (Morrison et al. 2000), and ATM deficient cells show abnormal kinetics of Rad54 focus formation following IR exposure (Morrison et al. 2000). Finally, studies from a variety of model systems have indicated a link between HR and NHEJ in the repair of single-ended DNA invasion events arising in the context of replication (Gloor and Lankenau 1998; Haber 1999). In this regard, our demonstration that NHEJ overlaps with Rad54-dependent HR may provide a mechanistic basis for the finding, in mammalian cells, that repair of an I-SceI-mediated DSB introduced into chromosomally integrated recombination substrates can proceed by a reaction in which HR and NHEJ are apparently coupled (Richardson and Jasin 2000). In that study, the homology available for HR-mediated recombination was constrained to disfavor recovery of short-tract gene conversion products (Richardson and Jasin 2000). As a result, the residual recombination pathway was initiated by homologous invasion and completed by NHEJ. Our finding of genetic interaction between Rad54 and Lig4 may indicate that such a coupled HR-NHEJ reaction is also relevant for maintenance of genomic stability following spontaneously occurring DNA damage. Furthermore, our results suggest that Rad54 and Lig4 can participate in such a coupled HR-NHEJ reaction.
Rad54 interaction with Lig4 versus DNA-PKcs
Our findings also bear significantly on the interpretation of results previously obtained by crossing Rad54 deficiency onto a DNA-PKcs mutant (SCID) or DNA-PKcs null background (Essers et al. 2000; Jaco et al. 2003). Rad54-deficient SCID mice are developmentally normal, but are extremely sensitive to IR, much more so than either single mutant (Essers et al. 2000). Additionally, Rad54-/- DNA-PKcs-/- double mutant (RD) cells are not significantly different from Rad54 single mutant cells with respect to proliferation or genomic instability (Jaco et al. 2003) In striking contrast to the lack of synergy between Rad54 and DNA-PKcs deficiency, we show a marked proliferative defect, a high level of unrepaired DSBs, and substantial chromatid instability in RL or RLP cells, demonstrating that for DSBs occurring spontaneously during DNA replication, HR and NHEJ do in fact have overlapping functions. Given the mild phenotypes associated with Rad54 inactivation and the lack of synergy between the Rad54 and DNA-PKcs mutations, this was an unanticipated finding.
The differences in RL and RD cellular phenotypes may reflect functional differences between DNA-PKcs and Lig4 in the repair of DSBs arising in S or G2 phases. It is well documented that DNA-PKcs is required for some but not all end-joining activities in mouse (for a review, see Jung and Alt 2004). During the resolution step of V(D)J recombination, DNA-PKcs and Artemis are necessary for coding joint formation, where end processing is required, but are relatively dispensable for ligation of blunt signal ends (for a review, see Jung and Alt 2004). A functional difference in the context of replication-induced breaks is supported by the observation that CHO cells deficient for Ku86 or XRCC4 are hypersensitive to the replication inhibitor aphidicolin (Saintigny et al. 2001), whereas DNA-PKcs-deficient CHO cells are not (Rothkamm et al. 2003). These observations, together with our current findings, support the notion that DNAPKcs and Lig4 act differentially in postreplication DSB repair and indicate an interplay between Rad54 and Lig4, whereas none is apparent between Rad54 and DNAPKcs.
Model for HR-NHEJ interplay
Our observation that both Lig4 and Rad54 play roles in maintaining genome stability in G2 demonstrate that NHEJ and HR pathways can overlap during (post)replication phases of the cell cycle. This is further supported by recent reports concerning the repair of IR or aphidicolin-induced breaks, which have suggested that NHEJ may also participate in the repair of a subset of these exogenously induced post-G1 breaks. The slightly higher overall rate of genomic instability in Lig4, versus Rad54, single mutant cells indicates that Lig4-dependent DSB repair is favored over Rad54-dependent repair, even in postreplication phases of the cell cycle and suggests two possible models (Fig. 5). The first model (Fig. 5A) indicates that Rad54- and Lig4-dependent pathways, although possibly remaining mechanistically separate, may also overlap in the repair of a subset of DSBs arising during or after DNA replication. Such a functional interrelationship would result in a synergistic effect on chromosome or chromatid instability when both pathways are deficient. In addition, Lig4- and Rad54-dependent pathways may directly interact in the repair of some DSBs, possibly by a coupled NHEJ-HR mechanism, although this remains speculative (shaded box in Fig. 5A). A second model (Fig. 5B) holds that Rad54 and Lig4 can both function during postreplication, but remain functionally distinct, with each addressing a subset of overall DSBs. By this model, double deficiency would result in an additive but not synthetic phenotype. We now show clear evidence that loss of Rad54 and Lig4 produces, in addition to a synergistic increase in instability, a synthetic shift toward chromatid breaks, relative to either single mutant. Thus, the findings described here support the first model and indicate an interplay between Rad54 and Lig4 in the repair of postreplication DSBs.
Figure 5.
Model for NHEJ/HR cooperation in genome stability.
Materials and methods
Mouse breeding and cell culture
C57/BL6 mice homozygous for a Rad54-deficient allele (Essers et al. 1997) were crossed with Lig4+/- p53+/- mice to generate Rad54+/- Lig4+/- (RL double heterozygous) or Rad54+/- Lig4+/- p53+/- (RLP triple heterozygous) mice. RL double heterozygous or RLP triple heterozygous mice were backcrosses to Rad54-/- to generated Rad54-/- Lig4+/- or Rad54-/- Lig4+/- p53+/-, respectively, which were subsequently intercrossed to generate either Rad54-/- Lig4-/- (RL) embryos or Rad54-/- Lig4-/- p53-/- (RLP) mice, for further analysis.
Passage 0 (P0) MEFs were obtained from embryos at E13.5 by cell dispersal following removal of organ block tissue. Cells were cultures in DMEM medium supplemented with 10% serum in a humidified 37°C incubator maintained at 5% CO2. To perform growth assays 0.5 × 106 cells were seeded onto 10-cm plates in duplicate. Plates were tryspinized and cell counts determined after 2, 4, 6, 8, 10, or 12 d of culture.
Flow cytometric analysis
For cell cycle analysis, MEFs were seeded at low density onto 6-cm dishes and cultured, as above, to ∼50%-60% confluence. Cells were trypsinized and fixed by addition of cold 70% ethanol. Fixed cells were stained with propidium iodide in PBS + 0.1% Triton X-100 and analyzed for DNA content on a Becton Dickinson FACSCalibur. Cell cycle analysis for each genotype was performed in duplicate.
Immunofluorescence
Immunostaining to detect γ-H2AX foci was performed using a phospho-specific anti-H2AX rabbit polyclonal antibody (Bassing et al. 2002a). Immunostaining to detect cleaved caspase-3 was performed using a rabbit monoclonal antibody (5A1) obtained from Cell Signaling Technology. Prior to immunostaining, cells were cultured on sterilized, gelatinized coverslips. Cells were rinsed with PBS, fixed by incubation for 10 min in 3% formaldehyde/2% glucose, and permeabilized with 0.1% Triton X-100. Fixed, permeabilized cells were incubated with polyclonal rabbit α-γH2AX antibody in 1× PBS/2% fetal calf serum overnight at 4°C. Cells were rinsed with three changes of 1× PBS and incubated for 1 h at room temperature with FITC-conjugated goat α-rabbit secondary antibody. Coverslips were mounted using Vectashield mounting medium with DAPI to visualize nuclear DNA. All images were captured on a Nikon microscope using a black-and-white CCD camera.
Cytological analyses
Spreads of metaphase chromosomes were prepared from MEFs for subsequent cytological analyses. Metaphase spreads were prepared by swelling trypsinized cells in a hypotonic (0.4%) potassium chloride solution for 10 min at 37°C. Swelled cells were fixed by two changes of 3:1 methanol/acetic acid, dropped onto microscopy slides, passed through steam, and air dried. For quantification of chromosome and chromatid breakage, metaphase spreads were covered with Vectashield mounting medium containing DAPI and imaged using a Nikon microscope and black and white CCD camera. All images were analyzed in blind fashion to eliminate counting bias. For SKY analysis, metaphase spreads were prepared exactly as for DAPI staining. SKY probe hybridization and analysis were performed as previously described (Ferguson et al. 2000).
Sister chromatid exchange
Cycling cells were labeled for 36 h with BrdU, metaphase spreads were prepared, and slides were stained with acridine orange to visualize differentially labeled chromatids. All assays were performed in triplicate, and SCE rate was determined as a function of the total number of chromosomes. All scoring was performed blind to eliminate counting bias.
Acknowledgments
We thank Chrystelle Couedel and Maria Jasin for thoughtful discussions and sharing of unpublished results. We also thank Meagan Gleason, Eric Pinaud, Sean Rooney, and Craig Bassing for critical reading, provocative suggestions, and graphic art assistance. This work was supported by NIH grants (AI35714, AI20047, and AI31541). F.W.A. is an Investigator of the Howard Hughes Medical Institutes. K.D.M. was supported by the Irvington Institute for Immunology and by the Lymphoma Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1204304.
References
- Abraham, R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 15: 2177-2196. [DOI] [PubMed] [Google Scholar]
- Arnaudeau, C., Lundin, C., and Helleday, T. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307: 1235-1245. [DOI] [PubMed] [Google Scholar]
- Barnes, D.E., Stamp, G., Rosewell, I., Denzel, A., and Lindahl, T. 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8: 1395-1398. [DOI] [PubMed] [Google Scholar]
- Bassing, C.H., Chua, K.F., Sekiguchi, J., Suh, H., Whitlow, S.R., Fleming, J.C., Monroe, B.C., Ciccone, D.N., Yan, C., Vlasakova, K., et al. 2002a. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. 99: 8173-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing, C.H., Swat, W., and Alt, F.W. 2002b. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109: S45-S55. [DOI] [PubMed] [Google Scholar]
- Bezzubova, O., Silbergleit, A., Yamaguchi-Iwai, Y., Takeda, S., and Buerstedde, J.M. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54-/- mutant of the chicken DT40 cell line. Cell 89: 185-193. [DOI] [PubMed] [Google Scholar]
- Difilippantonio, M.J., Petersen, S., Chen, H.T., Johnson, R., Jasin, M., Kanaar, R., Ried, T., and Nussenzweig, A. 2002. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196: 469-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert, M.L., Beverloo, H.B., Johnson, R.D., Hoeijmakers, J.H., Jasin, M., and Kanaar, R. 2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20: 3147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, J., Hendriks, R.W., Swagemakers, S.M., Troelstra, C., de Wit, J., Bootsma, D., Hoeijmakers, J.H., and Kanaar, R. 1997. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89: 195-204. [DOI] [PubMed] [Google Scholar]
- Essers, J., van Steeg, H., de Wit, J., Swagemakers, S.M., Vermeij, M., Hoeijmakers, J.H., and Kanaar, R. 2000. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 19: 1703-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, D.O. and Alt, F.W. 2001. DNA double strand break repair and chromosomal translocation: Lessons from animal models. Oncogene 20: 5572-5579. [DOI] [PubMed] [Google Scholar]
- Ferguson, D.O., Sekiguchi, J.M., Chang, S., Frank, K.M., Gao, Y., DePinho, R.A., and Alt, F.W. 2000. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. 97: 6630-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, K.M., Sekiguchi, J.M., Seidl, K.J., Swat, W., Rathbun, G.A., Cheng, H.L., Davidson, L., Kangaloo, L., and Alt, F.W. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396: 173-177. [DOI] [PubMed] [Google Scholar]
- Frank, K.M., Sharpless, N.E., Gao, Y., Sekiguchi, J.M., Ferguson, D.O., Zhu, C., Manis, J.P., Horner, J., DePinho, R.A., and Alt, F.W. 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 5: 993-1002. [DOI] [PubMed] [Google Scholar]
- Fukushima, T., Takata, M., Morrison, C., Araki, R., Fujimori, A., Abe, M., Tatsumi, K., Jasin, M., Dhar, P.K., Sonoda, E., et al. 2001. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 276: 44413-44418. [DOI] [PubMed] [Google Scholar]
- Gao, Y., Chaudhuri, J., Zhu, C., Davidson, L., Weaver, D.T., and Alt, F.W. 1998a. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity 9: 367-376. [DOI] [PubMed] [Google Scholar]
- Gao, Y., Sun, Y., Frank, K.M., Dikkes, P., Fujiwara, Y., Seidl, K.J., Sekiguchi, J.M., Rathbun, G.A., Swat, W., Wang, J., et al. 1998b. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95: 891-902. [DOI] [PubMed] [Google Scholar]
- Gao, Y., Ferguson, D.O., Xie, W., Manis, J., Sekiguchi, J., Frank, K. M., Chaudhuri, J., Horner, J., DePinho, R.A., and Alt, F.W. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404: 897-900. [DOI] [PubMed] [Google Scholar]
- Gloor, G.B. and Lankenau, D.H. 1998. Gene conversion in mitotically dividing cells: A view from Drosophila. Trends Genet. 14: 43-46. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Jin, S., Gao, Y., Weaver, D.T., and Alt, F.W. 1997a. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. 94: 8076-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Seidl, K.J., Rathbun, G.A., Zhu, C., Manis, J.P., van der Stoep, N., Davidson, L., Cheng, H.L., Sekiguchi, J.M., Frank, K., et al. 1997b. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7: 653-665. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Sekiguchi, J., Gao, Y., Dikkes, P., Frank, K., Ferguson, D., Hasty, P., Chun, J., and Alt, F.W. 2000. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc. Natl. Acad. Sci. 97: 2668-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J.E. 1999. DNA recombination: The replication connection. Trends Biochem. Sci. 24: 271-275. [DOI] [PubMed] [Google Scholar]
- Ivanov, E.L. and Haber, J.E. 1997. DNA repair: RAD alert. Curr. Biol. 7: R492-R495. [DOI] [PubMed] [Google Scholar]
- Jaco, I., Munoz, P., Goytisolo, F., Wesoly, J., Bailey, S., Taccioli, G., and Blasco, M.A. 2003. Role of mammalian Rad54 in telomere length maintenance. Mol. Cell. Biol. 23: 5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo, P. and O'Neill, P. 2002. The Greek goddess, Artemis, reveals the secrets of her cleavage. DNA Repair (Amst.) 1: 771-777. [DOI] [PubMed] [Google Scholar]
- Jeggo, P.A., Carr, A.M., and Lehmann, A.R. 1998. Splitting the ATM: Distinct repair and checkpoint defects in ataxia-telangiectasia. Trends Genet. 14: 312-316. [DOI] [PubMed] [Google Scholar]
- Jung, D. and Alt, F.W. 2004. Unraveling V(D)J recombination: Insights into gene regulation. Cell 116: 299-311. [DOI] [PubMed] [Google Scholar]
- Karanjawala, Z.E., Grawunder, U., Hsieh, C.L., and Lieber, M.R. 1999. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 9: 1501-1504. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Barnes, D.E., Lindahl, T., and McKinnon, P.J. 2000. Defective neurogenesis resulting from DNA ligase IV deficiency requires Atm. Genes & Dev. 14: 2576-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt, N.C., and Hickson, I.D. 2002. Caretaker tumour suppressor genes that defend genome integrity. Trends Mol. Med. 8: 179-186. [DOI] [PubMed] [Google Scholar]
- Lieber, M.R., Ma, Y., Pannicke, U., and Schwarz, K. 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 4: 712-720. [DOI] [PubMed] [Google Scholar]
- Lundin, C., Erixon, K., Arnaudeau, C., Schultz, N., Jenssen, D., Meuth, M., and Helleday, T. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 22: 5869-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, K.D., Ferguson, D.O., and Alt, F.W. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194: 77-95. [DOI] [PubMed] [Google Scholar]
- Morrison, C., Sonoda, E., Takao, N., Shinohara, A., Yamamoto, K., and Takeda, S. 2000. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 19: 463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig, A., Chen, C., da Costa Soares, V., Sanchez, M., Sokol, K., Nussenzweig, M.C., and Li, G.C. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382: 551-555. [DOI] [PubMed] [Google Scholar]
- Paull, T.T., Rogakou, E.P., Yamazaki, V., Kirchgessner, C.U., Gellert, M., and Bonner, W.M. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10: 886-895. [DOI] [PubMed] [Google Scholar]
- Richardson, C. and Jasin, M. 2000. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 20: 9068-9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou, E.P., Pilch, D.R., Orr, A.H., Ivanova, V.S., and Bonner, W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273: 5858-5868. [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., Boon, C., Redon, C., and Bonner, W.M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146: 905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, S., Sekiguchi, J., Zhu, C., Cheng, H.L., Manis, J., Whitlow, S., DeVido, J., Foy, D., Chaudhuri, J., Lombard, D., et al. 2002. Leaky scid phenotype associated with defective v(d)j coding end processing in artemis-deficient mice. Mol. Cell 10: 1379-1390. [DOI] [PubMed] [Google Scholar]
- Rooney, S., Alt, F.W., Lombard, D., Whitlow, S., Eckersdorff, M., Fleming, J., Fugmann, S., Ferguson, D.O., Schatz, D.G., and Sekiguchi, J. 2003. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 197: 553-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm, K., Kruger, I., Thompson, L.H., and Lobrich, M. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell Biol. 23: 5706-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman, G. and Shiloh, Y. 1999. ATM: A mediator of multiple responses to genotoxic stress. Oncogene 18: 6135-6144. [DOI] [PubMed] [Google Scholar]
- Saintigny, Y., Delacote, F., Vares, G., Petitot, F., Lambert, S., Averbeck, D., and Lopez, B.S. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20: 3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi, J., Ferguson, D.O., Chen, H.T., Yang, E.M., Earle, J., Frank, K., Whitlow, S., Gu, Y., Xu, Y., Nussenzweig, A., et al. 2001. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl. Acad. Sci. 98: 3243-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh, Y. 2001. ATM and ATR: Networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11: 71-77. [DOI] [PubMed] [Google Scholar]
- Symington, L.S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli, G.E., Amatucci, A.G., Beamish, H.J., Gell, D., Xiang, X.H., Torres Arzayus, M.I., Priestley, A., Jackson, S.P., Marshak Rothstein, A., Jeggo, P.A., et al. 1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 9: 355-366. [DOI] [PubMed] [Google Scholar]
- Takata, M., Sasaki, M.S., Sonoda, E., Morrison, C., Hashimoto, M., Utsumi, H., Yamaguchi-Iwai, Y., Shinohara, A., and Takeda, S. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17: 5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, T.L., Kanaar, R., and Wyman, C. 2003. Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amst.) 2: 787-794. [DOI] [PubMed] [Google Scholar]
- Thompson, L.H. and Schild, D. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509: 49-78. [DOI] [PubMed] [Google Scholar]
- Valerie, K. and Povirk, L.F. 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22: 5792-5812. [DOI] [PubMed] [Google Scholar]
- van den Bosch, M., Lohman, P.H., and Pastink, A. 2002. DNA double-strand break repair by homologous recombination. Biol. Chem. 383: 873-892. [DOI] [PubMed] [Google Scholar]
- Ward, I.M. and Chen, J. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276: 47759-47762. [DOI] [PubMed] [Google Scholar]
- Yang, J., Yu, Y., Hamrick, H.E., and Duerksen-Hughes, P.J. 2003. ATM, ATR and DNA-PK: Initiators of the cellular genotoxic stress responses. Carcinogenesis 24: 1571-1580. [DOI] [PubMed] [Google Scholar]
- Zhu, C., Mills, K.D., Ferguson, D.O., Lee, C., Manis, J., Fleming, J., Gao, Y., Morton, C.C., and Alt, F.W. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109: 811-821. [DOI] [PubMed] [Google Scholar]