Abstract
Recent in vitro studies have suggested that autophagy may play a role in both HIV-1 replication and disease progression. In this study we investigated whether autophagy protects the small proportion of HIV-1 infected individuals who remain clinically stable for years in the absence of antiretroviral therapy, these named long-term nonprogressors (LTNP) and elite controllers (EC). We found that peripheral blood mononuclear cells (PBMC) of the HIV-1 controllers present a significantly higher amount of autophagic vesicles associated with an increased expression of autophagic markers with respect to normal progressors. Of note, ex vivo treatment of PBMC from the HIV-1 controllers with the MTOR inhibitor rapamycin results in a more efficient autophagic response, leading to a reduced viral production. These data lead us to propose that autophagy contributes to limiting viral pathogenesis in HIV-1 controllers by targeting viral components for degradation.
Keywords: HIV-1, autophagy, long-term nonprogressors, elite controllers, cell death, AMBRA1, BECN1, ATG5
Introduction
In spite of significant improvements achieved with the introduction of combination antiretroviral drug therapy for the management of HIV-1 infection, we are still far from being able to prevent infection or to eradicate the virus from its reservoirs.1,2 Infection by HIV-1 is associated with a progressive decrease in CD4 T cell numbers and the consequent collapse of host immune defenses. However, a small proportion of infected individuals (5% to 15%) remain asymptomatic for more than 10 y (long-term nonprogressors) and a subset of these individuals (elite controllers) are able to maintain the viral load below the limits of detection in the absence of any antiretroviral therapy.3,4 The mechanism of viral containment in these HIV-1 controllers is unknown. So far, a limited number of studies involving a relative low number of subjects have shown that in these peculiar HIV-1 controllers (LTNP and EC) the slow-progression state is usually not driven by virus gross genetic defects but is frequently determined by the host's genetic factors permitting robust cell-mediated immunity able to control viral replication and reservoir generation.5-7 Recent findings suggest that autophagy, a key physiological process for eukaryotic cell homeostasis, also represents an essential mechanism in controlling viral infections.8-10 Autophagy encompasses various pathways by which cytoplasmic material, including soluble macromolecules and organelles, is delivered to lysosomes for degradation. Autophagy is initiated by signaling pathways centered around ULK1 (unc-51 like autophagy activating kinase 1) and ULK2, as well as the BECN1/Beclin 1-PIK3C3 (phosphatidylinositol 3-kinase, catalytic subunit type 3) complex. The activity of the BECN1-PIK3C3 complex is regulated by the interaction of a series of cofactors such as UVRAG (UV radiation-resistance-associated) ATG14 (autophagy-related 14), AMBRA1 (autophagy/Beclin 1 regulator 1) and KIAA0226.11 Autophagy also plays an important role for cell survival under stressful conditions induced by the accumulation of mutated/misfolded aggregated proteins and/or damaged organelles.12-15 The immune system utilizes autophagy to eliminate intracellular pathogens and to regulate adaptive immunity.16,17 Several viruses have evolved molecular mechanisms to evade this process and even to profit from it.10,18 Recently, the physical interaction of HIV-1 proteins and poly-proteins with the host's proteome has been described.19,20 The binding of HIV-1 Vif protein with various proteins involved in the regulation of autophagy has been described,21 including MAP1LC3A (microtubule-associated protein 1 light chain 3 α) SQSTM1 (sequestosome 1), and AMBRA1, an essential factor of the autophagic core machinery identified by our group.22,23 Moreover, large-scale siRNA screening studies of the host cell factors required for HIV-1 replication have identified several autophagic factors.24
It is increasingly evident that autophagy plays a role both in HIV-1 replication and disease progression.18,25-27 In the early phase of infection, HIV-1 is able to modulate autophagy to maximize virus production, thus playing an important role in the HIV-1 replication.28,29 It has been proposed that, during the early nondegradative phases, autophagy promotes HIV-1 replication. However, when autophagy progresses through maturation phases, the HIV-1 protein Nef acts as an anti-autophagic maturation factor through its interaction with BECN1, thus preventing HIV-1 from degradation.9,10 Starting from these observations, we hypothesized a protective role for autophagy in the control of HIV-1 infection in LTNP and EC. To verify this hypothesis, we compared the level of autophagy and its possible modulation in HIV-1-infected normal progressors patients (NP) and in HIV-1 controllers, namely LTNP and EC.
Results
Enhanced autophagy levels in LTNP and EC PBMC
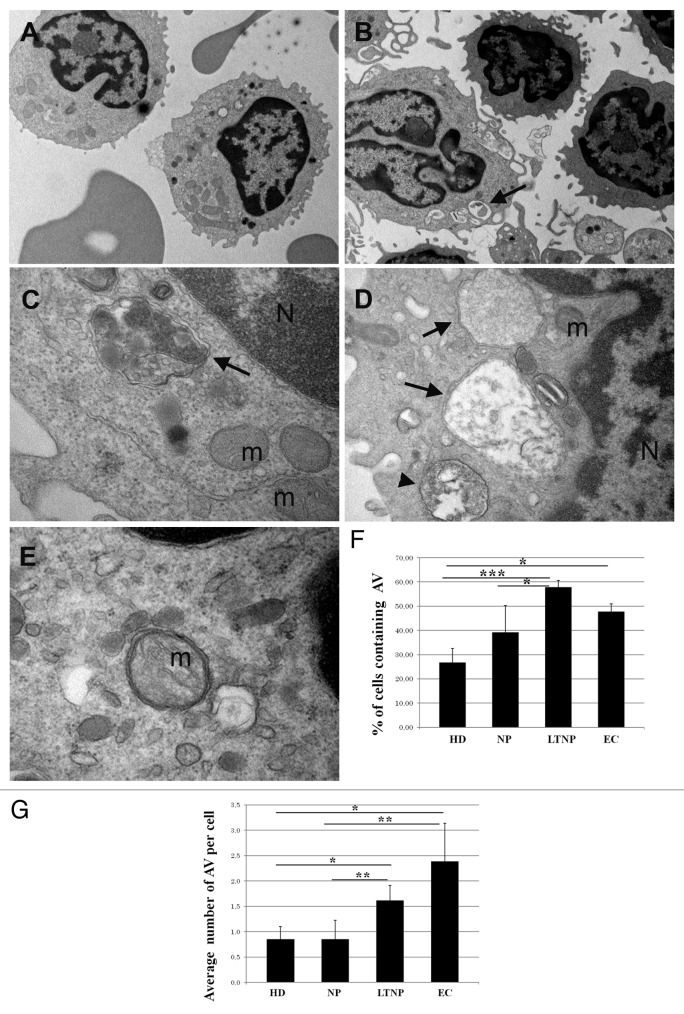
To gain an insight into the involvement of autophagy in HIV-1-induced pathogenesis, we first analyzed freshly isolated PBMC from HIV-1-infected patients and HD at the ultrastructural level.30 AV were clearly identified as a double-membrane structure containing undigested cytoplasmatic material (Fig. 1A–E). We also evaluated the presence of autolysosomes, generated by the fusion of the autophagic vacuoles with the lysosomes, which are limited by a single membrane and are indicative of active autophagy. AV containing undigested material surrounded by a double membrane were observed in HIV-1-infected patients (Fig. 1B–E). The quantitative analyses, aimed at evaluating possible differences between the HIV-1-infected patient categories, revealed that the PBMC from LTNP and EC displayed a significantly higher percentage of cells containing AV with the respect to NP (P < 0.05) (Fig. 1F). By analyzing the average number of AV per cell, we confirmed the significant increase observed between NP and LTNP (P < 0.01) (Fig. 1G).
Figure 1. Ultrastructural analysis of PBMC from HIV-infected patients. (A) PBMC from NP, (B–E) from LTNP. Cells from HIV-1 controllers showed numerous AV containing undigested material, surrounded by a double membrane (arrows) or limited by a single membrane (arrowhead). N, nucleus; m, mitochondria. (F) Quantitative analysis of the percentage of cells containing AV in HD, NP, LTNP, and EC. (G) Quantitative analysis of the average number of AV per cell in HD, NP, LTNP, and EC. Original magnification: (A and B) 7.000×, (C and E) 50.000×, (D) 30.000×. *P < 0.05, **P < 0.01, ***P < 0.001.
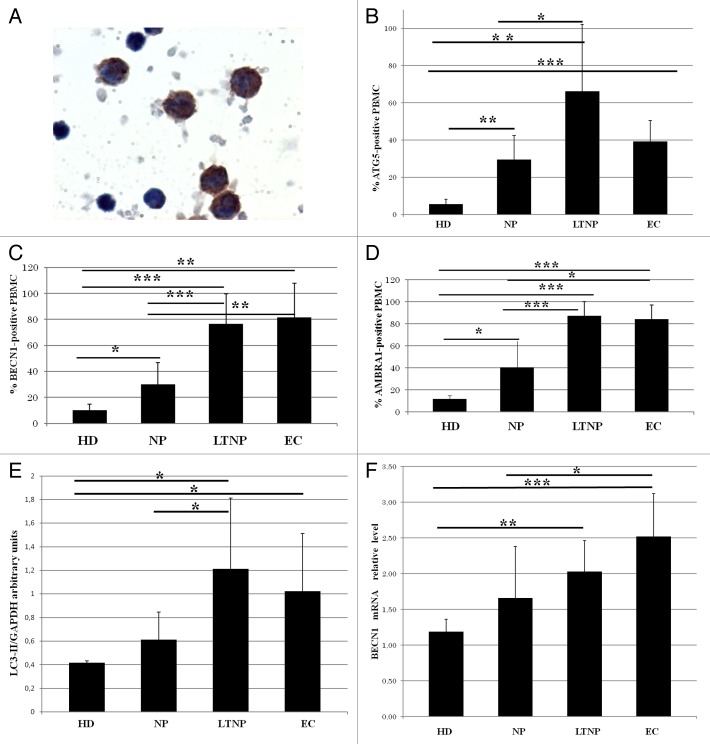
Differences in autophagy gene expression among the NP, LTNP, EC, and HD were detected in freshly isolated PBMC by immunocytochemistry (Fig. 2A–D) by evaluating the percentage of ATG5 (autophagy-related 5)-, BECN1- and AMBRA1-positive cells. In fact, a significant increase in the percentage of labeled cells for all the analyzed autophagic markers was detected in LTNP and EC compared with NP and HD. In HIV-1 controllers more than 60% of PBMC express ATG5, while in the NP only 30% of cells were positive (Fig. 2B). A similar trend was observed with BECN1 and AMBRA1 expression, with the percentage of positive cells significantly higher in HIV-1 controllers with the respect to NP (Fig. 2C and D). AMBRA1 increase was not restricted to a particular PBMC subpopulation in HIV-1 controllers (Fig. S1).
Figure 2. Enhanced autophagy in PBMC from LTNP and EC. (A) Immunocytochemical localization of BECN1 on PBMC from LTNP. (B–D) The percentage of cells expressing ATG5, BECN1, and AMBRA1 was quantified. (E) WB analysis of LC3-II levels on PBMC. (F) qRT-PCR of BECN1 mRNA relative level. Original magnification: (A) 100×. *P < 0.05, **P < 0.01, ***P < 0.001.
We also analyzed the relationship existing between these data and the viremia, showing that there is no linear correlation between these parameters (Fig. S2).
Quantitative analysis of the levels of autophagic factors on PBMC
Several methods are commonly used to quantify autophagy in vivo.30,31 One approach is to measure, by western blot (WB), the intracellular levels of the cleaved and lipidated MAP1LC3A-II (LC3-II). We analyzed this marker by WB in the PBMC from all the analyzed categories of HIV patients. The densitometric analysis confirmed a significantly enhanced level of LC3-II protein in LTNP and EC with respect to NP and HD (Fig. 2E). In keeping with these data, WB analysis of AMBRA1 and BECN1 showed an increased expression in LTNP with respect to NP (data not shown).
In order to confirm the steady-state upregulation of autophagy in HIV-infected individuals we also performed a qRT-PCR analysis of BECN1 expression. As shown in Figure 2F, this key upstream regulator of autophagy showed a significant higher level in the PBMC obtained from HIV-1 controllers with respect to HD. The mRNA levels of AMBRA1, an important BECN1 cofactor, were also higher in HIV-1-infected patients; by contrast, the level of ATG5, a downstream element of the autophagic program, was not significantly modified in PBMC from HIV-infected individuals when compared with those from noninfected controls (data not shown).
Autophagy in axillary lymph nodes from HIV-1-infected patients
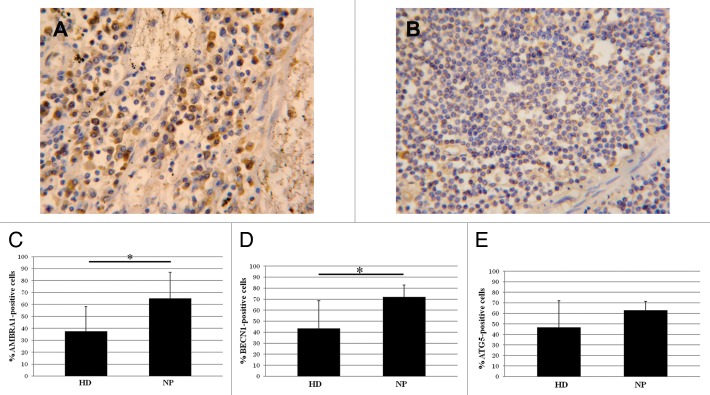
Axillary lymph nodes from HIV-1-infected NP and HD were analyzed to evaluate the in vivo expression of AMBRA1, BECN1, and ATG5; their expression was significantly higher in lymph nodes from HIV-1-infected donors (Fig. 3A) compared with uninfected ones (Fig. 3B). The quantification of AMBRA1, BECN1, and ATG5 positive cells showed a significant increase in NP vs. HD (Fig. 3C–E). In order to define the cell type specificity we performed the immune-characterization in consecutive sections of the lymph node by analyzing the phenotypic markers CD3, CD68, CD4, MS4A1/CD20, FCR2/CD23. This analysis showed that the autophagic proteins markers were preferentially expressed in macrophages, with the labeled cells also CD68 positive (Fig. S3).
Figure 3. Autophagy in axillary lymph nodes. (A) AMBRA1 immunostaining on lymph nodes from HIV-1-infected patients respect to noninfected ones (B). (C–E) Quantification of the AMBRA1, BECN1, and ATG5 labeling. Original magnification: (A and B) 40×. *P < 0.05, **P < 0.01, ***P < 0.001.
Modulation of autophagy and viral production in LTNP and NP
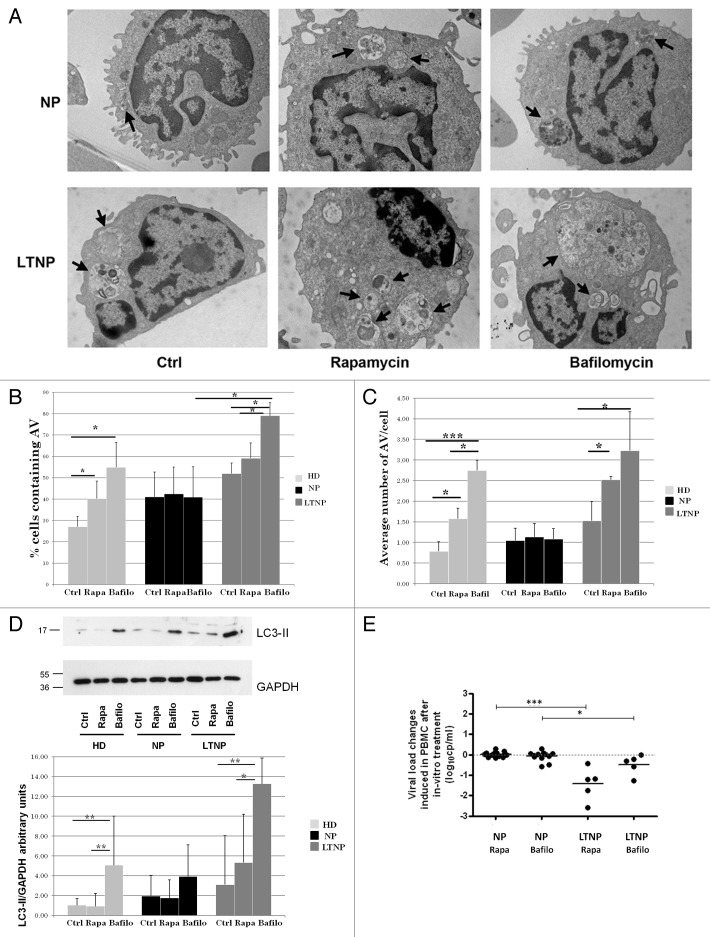
To investigate whether the increased expression levels of autophagic proteins correlates with a higher autophagic activity, we tested the effects of the autophagy inducer rapamycin in the presence of the autophagy inhibitor bafilomycin A1 on ex vivo PBMC from HIV-1-infected patients. Autophagy was first monitored by ultrastructural analysis. Both rapamycin and bafilomycin A1 have no significant effects on autophagosome number in NP (Fig. 4A–C). By contrast, the cells from LTNP proved to be highly and specifically responsive to these treatments. In particular, the treatment with bafilomycin A1, which inhibits vacuolar acidification, leads to the formation of very large AV containing a lot of undigested material (Fig. 4A–C). As a positive control, we analyzed cells from HD that showed a significant increase in the number of AV, after both rapamycin and bafilomycin A1 treatments as expected (Fig. 4B and C).
Figure 4. Pharmacological modulation of autophagy. (A) Electron microscopy analysis of PBMC from NP and LTNP treated with the autophagy inducer rapamycin, and the autophagy inhibitor bafilomycin A1. Arrows indicate AV containing undigested material. (B) Electron microscopy quantification of the percentage of cells containing AV in HD, NP, and LTNP. (C) Quantitative analysis of the average number of AV per cell in HD, NP, and LTNP. (D) WB analysis of LC3-II protein. In the graph were reported the mean values of 4 different experiments. (E) Changes in HIV RNA release in culture supernatants of NP and LTNP PBMC, after in vitro treatment with autophagy modulators rapamycin and bafilomycin A1. The difference in HIV RNA concentration between untreated and rapamycin- or bafilomycin A1-treated PBMC was calculated for each sample and reported in the figure. Original magnification: (A) 12.000×. *P < 0.05, **P < 0.01, ***P < 0.001.
The occurrence of autophagic flux was also analyzed by comparing the levels of LC3-II by WB analysis in HD, LTNP and NP in the presence or absence of the bafilomycin A1. In line with the increase of autophagosomes observed by ultrastructural analysis, we observed a greater increase of LC3-II in LTNP upon inhibition of lysosomal activity, indicating that PBMC from these patients have a higher autophagic flux than NP (Fig. 4D). The autophagic flux was also analyzed by detecting the levels of SQSTM1 in LTNP with respect to NP upon autophagy induction. Rapamycin treatment led to a decrease of SQSTM1 in LTNP while this modulation was significantly reduced in NP (Fig. S4A and S4B), thus confirming a more active autophagic flux in LTNP. We also observed a higher basal level of SQSTM1 in LTNP when compared with NP. However, this accumulation seems to be due to an increased expression of SQSTM1 gene in LTNP, as shown by qRT-PCR analysis (Fig. S4C), rather than a block of autophagy-mediated degradation. Altogether, these results indicate that autophagy flux is increased in PBMC from HIV-1 controllers.
Autophagy-mediated degradation of HIV-1 in LTNP
In order to analyze whether the modulation of autophagy could affect viral production, we measured the spontaneous HIV RNA yield in the cell culture supernatant fraction at the end of treatment. In NP, PBMC viral yield was not significantly modified by the autophagy modulators treatment, similar to levels measured in the untreated PBMC. Differently, PBMC from LTNP were sensitive to rapamycin and bafilomycin A1 treatments, the viral yield after treatment turning out to be significantly lower in comparison to PBMC from NP (Fig. 4E). We also analyzed EC, but considering the low level of viremia typically present in these patients, we were not able to assess whether changes in viral production occurred.
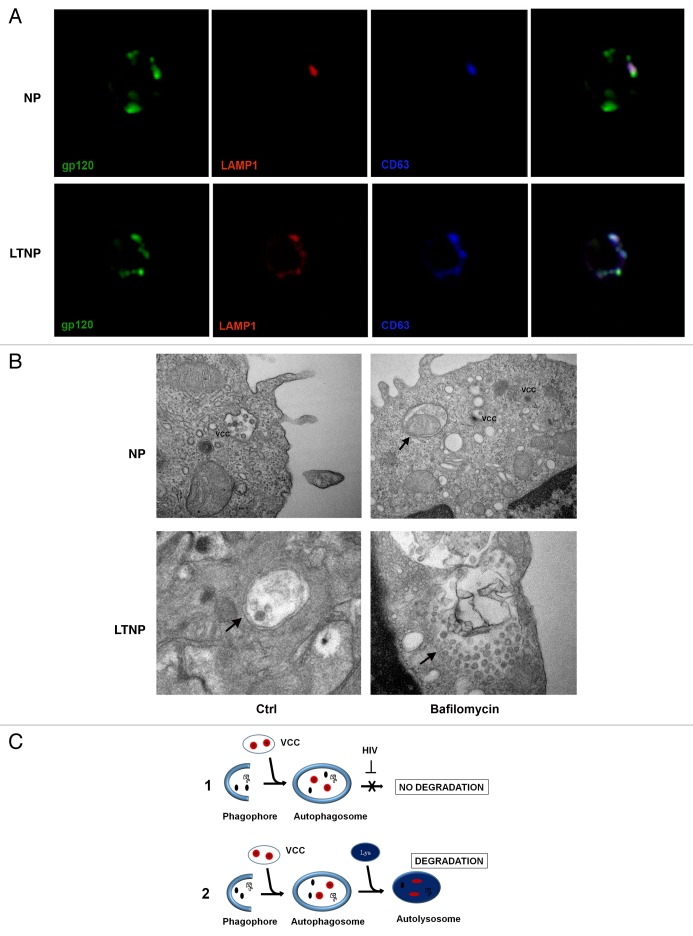
In order to confirm that in LTNP viral proteins are degraded within autolysosomes, we immunolocalized the HIV-1 protein gp120 in conjunction with LAMP1/lysosomal-associated membrane protein 1 to label the lysosomal membranes. Moreover, since HIV-1 assembles both at the plasma membrane of infected cells and in intracellular virus-containing compartments (VCCs),32 immunolocalizations were also performed using an anti-tetraspanin CD63 antibody to label VCCs. The results showed a colocalization of these proteins in LTNP cells, thus supporting the notion that HIV-1 could be degraded into autophagosomes (Fig. 5A). In contrast, the PBMC from NP showed only a few spots of colocalization (Fig. 5A). Double immunolocalizations have been also performed on PBMC from LTNP utilizing antibodies against to HIV-1 protein gp41 in conjunction with LC3-B (to label AV membranes) or lysosomal-associated membrane protein 2 (LAMP2; to label the lysosomal membranes) (Fig. S5A and S5B). We performed, also, a double immunofluorescence to detect the HIV-1 gp41 and the tetraspanins CD63 and CD81, these being considered as markers of VCCs (Fig. S5C and S5D). Finally, by utilizing an ultrastructural approach, we searched for “bona fide” viral particles inside VCCs or AV in PBMC from NP- and LTNP-untreated or bafilomycin A1-treated (Fig. 5B). The images showed viral particles or viral components preferentially localized in VCCs in PBMC from NP. Cells from LTNP showed viral particles inside AV; in particular bafilomycin A1-treated cells showed a very large AV containing a lot of undigested material and several viral particles.
Figure 5. HIV-1 particles inside AV. (A) Confocal microscopy immunolocalization of the HIV-1 protein gp120 (green), LAMP1 (red) and the tetraspanin CD63 (blue) on PBMC from NP and LTNP. (B) Ultrastructural images of PBMC from NP and LTNP patients showing VCCs that carried viral particles or viral components. An AV containing a mitochondrion was visible in bafilomycin A1-treated PBMC from NP (arrow). Control cells from LTNP showed a double-membrane AV containing viral particles or viral components (arrow). Bafilomycin-A1-treated LTNP PBMC showed a very large AV containing undigested material and viral particles or viral components (arrow). (C) Hypothetical mechanism of HIV-1 removal by autophagy. HIV-1 particles (red), budding into VCC, or viral components (black), can be captured by autophagosomes. Then the viral components degradation can be blocked by viral-specific protein/s in NP (C1). In contrast, HIV-1 components can get digested in autolysosomes in HIV-1 controllers (C2). Original magnification: (A) 63×; (B) NP Ctrl 30,000×, NP bafilomycin A1 30,000×, LTNP Ctrl 50,000×, LTNP bafilomycin A1 50,000×.
All these data enabled us to propose that in LTNP the increased and “functional” autophagy contribute to control viral yield by removing viral components in the cell cytoplasm or contained in VCCs (Fig. 5C, 2) differently from NP in which HIV-1 is able to block this process (Fig. 5C, 1).
Discussion
Infection by HIV-1 is associated with a progressive decrease in CD4 T-cell numbers and the consequent collapse and demise of host immune defenses.1 Recently, data from in vitro studies suggest that autophagy may play a complex role in HIV-1-induced pathogenesis by regulating both viral replication and the fate of host cells, however no data are available about its modulation in vivo and the consequences on pathogenesis.16,25-27,29,33
Kyei et al.9 show evidence that the initial stages of nondegradative autophagy are utilized by the virus to promote its replication in macrophages and, in addition, the HIV-1 protein Nef acts as an anti-autophagic maturation factor protecting the virus from the degradation by physically blocking BECN1, one of the key regulatory elements of autophagy.9,10,29 It is interesting to note that people infected with a virus containing a deletion in the Nef gene maintain low viremia for decades,34,35 thus showing a slow disease progression comparable to HIV-1 nonprogressors who are able to control the viral-induced pathogenesis. Viral factors, host genetics, and immune responses have been associated with the control of HIV-1 replication and lack of or slow disease progression in these particular patients.36-38 However, despite numerous studies, the exact cellular mechanisms responsible for viral containment in HIV-1 controllers are largely unknown. The identification and the characterization of the mechanisms regulating the HIV-1-induced “slow” pathogenesis in these exceptional individuals might help to identify new therapeutic strategies.
Based upon these premises, we verified the hypothesis of an involvement of autophagy in the control of HIV-1-induced pathogenesis in LTNP and EC. We compared the expression of several proautophagic proteins on PBMC from HIV-1 infected individuals comparing NP, LTNP, EC, and HD. All the autophagic markers analyzed, namely ATG5, BECN1, AMBRA1, and LC3-II, showed significantly higher expression levels in HIV-1-infected patients with respect to HD.
Moreover, the analysis of the same proteins on the lymph nodes suggests that autophagy is upregulated in HIV-1-infected samples predominantly in CD68-positive cells (macrophage lineage). These results are in line with previously published data suggesting that several autophagy proteins (such as BECN1, ATG5, ATG7, and ATG12) are required for HIV-1 replication.9,33,39,40
The ultrastructural analysis of PBMC clearly showed a higher number of cells containing AV in LTNP and EC with the respect to HD and NP. Also the number of AV contained in a single cell showed a significant increase in LTNP and EC respect to NP. The analysis of autophagic proteins by immunohistochemistry and WB showed a significantly increased expression in cells from HIV-1 controllers with the respect to NP. These data were confirmed by the analysis of BECN1 mRNA. It is interesting to note that the highest increase in the mRNA level of BECN1 was detected in EC that are able to fully control the viremia. This result matches several published data highlighting this protein’s key role in controlling viral yield,9,10,29,40 and represents the first evidence of the increased induction of BECN1 in HIV-1 controllers in respect to NP. However, it must be noted that no correlation was observed between increased levels of autophagic proteins and plasma HIV-1 RNA levels, both in chronic and in LTNP patients, suggesting that prevention of viral pathogenesis by autophagy is not related only to the control of patients’ viremia. HIV-1 controllers constitute a population spontaneously capable of controlling the 2 main parameters of HIV-1 disease progression: loss of CD4− T cells and viremia. In fact, they maintain normal CD4+ T cell counts (> 500 cells/μl) for years. There are several experimental findings that hypothesize a control of apoptotic cell death, normally associated with the disease, in these patients. It is now becoming clear that there is a crosstalk between apoptosis and autophagy and that in many different biological settings active autophagy can limit apoptosis induction.41 Thus, the upregulation of autophagy, acting as a cell survival mechanism, might play a role in limiting the drastic cell depletion induced by HIV-1 in LTNP and EC patients. In keeping with this assumption it is important to mention that some of the key genes described as being involved in HIV-1-induced apoptotic cell death TP53 (tumor protein p53) and MTOR (mechanistic target of rapamycin [serine/threonine kinase]) are also key regulators of autophagy.42-45
It remains unclear how autophagy prevents cells from undergoing apoptosis; one study has suggested that the autophagy-mediated sequestration of damaged mitochondria preventing cytochrome c release could limit the formation of a functional apoptosome in the cytoplasm.46 Interestingly, our ultrastructural analysis of PBMC from HIV-1 controllers revealed the presence of many mitochondria inside double-membrane-bound autophagic vacuoles. Thus the active removal of damaged mitochondria could indeed contribute to the resistance of the LTNP and EC cells to apoptosis.
By pharmacologically modulating autophagy in PBMC ex vivo we showed an increased autophagy in LTNP, whereas NP did not responded to treatments. Relying on previously published data, we hypothesize that in NP, HIV-1 may subvert autophagy to maximize virus production blocking the final steps of the degradation.9,10 By contrast, the increased functional autophagy could hamper the accumulation and release of viral particles in LTNP. In keeping with this hypothesis, the treatment with rapamycin was unable to reduce viral production in the PBMC obtained from NP, thus supporting the hypothesis that in NP the degradative steps of autophagy are blocked. On the contrary, LTNP showed an increased “functional” autophagy able to counteract HIV-1-yield by trapping viral components or viral particles or viral components inside AV. Indeed, in PBMC from HIV-1 controllers we identified a prominent presence of autophagic vacuoles many of which are positive to HIV-1 proteins staining. Unexpectedly, an increase in viral load was not detected when lysosomal activity was inhibited. However, this result could be explained by the fact that viral particles remained entrapped within nondegradative AV.
In conclusion, in this study we have shown for the first time that in vivo the resistance to HIV-1-induced pathogenesis in nonprogressors is accompanied by a significant increase in the autophagic activity in the PBMC of these individuals. This finding is clinically relevant since many autophagy inducers are currently under clinical investigation for the treatment of several diseases and aging.47-49 Interestingly, HIV-1 protease inhibitors, nelfinavir and saquinavir, and vitamin D are being evaluated for their autophagy-enhancing activities.33,50,51 Understanding the cellular and molecular mechanisms at the basis of the successful control of the chronic viral infection in vivo, in LTNP and EC, represent the most promising avenue for the development of novel therapeutic strategies to treat HIV-1 infection.
Materials and Methods
Patients and normal donors
Patients enrolled in the study, 18 LTNP, 6 EC, 18 NP, and 17 healthy donors (HD), were cared for the Outpatient HIV clinic of the National Institute for Infectious Disease (INMI) and provided written informed consent to participate in the study (Ethics Committee approval n° 49/2010).
LTNP plasma HIV RNA (viral load) was > 200 copies/ml, with CD4 T cells > 500/μl; EC viral load was < 50 copies/ml and CD4 T cells > 500/μl. LTNP and EC were infected for an average of 8 y. NP were not on antiretroviral therapy and the median values of viral load was 4,5 ± 0,6 log10 cp/ml and < 500 CD4 T cells/μl. HD were HIV-1 negative blood donors. Plasma HIV-1 RNA levels were measured with Abbott Real-time HIV-1 assay according to the manufacturer’s instructions (Abbott Molecular, RealTime HIV-1). PBMC were obtained from residual blood samples intended for diagnostic tests. PBMC were isolated by Ficoll/Hypaque (GE Healthcare, 17-1440-02) centrifugation of heparinized blood and utilized immediately for electron microscopy morphological analysis, immonocytochemical analysis, and frozen at −80 °C until western blot, qRT-PCR and for in vitro cultures. Axillary lymph node autoptic samples were obtained from healthy and HIV-1-infected individuals (all males, mean age 36 y).
Electron microscopy
PBMC were fixed with 2.5% glutaraldehyde (Sigma-Aldrich, R1012) in 0.1 M cacodylate buffer, pH 7.4, for 45 min at 4 °C, rinsed in buffer, postfixed in 1% OsO4 in 0.1 M cacodylate buffer, pH 7.4, dehydrated, and embedded in Epon resin (Agar Scientific, 45359-1EA-F). Grids were thoroughly rinsed in distilled water, stained with aqueous 2% uranyl acetate for 20 min and photographed in a Zeiss EM 900 electron microscope (Carl-Zeiss-Straße 56 73447 Oberkochen, Germany). The percentage of cells containing autophagic vacuoles (AV) vs. the total cell number and the average number of AV per cell was evaluated. A minimum of 50 cells/patient were observed. Cell counting was done by 3 independent individuals; data are presented as mean ± SD.
Immunodetection of proteins involved in autophagy
PBMC and lymph nodes from HIV-1-infected patients and HD were isolated and fixed in 4% freshly depolymerized paraformaldehyde (Sigma-Aldrich, P-6148) in PBS (Sigma-Aldrich, P-4417) pH 7.2.
Immunohistochemistry
Lymph nodes were embedded in paraffin and sections were deparaffinized, rehydrated, and subjected to high temperature antigen retrieval in 10 mM sodium citrate buffer pH 6,0. Immunohistochemistry was performed as previously reported,52 in tissue sections and fixed PBMC. The primary antibodies utilized were: rabbit polyclonal anti-AMBRA1 (1:100, ProSci, 4557) rabbit polyclonal anti-BECN1 (1:50, Santa Cruz Biotechnology, sc-11427), rabbit polyclonal anti-ATG5 (1:50, Santa Cruz Biotechnology, sc-33210).
The percentage of positive PBMC/total PBMC, was counted for AMBRA1, BECN1, and ATG5 stainings. Three independent observers evaluated the number of positive cells by using a light microscope without the knowledge of clinical diagnosis. A minimum of 500 PBMC/patient were analyzed. The percentage of positive lymph node cells/total cells, was counted for AMBRA1, BECN1 and ATG5 stainings. For each slide, a minimum of 10 fields was examined at 40× magnification.
Immunofluorescence
For immunofluorescence experiments, PBMC were incubated with the following antibodies: human monoclonal anti-HIV-1 gp41 (1:25, kindly provided by Dr Marie-Lise Gougeon, Paris); human monoclonal anti-gp120 (1:200, kindly provided by Dr Jean-Luc Perfettini, Paris); rabbit polyclonal anti-LC3B (1:200, Sigma-Aldrich, L7543); mouse monoclonal anti-LAMP2 (1:100, Santa Cruz Biotechnology, sc-18822); rabbit polyclonal anti-LAMP1 (1:100, abcam, ab24170); mouse monoclonal anti-CD63 (1:200, abcam, ab8219); mouse monoclonal anti-CD81 (1:200, abcam, ab59477). Sections were thoroughly rinsed with PBS, then incubated for 1 h at RT with 1:400 Alexa488 conjugated goat anti-human IgG (Molecular Probes MP 11013), 1:400 Alexa594 conjugated goat anti-mouse IgG (Invitrogen, Life Technologies, A-11020) or 1:400 Alexa594 conjugated goat anti-rabbit IgG (Invitrogen, Life Technologies, A-11037), 1:400 donkey anti-mouse AlexaFluor 647 (Jackson Immunoresearch, 715-606-151). Controls were performed by omitting the primary antibodies. Slides were observed and photographed in a Leica TCS SP2 confocal microscope (Leica Microsystems GmgH, Ernst-Leitz-trasse 17-37 35578 Wetzlar Germany).
Western blot analysis of proteins involved in autophagy
Total proteins were extracted from PBMC, isolated by Ficoll-Hypaque isolated PBMC, by using the Cell Lytic buffer (Sigma-Aldrich, C3228) following addition of protease inhibitors and resolved by electrophoresis through NuPAGE Bis-Tris gel (Invitrogen, NP0321BOX) and electroblotted onto nitrocellulose (Protran, 10402062) or PVDF (Millipore, IPVH20200) membranes. Blots were incubated with indicated primary antibodies in 5% nonfat dry milk in PBS plus 0.1% Tween 20, overnight at 4 °C. Primary antibodies were: rabbit anti-LC3B (1:2000; Cell Signaling Technology, 2775), rabbit anti-SQSTM1 (1:2000; MBL, PM045) anti-GAPDH (1:60000; Calbiochem, CM1001). Detection was achieved using horseradish peroxidase-conjugate secondary antibody (1:5000; Jackson ImmunoResearch, 715-036-150) and visualized with ECL Prime (GE Healthcare, RPN2232) using ECL-Hyperfilm (GE Healthcare, 28-9068-40). Mouse anti-GAPDH antibody was used to monitor equal protein loading. Western blot images were analyzed densitometrically using a charge-coupled device camera (GelDoc 2000, Bio-Rad, Hercules, CA, USA) and processed with the QuantyOne software (Bio-Rad) in order to quantify the amount of LC3-II band intensity.
qRT-PCR
RNA was extracted from PBMC by using Trizol reagent (Invitrogen, 15596-026) as indicated by the supplier. cDNA synthesis was generated using a reverse transcription kit (Promega, M5101) according to the manufacturer’s recommendations. Quantitative RT-PCR reactions were performed with the LightCycler (Roche, Nutley, NJ, USA) thermocycler, as previously described.53 Primer sets for all amplicons were designed using the Primer-Express 1.0 software system.
L34/ribosomal protein L34 (RPL34) forward: 5′-GTCCCGAACC CCTGGTAATA GA-3′
RPL34 reverse: 5′-GGCCCTGCTG ACATGTTTCT T-3′
BECN1 forward: TCTCGCAGAT TCATCCCCC
BECN1 reverse: TCTTCGGCTG AGGTTCTCCA T
SQSTM1 forward: ACAGATGCCA GAATCCGAAG
SQSTM1 reverse: TGGGAGAGGG ACTCAATCAG
The RPL34 mRNA level was used as an internal control and results were expressed as previously described.53
Pharmacological modulation of autophagy in cultured PBMC
The chemical compounds rapamycin and bafilomycin A1, were used to try to modulate autophagy in PBMC by in vitro treatment. Rapamycin inhibits MTOR, thus inducing autophagy; instead, bafilomycin A1, is a specific inhibitor of vacuolar ATP6V1H (ATPase, H+ transporting, lysosomal 50/57 kDa, V1 subunit H), and inhibits the acidification of organelles containing this enzyme, such as lysosomes and endosomes.30 PBMC from 7 LTNP, 3EC, 17 NP and 5 HD were thawed, washed 3 times with PBS and cultured (2 × 106 cells/ml) in RPMI 1640 medium (GIBCO, 12633-012) containing 10% fetal calf serum, 2 mM L-glutamine, 100 U penicillin/ml and 100 μg streptomycin/ml at 37 °C. After 24 h, PBMC were divided into 3 wells and cultured for an additional 20 h in the presence of 100 nM rapamycin (Calbiochem, 553210), or 100 nM bafilomycin A1 (Sigma-Aldrich, B1793), or medium as control. Culture supernatant fractions underwent HIV-1 RNA quantification with Abbott Real-time HIV-1 assay according to the manufacturer’s instructions, while treated PBMC were used for WB or qRT-PCR analysis.
Statistical analysis
To determine statistical significance, the Student t test and Pearson coefficient correlation were used. Statistical significance was set at P < 0.05.
Supplementary Material
Disclosure of Potential Conflict of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr Marie-Lise Gougeon (Institute Pasteur, Paris, France) and Dr Jean-Luc Perfettini (Institut Gustave Roussy, Villejuif, France) for stimulating discussions and for the kind gift of the anti-gp41 and anti-gp120 antibodies. We thank the U.O.C. Microbiologia e Banca Biologica (National Institute for Infectious Diseases, IRCCS “L. Spallanzani” Rome) for the recruitment of patient samples and Alessia Brenna, Mario Moauro, and Diletta Collalto for technical assistance. A special thanks to the patients enrolled in this study, for their availability and participation. This work was supported by grants from the Ministry for Health of Italy to MP (“Ricerca Corrente” and Ricerca AIDS RF-IMI-2009-1303225) and the Italian Ministry of University and Research (FIRB 2011).
Glossary
Abbreviations:
- AMBRA1
autophagy/Beclin 1 regulator 1
- ATG5
autophagy-related 5
- AV
autophagic vacuoles
- BECN1
Beclin 1, autophagy-related
- EC
elite controllers
- HD
healthy donors
- LAMP1
lysosomal-associated membrane protein 1
- LAMP2
lysosomal-associated membrane protein 2
- LTNP
long-term nonprogressors
- MAP1LC3A
microtubule-associated protein 1 light chain 3 alpha
- NP
HIV-1-infected normal progressor patients
- PBMC
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative RT-PCR
- SQSTM1
sequestosome 1
- VCC
virus-containing compartment
- WB
western blot
References
- 1.Pierson T, McArthur J, Siliciano RF. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG. HIV: How to escape treatment. Nature. 2011;477:36–7. doi: 10.1038/477036a. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–16. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Autran B, Descours B, Avettand-Fenoel V, Rouzioux C. Elite controllers as a model of functional cure. Curr Opin HIV AIDS. 2011;6:181–7. doi: 10.1097/COH.0b013e328345a328. [DOI] [PubMed] [Google Scholar]
- 5.Whittall T, Peters B, Rahman D, Kingsley CI, Vaughan R, Lehner T. Immunogenic and tolerogenic signatures in human immunodeficiency virus (HIV)-infected controllers compared with progressors and a conversion strategy of virus control. Clin Exp Immunol. 2011;166:208–17. doi: 10.1111/j.1365-2249.2011.04463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poropatich K, Sullivan DJ., Jr. Human immunodeficiency virus type 1 long-term non-progressors: the viral, genetic and immunological basis for disease non-progression. J Gen Virol. 2011;92:247–68. doi: 10.1099/vir.0.027102-0. [DOI] [PubMed] [Google Scholar]
- 7.Buckheit RW, 3rd, Allen TG, Alme A, Salgado M, O’Connell KA, Huculak S, Falade-Nwulia O, Williams TM, Gallant JE, Siliciano RF, et al. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat Commun. 2012;3:716. doi: 10.1038/ncomms1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 9.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–68. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gannagé M, Rämer PC, Münz C. Targeting Beclin 1 for viral subversion of macroautophagy. Autophagy. 2010;6:166–7. doi: 10.4161/auto.6.1.10624. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 12.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–49. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009;16:57–69. doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deretic V. Autophagy: an emerging immunological paradigm. J Immunol. 2012;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orvedahl A, Levine B. Viral evasion of autophagy. Autophagy. 2008;4:280–5. doi: 10.4161/auto.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–70. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grégoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7:e1002422. doi: 10.1371/journal.ppat.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–5. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 23.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–68. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eekels JJ, Sagnier S, Geerts D, Jeeninga RE, Biard-Piechaczyk M, Berkhout B. Inhibition of HIV-1 replication with stable RNAi-mediated knockdown of autophagy factors. Virol J. 2012;9:69. doi: 10.1186/1743-422X-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espert L, Codogno P, Biard-Piechaczyk M. What is the role of autophagy in HIV-1 infection? Autophagy. 2008;4:273–5. doi: 10.4161/auto.5211. [DOI] [PubMed] [Google Scholar]
- 26.Gougeon ML, Piacentini M. New insights on the role of apoptosis and autophagy in HIV pathogenesis. Apoptosis. 2009;14:501–8. doi: 10.1007/s10495-009-0314-1. [DOI] [PubMed] [Google Scholar]
- 27.Dinkins C, Arko-Mensah J, Deretic V. Autophagy and HIV. Semin Cell Dev Biol. 2010;21:712–8. doi: 10.1016/j.semcdb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–69. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Killian MS. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res Ther. 2012;9:16. doi: 10.1186/1742-6405-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan J, Sattentau QJ. The HIV-1-containing macrophage compartment: a perfect cellular niche? Trends Microbiol. 2013;21:405–12. doi: 10.1016/j.tim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Campbell GR, Spector SA. Hormonally active vitamin D3 (1alpha,25-dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection. J Biol Chem. 2011;286:18890–902. doi: 10.1074/jbc.M110.206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–91. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 35.Churchill MJ, Rhodes DI, Learmont JC, Sullivan JS, Wesselingh SL, Cooke IR, Deacon NJ, Gorry PR. Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J Virol. 2006;80:1047–52. doi: 10.1128/JVI.80.2.1047-1052.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casado C, Colombo S, Rauch A, Martínez R, Günthard HF, Garcia S, Rodríguez C, Del Romero J, Telenti A, López-Galíndez C. Host and viral genetic correlates of clinical definitions of HIV-1 disease progression. PLoS One. 2010;5:e11079. doi: 10.1371/journal.pone.0011079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pernas M, Casado C, Arcones C, Llano A, Sánchez-Merino V, Mothe B, Vicario JL, Grau E, Ruiz L, Sánchez J, et al. Low-replicating viruses and strong anti-viral immune response associated with prolonged disease control in a superinfected HIV-1 LTNP elite controller. PLoS One. 2012;7:e31928. doi: 10.1371/journal.pone.0031928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buckheit RW, 3rd, Allen TG, Alme A, Salgado M, O’Connell KA, Huculak S, Falade-Nwulia O, Williams TM, Gallant JE, Siliciano RF, et al. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat Commun. 2012;3:716. doi: 10.1038/ncomms1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Gao Y, Tan J, Devadas K, Ragupathy V, Takeda K, Zhao J, Hewlett I. HIV-1 and HIV-2 infections induce autophagy in Jurkat and CD4+ T cells. Cell Signal. 2012;24:1414–9. doi: 10.1016/j.cellsig.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 40.Zhou D, Masliah E, Spector SA. Autophagy is increased in postmortem brains of persons with HIV-1-associated encephalitis. J Infect Dis. 2011;203:1647–57. doi: 10.1093/infdis/jir163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fimia GM, Piacentini M. Regulation of autophagy in mammals and its interplay with apoptosis. Cell Mol Life Sci. 2010;67:1581–8. doi: 10.1007/s00018-010-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castedo M, Roumier T, Blanco J, Ferri KF, Barretina J, Tintignac LA, Andreau K, Perfettini JL, Amendola A, Nardacci R, et al. Sequential involvement of Cdk1, mTOR and p53 in apoptosis induced by the HIV-1 envelope. EMBO J. 2002;21:4070–80. doi: 10.1093/emboj/cdf391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perfettini JL, Roumier T, Castedo M, Larochette N, Boya P, Raynal B, Lazar V, Ciccosanti F, Nardacci R, Penninger J, et al. NF-kappaB and p53 are the dominant apoptosis-inducing transcription factors elicited by the HIV-1 envelope. J Exp Med. 2004;199:629–40. doi: 10.1084/jem.20031216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nardacci R, Antinori A, Kroemer G, Piacentini M. Cell death mechanisms in HIV-associated dementia: the involvement of syncytia. Cell Death Differ. 2005;12(Suppl 1):855–8. doi: 10.1038/sj.cdd.4401590. [DOI] [PubMed] [Google Scholar]
- 45.Perfettini JL, Castedo M, Roumier T, Andreau K, Nardacci R, Piacentini M, Kroemer G. Mechanisms of apoptosis induction by the HIV-1 envelope. Cell Death Differ. 2005;12(Suppl 1):916–23. doi: 10.1038/sj.cdd.4401584. [DOI] [PubMed] [Google Scholar]
- 46.Ravikumar B, Berger Z, Vacher C, O’Kane CJ, Rubinsztein DC. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006;15:1209–16. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 47.Bové J, Martínez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12:437–52. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 48.García-Mauriño S, Alcaide A, Domínguez C. Pharmacological control of autophagy: therapeutic perspectives in inflammatory bowel disease and colorectal cancer. Curr Pharm Des. 2012;18:3853–73. doi: 10.2174/138161212802083653. [DOI] [PubMed] [Google Scholar]
- 49.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–95. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 50.McLean K, VanDeVen NA, Sorenson DR, Daudi S, Liu JR. The HIV protease inhibitor saquinavir induces endoplasmic reticulum stress, autophagy, and apoptosis in ovarian cancer cells. Gynecol Oncol. 2009;112:623–30. doi: 10.1016/j.ygyno.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 51.Gills JJ, Lopiccolo J, Dennis PA. Nelfinavir, a new anti-cancer drug with pleiotropic effects and many paths to autophagy. Autophagy. 2008;4:107–9. doi: 10.4161/auto.5224. [DOI] [PubMed] [Google Scholar]
- 52.Nardacci R, Antinori A, Larocca LM, Arena V, Amendola A, Perfettini JL, Kroemer G, Piacentini M. Characterization of cell death pathways in human immunodeficiency virus-associated encephalitis. Am J Pathol. 2005;167:695–704. doi: 10.1016/S0002-9440(10)62044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corazzari M, Lovat PE, Armstrong JL, Fimia GM, Hill DS, Birch-Machin M, Redfern CP, Piacentini M. Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: the role of stress proteins ERdj5 and ERp57. Br J Cancer. 2007;96:1062–71. doi: 10.1038/sj.bjc.6603672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.