Abstract
Asymmetric cell divisions combine cell division with fate specification and one general model of how this is achieved was proposed already decades ago1,2: During interphase, the cell polarity axis is specified, followed by orientation of the spindle along the polarity axis and segregation of fate determinants along the polarity axis during mitosis. In most cells, the polarity axis and the spindle will usually align with the long axis that the cell had before division, also called Hertwig’s rule3–6. In the C. elegans embryo, the first polarity axis also forms along the long axis of the embryo by enrichment of myosin in the anterior7 and formation of mutually exclusive anterior and posterior cortical polarity domains, mediated through directional cortical contractile flow8–10. The directionality of this flow is determined by an extrinsic cue, the entry of the sperm, which inhibits Rho-dependent myosin activation at the future posterior pole by bringing with it the Rho GTPase activating protein CYK-411,12. Moreover, since there is no previous division ‘history’ before the first cleavage, mechanisms have to ensure that the nucleus-centrosome complex undergoes a 90 degree rotation so that the spindle can subsequently elongate along the long axis13–15. Additional mechanisms ensure that the site of cleavage is perpendicular to the long axis16,17. Hence, tight coupling of an extrinsic cue to intrinsic polarity formation and spindle elongation enables alignment of the division orientation with the long axis of the organism and successful segregation of fate determinants.
Keywords: midbody, asymmetry, axis formation, patterning, development, C. elegans, cytokinesis
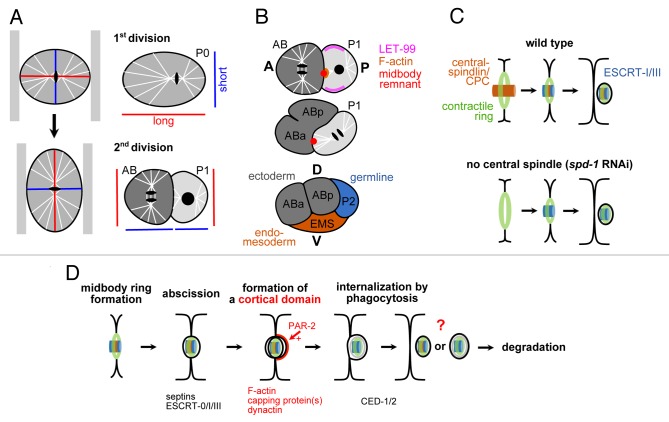
The key event in dorsoventral axis formation during the next division in the C. elegans embryo is a similar 90 degree rotation of the nucleus-centrosome complex during prophase in the germline blastomere onto the anteroposterior axis.14 In contrast to the first division, this rotation is an alignment along the short axis of the cell, a violation of Hertwig’s rule (Fig. 1A). Our recent work18 suggests that a combination of two previously proposed models might best explain the interplay between extrinsic and intrinsic polarity cues necessary for this violation: The 'cortical-site model' suggested the existence of a special landmark at the cell membrane, the midbody remnant of the first division, which combined with the spatial constraints of the oblong egg, directs nucleus-centrosome complex rotation.19-22 The midbody remnant is an organelle that forms at the end of cytoplasmic division when the fully constricted actomyosin furrow embraces the condensed material of the spindle midzone.23 The role of the midbody remnant has been questioned by a differing model, in which the cortical protein LET-99, by its unique lateral localization pattern, reduces cortical pulling forces in two cortical domains along the anteroposterior axis, thereby leading to spindle orientation violating the geometric rule.24-26 Our findings unify these two models by showing that although LET-99 will bias spindle rotation onto the anteroposterior axis, the spindle needs to be tethered to the midbody remnant in order to become skewed ventrally during elongation (Fig. 1B). We find that tethering of the spindle to the midbody remnant requires formation of a transient, cortical, actin-rich structure at the site of the midbody remnant which depends on the germline blastomere having posterior polarity. This tethering function of the midbody remnant does not require the core of the midbody remnant (the condensed ‘remainder’ of the central spindle), neither does it require abscission or midbody remnant internalization. Interestingly, it has been shown that the midbody ring component of the midbody (the outer ‘shell’ derived from the actomyosin ring) is sufficient to recruit the molecular machinery that will mediate cytokinetic abscission27 (Fig. 1C). Additionally, a recent report shows that midbody remnant internalization during C. elegans embryogenesis is not required for patterning, morphogenesis or development28 and most remnants seem to simply represent ‘junk’ after abscission since they become cleared by the phagocytosis/engulfment machinery that usually takes care of cell corpses.28,29
Figure 1. A. Hertwig’s rule and its violation during the division of the P1 blastomere in C. elegans. Left: Exerting a compressive force along the long axis of a cell can transform this axis into the short axis and the spindle will re-orient along the new long axis. Right: Long and short cell axes during the first two cleavages in the C. elegans embryo. Note that all cells except P1 obey Hertwig’s rule. B. Function of the midbody remnant during dorsoventral axis formation in C. elegans. See text for details. A = anterior; P = posterior; D = dorsal; V = ventral. C. Midbody remnant internalization does not require the central spindle-derived part of the midbody. Schematic adapted from ref. 27. CPC = chromosomal passenger complex; ESCRT = endosomal sorting complex required for transport. D. Hypothetical scheme for a midbody remnant ‘pathway’ in C. elegans early embryogenesis. Bottom: Protein factors required for the respective step. See text for details.
Taking together these findings27,28 and our observations that midbody remnants are internalized by neighboring cells with low cortical tension that did not participate in the division that gave rise to the respective midbody remnant,18 allows us to propose the following sequence of processes for the C. elegans embryo (Fig. 1D): (1) depending on cell polarity, nascent midbody remnants can organize asymmetric cortical domains; (2) abscission seems to occur on both sides of the midbody remnant and only requires the midbody ring component; (3) remnants are internalized by cells in a stereotyped fashion and are subsequently degraded; (3) the internalization mechanism is phagocytosis/engulfment; (4) the stereotypy is probably explained by the fact that endocytosis and phagocytosis is facilitated in cells with low cortical tension.30
Although a common picture for midbody remnant inheritance seems to emerge from these recent studies of C. elegans embryogenesis, several mechanisms have been discussed in other systems.31-33 Recent studies in Drosophila seem to reconcile apparent conflicts by suggesting that inheritance regulation and developmental functions of midbody remnants seem to depend on tissue and cell type, cell polarity, regulation of centrosome duplication and inheritance, as well as on the extracellular environment.34-37 These findings very strongly disfavor a uniform inheritance mechanism for midbody remnants and probably also a uniform function in development. One main reason for this is that cytokinesis itself has dramatically different outcomes depending on the cellular and developmental context, e.g., instead of undergoing abscission, cells can also stabilize and remodel their midbodies to form long-lasting intercellular bridges. Thus, although there is ample evidence for developmental functions of cytokinesis, these functions have not been systematically analyzed and linked to alternative fates and functions of the midbody remnant. It will therefore be necessary to investigate how developmental programs differentially regulate cytokinesis and modulate specific processes such as trafficking to the midbody, abscission, phagocytosis, and regulation of cortical dynamics to obtain a clearer picture of midbody remnant functions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
The authors would like to acknowledge funding by the Cluster of Excellence ‘Macromolecular Complexes in Action’ (Deutsche Forschungsgemeinschaft project EXC115) and by the EU Framework Program 7 Marie Curie Actions (Project 326632) to CP.
References
- 1.Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992;68:237–55. doi: 10.1016/0092-8674(92)90468-R. [DOI] [PubMed] [Google Scholar]
- 2.Rhyu MS, Knoblich JA. Spindle orientation and asymmetric cell fate. Cell. 1995;82:523–6. doi: 10.1016/0092-8674(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 3.Hertwig O. Über den Werth der ersten Furchungszellen für die Organbildung des Embryos. Experimentelle Studien am Frosch und Tritonei. Archiv für mikroskopische Anatomie 1893; 42:662–807; doi:10.1007/BF02976796
- 4.Théry M, Racine V, Pépin A, Piel M, Chen Y, Sibarita JB, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–53. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 5.Théry M, Jiménez-Dalmaroni A, Racine V, Bornens M, Jülicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–6. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- 6.Gibson WT, Veldhuis JH, Rubinstein B, Cartwright HN, Perrimon N, Brodland GW, Nagpal R, Gibson MC. Control of the mitotic cleavage plane by local epithelial topology. Cell. 2011;144:427–38. doi: 10.1016/j.cell.2010.12.035. doi [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng NN, Kirby CM, Kemphues KJ. Control of cleavage spindle orientation in Caenorhabditis elegans: the role of the genes par-2 and par-3. Genetics. 1995;139:549–59. doi: 10.1093/genetics/139.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hird SN, White JG. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol. 1993;121:1343–55. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–24. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Mayer M, Depken M, Bois JS, Jülicher F, Grill SW. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature. 2010;467:617–21. doi: 10.1038/nature09376. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. Development. 1996;122:1467–74. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- 13.Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- 14.Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J Cell Biol. 1987;105:2123–35. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gönczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, Schnabel R. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol. 1999;144:927–46. doi: 10.1083/jcb.144.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–4. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- 17.Schenk C, Bringmann H, Hyman AA, Cowan CR. Cortical domain correction repositions the polarity boundary to match the cytokinesis furrow in C. elegans embryos. Development. 2010;137:1743–53. doi: 10.1242/dev.040436. doi [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh D, Pohl C. Coupling of rotational cortical flow, asymmetric midbody positioning, and spindle rotation mediates dorsoventral axis formation in C. elegans. Dev Cell. 2014;28:253–67. doi: 10.1016/j.devcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–93. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waddle JA, Cooper JA, Waterston RH. Transient localized accumulation of actin in Caenorhabditis elegans blastomeres with oriented asymmetric divisions. Development. 1994;120:2317–28. doi: 10.1242/dev.120.8.2317. [DOI] [PubMed] [Google Scholar]
- 21.Keating HH, White JG. Centrosome dynamics in early embryos of Caenorhabditis elegans. J Cell Sci. 1998;111:3027–33. doi: 10.1242/jcs.111.20.3027. [DOI] [PubMed] [Google Scholar]
- 22.Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr Biol. 1998;8:1110–6. doi: 10.1016/S0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- 24.Rose LS, Kemphues K. The let-99 gene is required for proper spindle orientation during cleavage of the C. elegans embryo. Development. 1998;125:1337–46. doi: 10.1242/dev.125.7.1337. [DOI] [PubMed] [Google Scholar]
- 25.Tsou MF, Hayashi A, DeBella LR, McGrath G, Rose LS. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development. 2002;129:4469–81. doi: 10.1242/dev.129.19.4469. [DOI] [PubMed] [Google Scholar]
- 26.Tsou MF, Ku W, Hayashi A, Rose LS. PAR-dependent and geometry-dependent mechanisms of spindle positioning. J Cell Biol. 2003;160:845–55. doi: 10.1083/jcb.200209079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green RA, Mayers JR, Wang S, Lewellyn L, Desai A, Audhya A, Oegema K. The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules. J Cell Biol. 2013;203:505–20. doi: 10.1083/jcb.201306036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chai Y, Tian D, Yang Y, Feng G, Cheng Z, Li W, Ou G. Apoptotic regulators promote cytokinetic midbody degradation in C. elegans. J Cell Biol. 2012;199:1047–55. doi: 10.1083/jcb.201209050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou G, Gentili C, Gönczy P. Stereotyped distribution of midbody remnants in early C. elegans embryos requires cell death genes and is dispensable for development. Cell Res. 2014;24:251–3. doi: 10.1038/cr.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur S, Fielding AB, Gassner G, Carter NJ, Royle SJ. An unmet actin requirement explains the mitotic inhibition of clathrin-mediated endocytosis. Elife. 2014;3:e00829. doi: 10.7554/eLife.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol 2009; 11:65-70; 10.1038/ncb1813.PMID: 19079246. [DOI] [PubMed]
- 32.Ettinger AW, Wilsch-Bräuninger M, Marzesco AM, Bickle M, Lohmann A, Maliga Z, Karbanová J, Corbeil D, Hyman AA, Huttner WB. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat Commun. 2011;2:503. doi: 10.1038/ncomms1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo TC, Chen CT, Baron D, Onder TT, Loewer S, Almeida S, Weismann CM, Xu P, Houghton JM, Gao FB, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–23. doi: 10.1038/ncb2332. doi [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollarolo G, Schulz JG, Munck S, Dotti CG. Cytokinesis remnants define first neuronal asymmetry in vivo. Nat Neurosci. 2011;14:1525–33. doi: 10.1038/nn.2976. [DOI] [PubMed] [Google Scholar]
- 35.Herszterg S, Leibfried A, Bosveld F, Martin C, Bellaiche Y. Interplay between the dividing cell and its neighbors regulates adherens junction formation during cytokinesis in epithelial tissue. Dev Cell. 2013;24:256–70. doi: 10.1016/j.devcel.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Morais-de-Sá E, Sunkel C. Adherens junctions determine the apical position of the midbody during follicular epithelial cell division. EMBO Rep. 2013;14:696–703. doi: 10.1038/embor.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzmann V, Chen C, Chiang CY, Tiyaboonchai A, Mayer M, Yamashita YM. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol Biol Cell. 2014;25:267–75. doi: 10.1091/mbc.E13-09-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]