Abstract
Recent studies have indicated that cancer stem-like cells (CSCs) exhibit a high resistance to current therapeutic strategies, including photodynamic therapy (PDT), leading to the recurrence and progression of colorectal cancer (CRC). In cancer, autophagy acts as both a tumor suppressor and a tumor promoter. However, the role of autophagy in the resistance of CSCs to PDT has not been reported. In this study, CSCs were isolated from colorectal cancer cells using PROM1/CD133 (prominin 1) expression, which is a surface marker commonly found on stem cells of various tissues. We demonstrated that PpIX-mediated PDT induced the formation of autophagosomes in PROM1/CD133+ cells, accompanied by the upregulation of autophagy-related proteins ATG3, ATG5, ATG7, and ATG12. The inhibition of PDT-induced autophagy by pharmacological inhibitors and silencing of the ATG5 gene substantially triggered apoptosis of PROM1/CD133+ cells and decreased the ability of colonosphere formation in vitro and tumorigenicity in vivo. In conclusion, our results revealed a protective role played by autophagy against PDT in CSCs and indicated that targeting autophagy could be used to elevate the PDT sensitivity of CSCs. These findings would aid in the development of novel therapeutic approaches for CSC treatment.
Keywords: apoptosis, autophagy, autophagy-related proteins, cancer stem-like cells, colonosphere, colorectal cancer, photodynamic therapy, prominin 1 (PROM1)/CD133, tumorigenicity
Introduction
Colorectal cancer (CRC) is the fourth most common cause of cancer-related mortality worldwide, with more than 1.2 million new cases and 600,000 deaths per year.1 The relative survival rate of patients with unresectable metastatic lesions drops to only 5%.2 In addition to surgery, radiation therapy, and chemotherapy, photodynamic therapy (PDT) is regarded as another anticancer modality for the treatment of advanced colorectal cancer. PDT is a promising therapeutic approach for malignant and premalignant tumors and other diseases.3,4 With this technique, a photosensitizer is irradiated and excited by light of a specific wavelength and then transfers this energy to oxygen to cause reactive oxygen species (ROS) formation. Subcellular damage subsequently occurs at the site of photosensitizer accumulation, which further drives either apoptosis or necrosis.5-8 Because PDT causes essentially no effect on connective tissues, the anatomical integrity of hollow organs such as the colon can be preserved in patients undergoing PDT. However, numerous studies have indicated that several resistance mechanisms cause a reduction in the efficacy of PDT on cancer cells.9-13 Therefore, to improve the therapeutic outcome of PDT in colorectal cancer, mechanisms of primary resistance to PDT must be clarified.
Recently, increasing evidence has suggested that a small subpopulation of cancer cells may play a resistant role during treatment, leading to tumor recurrence and progression. Accordingly, these malignant cells responsible for tumor development and sustained tumor growth have been termed “cancer stem-like cells” (CSCs).14 CSCs are thought to possess stem cell-like properties such as self-renewal and differentiation. The existence of CSCs has implications for novel cancer therapeutics, as the effective eradication of CSCs will likely be essential for successful cancer treatment.15 Currently, CSCs are identified by their expression of specific surface markers; recent investigations have identified CSCs isolated from colorectal tumors with PROM1/CD133, a surface marker commonly found on the stem cells of various tissues.16,17 Many studies propose that CSCs are resistant to chemotherapy and radiotherapy.18-21 It has also been reported that side population (SP) stem cell-like cancer cells, which are considered CSCs, are resistant to PDT compared with non-SP cells.22 However, a more detailed cellular mechanism underlying the resistance to PDT in CSCs remains poorly understood.
Autophagy is a dynamic process that involves the transport of cellular organelles and proteins through a lysosomal degradation pathway, and has been shown to play a variety of important roles related to cell survival, differentiation, and development.23,24 Autophagy is regulated by a series of autophagy-related genes (ATG) that mediate the formation of the autophagosome and the activity of the PI3K-MTOR (phosphoinositide 3-kinase-mechanistic target of rapamycin) pathway controls autophagic flux.25,26 Recent studies have shown that autophagy is involved in cancer development and progression and is activated by anticancer therapy.27-29 It has been reported that autophagy can be induced by PDT.30 Although the role played by autophagy in cell survival and cell death is still unclear, accumulating data show that an increase in autophagy levels can protect cancer cells against various apoptotic stimuli.31-34 In addition, further studies are needed to determine the function of autophagy in CSCs and if autophagy can be induced by PDT in CSCs.
In this study, we examined the induction of autophagy by PDT and its role in colorectal PROM1/CD133+ CSCs. We found that CSC-enriched colorectal cancer cells would have more autophagosome formation than non-CSC-enriched cancer cells as shown by higher expression of autophagy-related proteins in CSC-enriched colorectal cancer cells. We also found that the maintenance, self-renewal, and resistance of CSCs after PDT treatment are dependent on autophagy. Inhibition of autophagy, both by an autophagy inhibitor and silencing of ATGs, sensitized the CSCs to PDT in vitro and in vivo. These results suggest that autophagy inhibition might abrogate CSC’s resistance to PDT. Therefore, PDT combining autophagy inhibition might be a more efficient therapeutic approach for CSC treatment.
Results
PROM1/CD133+ cells of colorectal cancer exhibit the characteristics of CSCs and resistance to PDT
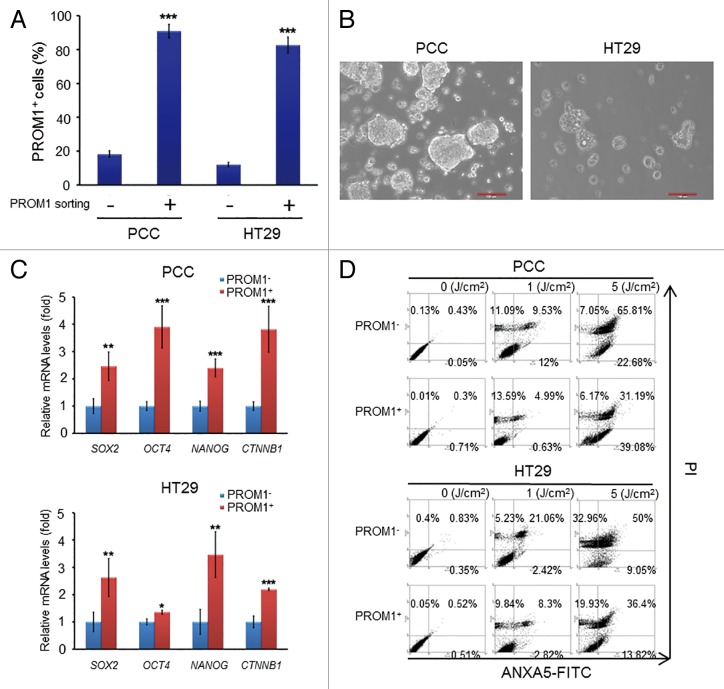
It has been shown that PROM1/CD133+ subpopulations in colorectal cancer cells exhibit colorectal cancer stem cell properties.16 To verify whether PROM1/CD133+ cells exhibit cancer stem cell properties, we separated PROM1/CD133+ and PROM1/CD133− subpopulations from primary colorectal tumor (PCC) and HT29 colorectal cancer cells. After cell sorting, 80% of the cell population was PROM1/CD133+ (Fig. 1A). To analyze the stemness-associated properties of the isolated PROM1/CD133+ cells, sphere forming, stemness gene expression and drug sensitization assays were performed. The isolated PROM1/CD133+ cells could form clonal nonadherent 3D spheres (Fig. 1B) and were more resistant to conventional chemotherapy (Fig. S1). In contrast, fewer PROM1/CD133− cells could form spheres than the PROM1/CD133+ cells. The expression of stemness-related genes, such as SOX2, POU5F1/OCT4, and NANOG, was higher in PROM1/CD133+ cells than PROM1/CD133− cells (Fig. 1C).
Figure 1. PROM1/CD133+ cells of colorectal cancer exhibit the characteristics of CSCs and resistance to PDT. (A) The PROM1/CD133+ cells were isolated from PCC or HT29 by FACS sorting. The percentage of PROM1/CD133+ cells was determined by FACS assay. (B) The morphology of PROM1/CD133+ cells was observed 10 d after FACS sorting. The PROM1/CD133+ cells formed floating tumorspheres under stem-cell conditions in the presence of EGF and FGF2. Scale bar: 100 μm. (C) Total RNA was isolated from PROM1/CD133− and PROM1/CD133+ cells from PCC and HT29 cells and then analyzed by qPCR for SOX2, OCT4, NANOG, and CTNNB1. (D) Following PpIX-mediated PDT at various light doses, PROM1/CD133+ and PROM1/CD133− cells viability was analyzed by ANXA5-FITC-PtdIns staining at 24 h after PDT. These results are expressed as the mean ± SE of 3 different experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Previous studies demonstrate that SP cells are more resistant to PDT than non-SP cells.22 To verify whether PROM1/CD133+ cells from PCC and HT29 cells exhibit similar resistance, PpIX-mediated PDT was utilized. PpIX is a naturally occurring photosensitizer used to detect and treat cancer. Cells were treated with PpIX in medium and then subjected to laser irradiation, which has a specific wavelength of 633 nm, at various doses. Cell survival after PDT treatment was analyzed by ANXA5/annexin V-FITC-PI staining. PpIX-mediated PDT caused significantly less PROM1/CD133+ cell death compared with PROM1/CD133- cells at the same irradiation dose (Fig. 1D; Fig. S2). These data showed that PROM1/CD133+ colorectal cancer cells were less sensitive to PDT than PROM1/CD133- cells. In addition, after PDT treatment, clonogenic survival fractions in both PCC and HT29 cell lines were reduced (Fig. S3) and suggested PDT resistance effect in PROM1/CD133+ cells. WST-1 assays also have similar results (Fig. S4). These data confirmed that PROM1/CD133+ cells have CSC-like properties, such as generation of spheres, high expression of stemness-associated genes, and high resistance to chemotherapeutic drugs and PDT.
PDT induces autophagy in PROM1/CD133+ cells
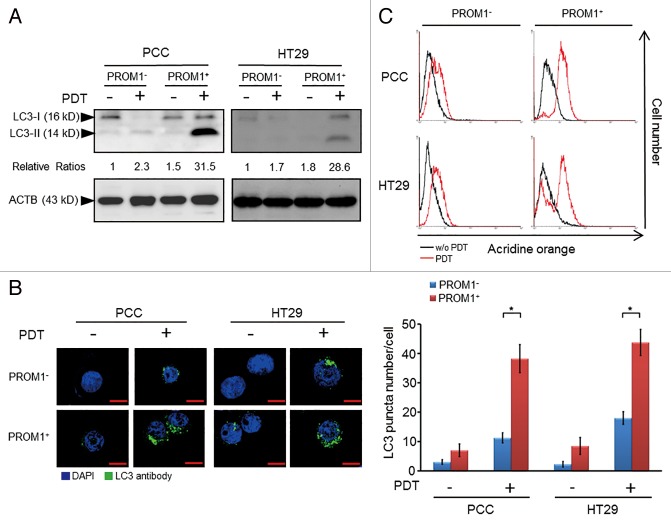
Numerous investigations indicated that autophagy, which is frequently activated in response to chemotherapy or radiotherapy, may contribute to resistance of different carcinoma cells.35-40 Recent findings have also shown that autophagy could be induced in response to PDT in cancers cell lines and acts as a defense mechanism against PDT-mediated cellular damage.41 To elucidate the extent of autophagy after treatment with PpIX-mediated PDT in colorectal CSCs, we analyzed the accumulation of LC3-II, the lipidated form of LC3 associated with the autophagosomal membrane, in the isolated PROM1/CD133+ cells. The amount of LC3-II reflects the number of autophagosomes. The accumulation of LC3-II markedly increased in PROM1/CD133+ cells at 24 h post-PDT with a laser irradiation dose of 1.3 J/cm2 (~IC50 of PROM1/CD133- cells) (Fig. 2A). The levels of autophagy were also observed by the formation of autophagosomes, as visualized using an anti-LC3 antibody. The number of LC3 puncta notably increased in PROM1/CD133+ PCC or HT29 cells after treatment with PDT (1.3 J/cm2) (Fig. 2B). In addition, acridine orange (AO) staining, a lysosomotropic dye that enters acidic compartments and emits bright red fluorescence, was used to assess the levels of autophagy by flow cytometry.42 PROM1/CD133+ cells treated with PDT (1.3 J/cm2) displayed intense AO fluorescence (Fig. 2C). These data demonstrated that PDT-stimulated autophagy is robustly active in PROM1/CD133+ cells.
Figure 2. PpIX-mediated PDT leads to an increased level of autophagy in PROM1/CD133+ cells. (A) PROM1/CD133− and PROM1/CD133+ cells were treated with PDT (1.3 J/cm2). After 24 h, total cell lysates were analyzed by western blot. The band intensities on films were analyzed by ImageJ software. The relative amounts of LC3-II were quantified as ratios to ACTB, indicated underneath each gel. The relative ratio of LC3-II in PROM1/CD133− cells without PDT treatment is arbitrarily presented as 1. (B) Cells were labeled with anti-LC3 primary antibody and DyLight 488 conjugated secondary antibody at 24 h after PDT (1.3 J/cm2). LC3 puncta were observed by immunofluorescence using confocal microscopy. Nuclei were counterstained with DAPI. Scale bar: 10 μm. The number of LC3 puncta was counted using 20 cells for each condition. (C) Flow cytometric analysis of AO staining for cells at 24 h post PDT treatment (1.3 J/cm2). These results are expressed as the mean ± SE of 3 different experiments. *P < 0.05.
PDT-induced autophagy protects PROM1/CD133+ cells from PDT-induced cell death
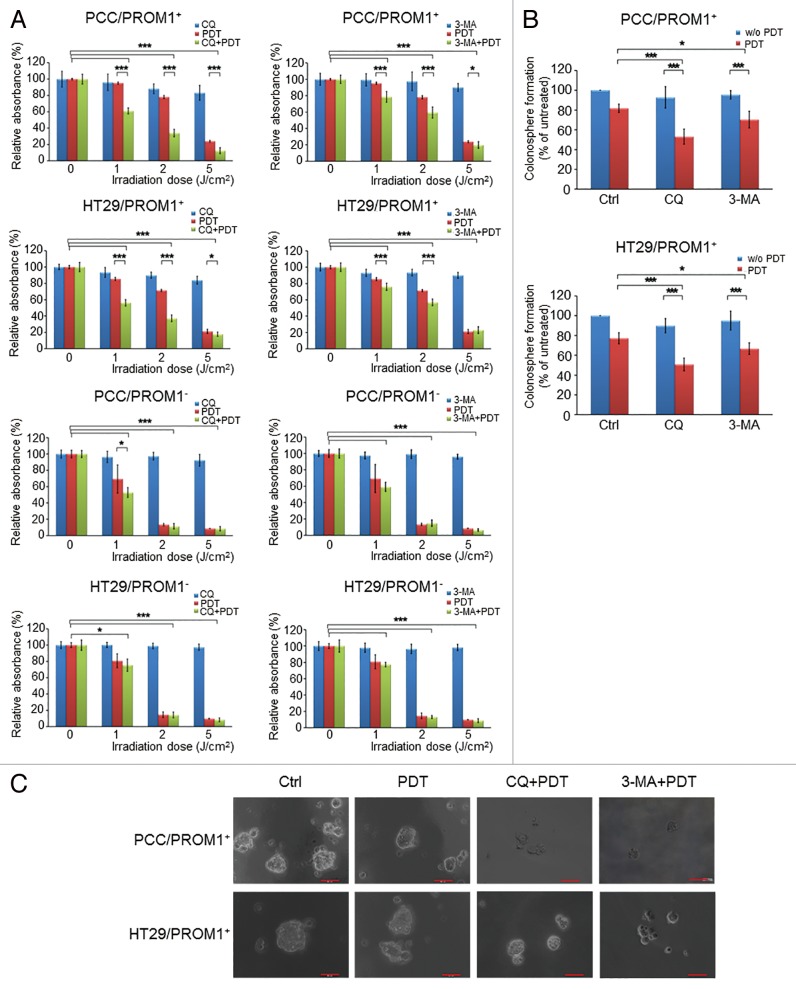
To understand whether the autophagy induced by PDT has a role in PROM1/CD133+ cell escape from PDT treatment, the autophagy inhibitors chloroquine (CQ) and 3-methyladenine (3-MA) were used to examine the cytotoxicity of PCC and HT-29 cells treated with PDT. To determine the concentrations that effectively suppress autophagy but have only minimal cytotoxicity, CQ and 3-MA were analyzed by WST-1 assays and western blotting (Fig. S5 and S6). The appropriate concentrations were found to be 10 µM and 5 mM for CQ and 3-MA, respectively. Treatment of PROM1/CD133+ PCCs with either CQ or 3-MA significantly increased cytotoxicity in the PROM1/CD133+ cells treated with PDT compared with cells that did not receive CQ or 3-MA (Fig. 3A). Similar results were observed in PROM1/CD133+ HT29 cells. In PROM1/CD133- cells, no differences were observed in the cytotoxicity for cells treated with PDT in the presence or absence of CQ or 3-MA.
Figure 3. PpIX-mediated PDT combined with autophagy inhibitors enhanced the cytotoxic effect and blocked colonosphere formation in CSCs. (A) The PROM1/CD133+ and PROM1/CD133− cells form PCCs and HT29 cells were treated with chloroquine (10 μM) or 3-MA (5 mM) for 24 h followed by PDT at various light doses. Cytotoxicity was measured using the WST-1 assay. (B) and (C) The disaggregated PROM1/CD133+ cells were plated at 100 cells/well in an ultra-low attachment plate and treated with PDT (1.3 J/cm2) in the presence of CQ or 3-MA. Colonospheres that contained at least 20 cells were counted in 8 different wells. Representative images show the sizes of the newly formed colonospheres after 7 d. Scale bar: 50 μm. The results are expressed as the mean ± SE of 3 different experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
The ability to form spheres in nonadherent culture is one of the characteristics of CSCs. Descriptions of sphere-forming cells play an important role in understanding the maintenance of tumor growth and the ability of cancers to develop resistance to conventional therapy. To examine whether autophagy has a role in the ability of self-renewing sphere-forming cells to resist PDT, the sensitivity of colonosphere-forming cells to PDT alone or in combination with autophagy inhibitors was assessed. Spheres containing more than 20 cells were scored, and the PDT irradiation dose of 1.3 J/cm2 was applied. We found that the number of colonospheres was reduced when they had been treated with CQ and subjected to PDT (Fig. 3B). Consistently, inhibition of autophagy with either CQ or 3-MA in PROM1/CD133+ cells significantly decreased the ability of PROM1/CD133+ cells to form colonospheres following PDT treatment. In addition, the size of the newly formed spheres was markedly smaller (sphere size is smaller than 50 µm) in the group treated with PDT (1.3 J/cm2) in combination with autophagy inhibitors than in the control group or the group treated with PDT alone (Fig. 3C). These data indicated that, in PROM1/CD133+ cells treated with PDT in combination with autophagy inhibitors, not only was the cytotoxicity increased, but sphere-forming capacity of colon CSCs was also affected. Collectively, these data showed that induction of autophagy by PDT treatment contributes to the ability of PROM1/CD133+ cells to proliferate and self-renew, which makes PROM1/CD133+ cells resistant to PDT.
Silencing of autophagy-related genes sensitized PROM1/CD133+ cells to PDT
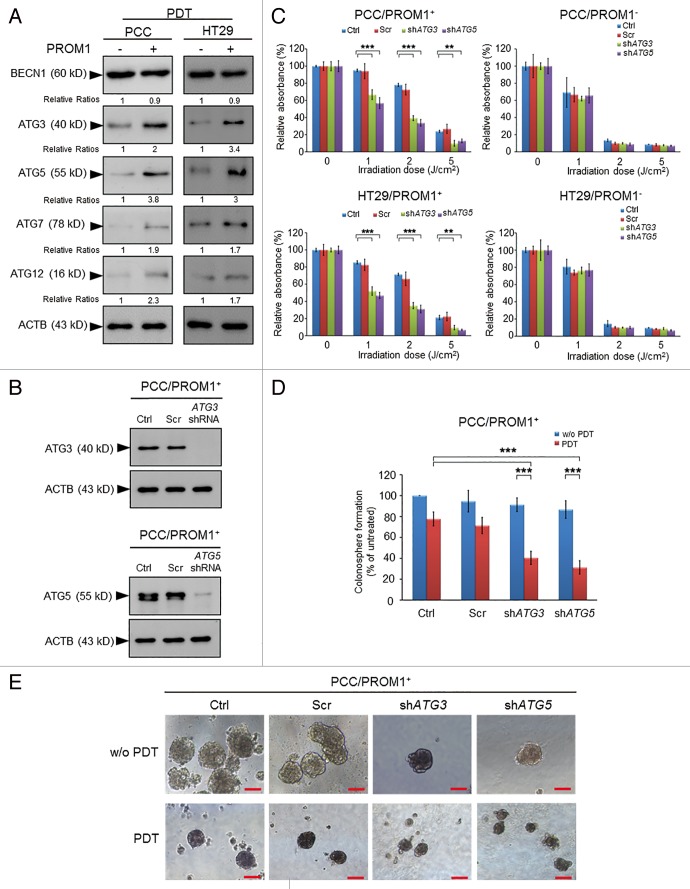
Autophagy-related proteins are involved in phagophore induction, cytoplasm engulfment, autophagic vesicle formation, and lysosomal fusion. The study demonstrated that PROM1/CD133+ cells express higher levels of the autophagy-related proteins LC3, ATG5, and ATG12, which are involved in autophagosome formation, than PROM1/CD133- cells after irradiation, and inhibition of autophagy preferentially sensitizes PROM1/CD133+ cells to irradiation and decreases their sphere-forming capacity.43 To further understand the molecular machinery of autophagy induced by PDT, western blotting was performed. The PROM1/CD133+ cells treated with PDT (1.3 J/cm2) expressed higher levels of the autophagy-related proteins, including ATG3, ATG5, ATG7, and ATG12, than PROM1/CD133- cells (Fig. 4A). This result indicated that PDT-induced ATG expression in PROM1/CD133+ cells may be responsible for the increase in the levels of autophagy after PDT.
Figure 4. Genetic silencing of autophagy enhances the cytotoxic effect of PpIX-mediated PDT in the CSCs. (A) The total extracts of PROM1/CD133− and PROM1/CD133+ cells were harvested at 24 h post-PDT (1.3 J/cm2). The levels of ACTB and autophagy-related proteins, including ATG3, ATG5, ATG7, ATG12, and BECN1, were assessed by western blot analysis. The band intensities on films were analyzed by ImageJ software. The relative amounts of each protein were quantified as ratios to ACTB, indicated underneath each gel. The relative ratio of each protein in PROM1/CD133− cells is arbitrarily presented as 1. (B) The PROM1/CD133+ PCCs were infected with pGIPZ lentivirus (scrambled), ATG3 or ATG5 shRNA lentivirus. The protein expression of ATG3 and ATG5 was examined by western blot analysis. (C) The PROM1/CD133+ PCCs and PROM1/CD133+ HT29 cells that expressed specific shRNA were treated with PDT at various light doses. The cytotoxicity was then measured using the WST-1 assay. (D and E) The disaggregated PROM1/CD133+ PCCs that expressed specific shRNA were plated at 100 cells/well in an ultra-low attachment plate and treated with PDT (1.3 J/cm2). The number and size of the colonospheres were assessed as described for Figure 3. Scale bar: 50 μm. The results are expressed as the mean ± SE of 3 different experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
To further elucidate whether ATG mediate PDT-induced autophagy and contribute to PDT resistance in PROM1/CD133+ cells, a knockdown experiment was performed. As chemical inhibitors of autophagy may lead to off-target effects, specific shRNAs of ATG3 and ATG5, which are genes whose protein products are involved in 2 essential ubiquitin-like conjugation systems in autophagy, were used. Western blot analysis showed efficient knockdown of ATG3 and ATG5 in PROM1/CD133+ cells (Fig. 4B). We further examined the influence of shATG3 and shATG5 on PDT-induced LC3-II accumulation in PROM1/CD133+ cells (Fig. S7). LC3-II accumulation was markedly decreased in ATG-silenced PROM1/CD133+ cells after PDT.
Next, we examined the cytotoxicity of the ATG-silenced PROM1/CD133+ PCCs subjected to PDT. Silencing of ATG3 or ATG5 significantly increased the cytotoxicity of PDT in PROM1/CD133+ cells compared with the effect observed by scrambled shRNA in PROM1/CD133+ cells, and silencing of ATG3 or ATG5 without PDT treatment did not cause significant cell death (Fig. 4C). In PROM1/CD133- PCCs, no differences were observed in the cytotoxicity for cells treated with PDT either in the presence or absence of silencing of ATG3 or ATG5. Similar results were observed in both the PROM1/CD133+ and PROM1/CD133- populations of HT29 cells. In addition, the ability of ATG-silenced PROM1/CD133+ PCCs to form colonospheres was also significantly reduced, resembling the results caused by the pharmacological inhibitors (Fig. 4D and E). The genetic silencing of ATGs remarkably improved the cytotoxicity of PDT and attenuated the stemness-associated properties of the PROM1/CD133+ cells. Together, these data further showed that after PDT treatment, PROM1/CD133+ cells not only expressed more ATGs but also activated higher levels of autophagy to lead to PDT resistance.
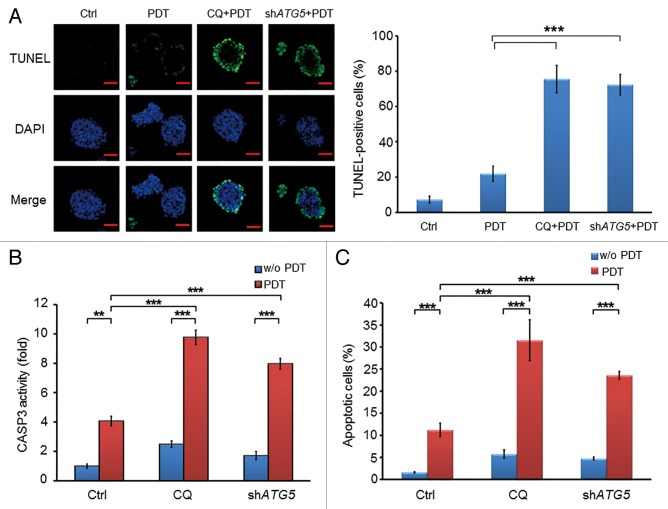
Inhibition of PDT-induced autophagy enhances the apoptotic effect of PDT in PROM1/CD133+ cells
Many previous reports have indicated that autophagy can perform 2 seemingly opposite roles, prosurvival or prodeath, and it can cause resistance to apoptosis when induced by anticancer therapy.44 It is well known that PDT can induce cell death through apoptosis or necrosis both in vitro and in vivo.45 Here, we investigated whether inhibition of autophagy enhances apoptotic effects in PROM1/CD133+ cells. Both chemical inhibition of autophagy and depletion of ATG5 in PROM1/CD133+ PCCs resulted in massive apoptosis following PDT detected by a TUNEL assay compared with the control group or the group treated with PDT alone (Fig. 5A). In addition, CASP3/caspase-3 activity and ANXA5-FITC-PI staining were performed to measure the apoptosis level in each treatment group. As presented in Figure 5B and C, both chemical inhibition of autophagy and depletion of ATG5 enhanced apoptosis after PDT treatment, as shown by a significant elevation of CASP3 activity and an increase in the percentage of ANXA5-positive cells. In contrast, without PDT treatment, CQ-treated or ATG5-silenced PROM1/CD133+ cells showed only slight increases in apoptosis. These results indicated that autophagy plays a protective role for PROM1/CD133+ cells by modulating apoptosis in response to PDT. Therefore, autophagy acts as an antiapoptotic mechanism for CSCs under stress induced by PDT.
Figure 5. Inhibition of autophagy by chloroquine or ATG5 shRNA enhanced the apoptotic effect of PDT in CSCs. (A) Apoptotic cells of PROM1/CD133+ PCCs were visualized by TUNEL assay at 24 h post-PDT (1.3 J/cm2). Nuclei were specifically labeled using DAPI staining. Scale bar: 50 μm. The percentage of TUNEL-positive cells was scored per 100 cells. (B) Whole cell lysates of PROM1/CD133+ PCCs were collected at 24 h post-PDT (1.3 J/cm2) and measured by CaspACE™ Assay System for CASP3 activities. (C) The percentage of apoptotic cells of PROM1/CD133+ PCCs was monitored by ANXA5-FITC-PtdIns staining and flow cytometry at 24 h post-PDT (1.3 J/cm2), and apoptotic ratio was calculated by plotting ANXA5-FITC-positive cell number against total cell number. These results are expressed as the mean ± SE of 3 different experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Blocking autophagy enhances the antitumorigenicity of PDT in a xenograft model
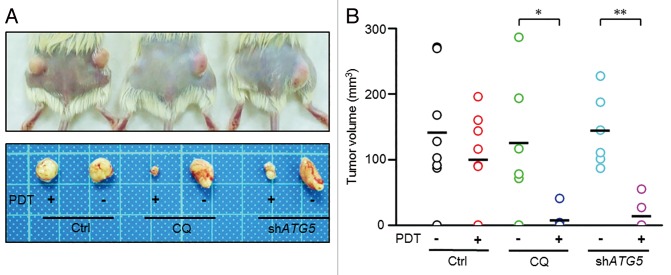
To assess the PDT resistance effect of autophagy in PROM1/CD133+ cells, in vivo tumorigenicity after PDT pretreatment in vitro was assayed. Specifically, the in vivo tumorigenicity of PROM1/CD133+ cells pretreated with CQ or shATG5 following PDT treatment (1.3 J/cm2) in vitro in NOD/SCID mice were analyzed. For the positive control, mice were inoculated with 1 × 103 PROM1/CD133+ cells, and 87.5% (7/8) of the mice in that group developed tumors. For the experimental group, mice were inoculated with 1 × 103 viable PROM1/CD133+ cells that were subjected to PDT, and 75% (6/8) of the mice in that group developed tumors. This result indicates PDT did not suppress in vivo tumorigenicity. The incidence of tumor formation was lower when CQ-treated or ATG5-silenced PROM1/CD133+ cells subjected to PDT were injected into NOD/SCID mice (both are 2/6) than when CQ-treated or ATG5-silenced PROM1/CD133+ cells that were not subjected to PDT were injected into NOD/SCID mice (Table 1). As expected, tumor volume was lowest with CQ-treated or ATG5-silenced PROM1/CD133+ cells following PDT; however, CQ-treated or ATG5-silenced PROM1/CD133+ cells without PDT did not affect tumor growth (Fig. 6A). These results were consistent with the findings of the in vitro assay of self-renewal potential (Fig. 3; Fig. 4).
Table 1. Tumor-initiating potential of PROM1/CD133+ PCCs in each treatment group.
| Cells injected | 1 × 103 |
|---|---|
| Control | 7/8 |
| PDT | 6/8 |
| CQ | 5/6 |
| CQ+PDT | 2/6 |
| shATG5 | 6/6 |
| shATG5+PDT | 2/6 |
A total of 1 × 103 viable PROM1/CD133+ PCCs, pretreated with PDT, CQ, shATG5 lentivirus, either alone or in the indicated combinations, were implanted subcutaneously into both flanks of NOD/SCID mice. Tumor-initiating potential in each treatment group was observed.
Figure 6. Inhibition of autophagy enhanced the antitumorigenicity of PDT in a xenograft model. (A) A total of 1 × 103 viable PROM1/CD133+ PCCs, pretreated with CQ (10 μM), ATG5 shRNA lentivirus, or PDT (1.3 J/cm2), either alone or in the indicated combinations, were implanted subcutaneously into both flanks of NOD/SCID mice. Representative pictures of each treatment group. Mice were sacrificed 12 wk after implantation. (B) Tumor sizes were measured at the end of the experiment. Data are presented as dot plots, and the short black lines indicate mean tumor size. Tumor volume was calculated as (L * W2)/2, where L is the length and W is the width of the tumor. The tumors in CQ-treated and ATG5-silenced groups were significantly distributed in lower tumor volume ranges with PDT compared with without PDT. *P < 0.05; **P < 0.01.
Subsequently, these tumor burdens were measured. The results showed that the tumors in CQ-treated and ATG5-silenced groups were significantly distributed in lower tumor volume ranges with PDT compared with without PDT (Fig. 6B). Taken together, our results suggested that PDT combined with autophagy inhibition was effective in reducing the tumorigenic potential of PROM1/CD133+ cells. These findings not only further verified that PDT-induced autophagy is responsible for PDT resistance in PROM1/CD133+ cells but also opened a new avenue for potentially treating CSC through combination therapy of PDT and autophagy inhibition.
Discussion
Currently, all existing therapeutic approaches for CRC have high risks of recurrence. The hypothesis of cancer stem-like cells (CSCs) identifies the possibility that a small subpopulation of cancer cells may result in therapeutic failure for cancer patients.14 Over the years, CSCs have been specifically shown to cause tumor relapse and therapeutic resistance;46 therefore, it is important to find the mechanisms of resistance to anticancer therapies in CSCs.
So far, CSCs are thought to demonstrate resistance to chemotherapeutic agents and radiation mainly due to their expressions of multidrug transporter family genes and DNA damage response genes.18-21 Numerous studies have indicated that ATP-binding cassette (ABC) transporters can affect PDT efficacy by pumping out photosensitizers.11,47 However, from our unpublished data, we have observed that PROM1/CD133+ cells, which are considered CSCs for CRC, can accumulate more PpIX in comparison to PROM1/CD133- cells. This result implies that there must be other mechanisms of resistance to PDT in CSCs. PDT can induce cell death by generating many ROS which are believed to be key regulators of autophagy in cancer cells.7,8,48-50 Autophagy is an important mechanism mediating the resistance of cancer cells to various therapeutic strategies.29,35-40 Therefore, we inferred that autophagy might be a major cause of PDT-resistance in CSCs.
In a previous study, the level of LC3-II and autophagosome synthesis are higher in mammospheres than in adherent cells under both basal and starvation conditions.51 In addition, PROM1/CD133+ cells show higher activity of autophagy by forming more autophagic puncta and causing more accumulation of LC3-II compared with PROM1/CD133- cells under low-nutrient conditions.52 Similarly, we had also observed increased autophagic characteristics in PROM1/CD133+ cells in response to PpIX-mediated PDT. This finding implied that PDT could result in high levels of autophagy activation in CSCs. Simultaneously, we found that PROM1/CD133+ cells with the stimulus of PDT expressed higher levels of the autophagy-related proteins, ATG3, ATG5, and ATG7. The increased expression of these autophagy-related proteins may enhance the levels of autophagy in PDT-treated CSCs. Furthermore, we demonstrated that autophagy inhibition with pharmacological inhibitors or silencing of ATG3 and ATG5 could significantly enhance the sensitivity of PROM1/CD133+ cells to PpIX-mediated PDT by a cytotoxicity and colonosphere formation assay. However, there were no differences in the cytotoxicity of PROM1/CD133- cells undergoing PDT treatments either in the presence or absence of CQ. This result suggested that autophagy most likely plays no role in influencing PROM1/CD133- cells’ response to PDT. Our result is consistent with a previous report demonstrating that autophagy exerts a protective role against chemotherapy and radiotherapy in CSCs.43,53 These findings all highlighted the fact that autophagy must contribute to the resistance of CSCs to PDT.
It is well known that PDT-generated ROS could concurrently induce both autophagy and apoptosis pathways in cancer cells.54,55 Accumulating data propose that autophagy may antagonize apoptosis.56-59 This evidence indicates that cancer cells could enhance autophagy to decrease apoptosis. On the other hand, inhibition of autophagic flux could accelerate the proapoptotic capacity of therapeutic modalities when treating CSCs.60 Our study also consistently showed that PROM1/CD133+ cells with autophagy inhibition were more susceptible to PDT-mediated apoptosis. To further elucidate the autophagy/apoptosis interconnection, we discussed the roles played by ATG3 and ATG5. ATG3 and ATG5 are both essential regulatory components of autophagosome biogenesis.61,62 A recent report shows that ATG3 degradation by CASP8 could lead to autophagy inactivation during receptor-triggered apoptosis.63 Full-length ATG5 can participate in autophagy activation.64,65 Under lethal stress, ATG5 is cleaved by CAPN/calpains, which triggers apoptosis via induction of MOMP (mitochondrial outer membrane permeabilization) or interaction with FADD [Fas (TNFRSF6)-associated via death domain].64,66
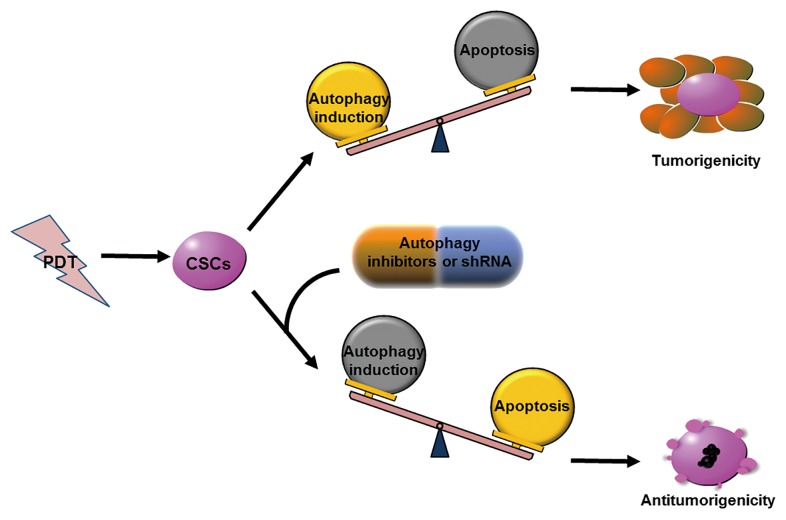
In both CSCs and malignant cells, autophagy could support tumor establishment and development.67 Our present study clearly showed that the therapeutic strategy of PDT combined with autophagy inhibition could decrease tumorigenicity of PROM1/CD133+ cells in a xenograft model. Even the tumors that grew under combination treatment had significantly suppressed size. Altogether, it is very probable that autophagy inhibition played a key role in increasing apoptosis of PDT-treated PROM1/CD133+ cells and the consequent tumorigenicity reduction. However, when PROM1/CD133+ cells underwent PDT treatment without autophagy inhibition, there was no effect on tumorigenicity or tumor volume. This most likely indicates that CSC properties are affected by autophagy inhibition. Our data support a model in which autophagy inhibition enhances CSC sensitivity to PDT and PDT-induced apoptosis, and dramatically reduces the tumorigenicity of CSCs under PDT treatment (Fig. 7).
Figure 7. Model of antitumorigenicity ability of PDT combined with autophagy inhibition. Autophagy inhibition enhances CSC sensitivity to PDT and PDT-induced apoptosis, and dramatically reduces the tumorigenicity of CSCs under PDT treatment.
Overall, we demonstrated that PpIX-mediated PDT could significantly upregulate the levels of autophagy in PROM1/CD133+ cells. Suppressing autophagy could sensitize PROM1/CD133+ cells to apoptosis induced by PDT. In vitro spheroid formation and the in vivo tumorigenicity of PROM1/CD133+ cells were decreased after autophagy had been blocked. These results all imply that enhanced autophagy levels are related to the increase of resistance to PDT in CSCs. Therefore, our study not only revealed the possible mechanisms executed by autophagy in the resistance and tumorigenicity of CSCs after PDT but also provided a new therapeutic avenue for targeting autophagy in anti-CSC treatment with PDT.
Materials and Methods
Materials
PpIX was obtained from Sigma-Aldrich (P8293), dissolved in 100% dimethyl sulfoxide (DMSO; Sigma-Aldrich, D2650) to a concentration of 2 mg/ml. Chloroquine (Sigma-Aldrich, C6628) and 3-MA (Calbiochem, 189490) were dissolved in deionized water to create 1 mM and 100 mM stock solutions, respectively. The antibodies used included CD133/2-phycoerythrin (PE) (Miltenyi Biotec, 130-090-853) and isotype control IgG2b-PE (Miltenyi Biotec, 130-092-215).
PROM1/CD133+ cell isolation and culture
The colorectal cancer primary cultured cells (PCCs) were kindly provided by SC Hung (Institute of Clinical Medicine, School of Medicine, National Yang-Ming University, Taipei, Taiwan). After the PCCs and the HT29 cells (purchased from American Type Culture Collection, HTB-38 ™) were collected and stained with a CD133/2-PE antibody for 30 min, the PROM1/CD133+ cells were sorted using a FACSAria-I cell sorter (Becton Dickinson, CA, USA). The isolated PROM1/CD133+ cells were cultured under stem-cell conditions in serum-free DMEM/F12 medium (Gibco, 12500-062) supplemented with 20 ng/mL of human recombinant epidermal growth factor (EGF; PeproTech, AF-100-15), 10 ng/mL of FGF2 [fibroblast growth factor 2 (basic)] (bFGF; PeproTech, 100-18B), and 1% N-2 supplement (Gibco, 17502-048). The cells were cultured on ultra-low attachment plates (Corning Inc., 3471) for subsequent organization into spheres.68
Quantitative real-time PCR
Total RNA was isolated from PROM1/CD133− and PROM1/CD133+ cells from PCC and HT29 cells using the PureLink Micro-to-Midi Total RNA Purification system (Invitrogen, 12183-018). The reverse transcription reaction was performed using the SuperScript III First-Strand Synthesis system for RT-PCR (Invitrogen, 18080-051). Expression of SOX2, OCT4, NANOG, and CTNNB1/β-catenin was quantified using the ABI Prism 7900 real-time system (Applied Biosystems, Foster City, CA, USA) and their expression is normalized to GAPDH. All reactions were performed in triplicate. The following primers were used: SOX2: forward 5′-GTATCAGGAG TTGTCAAGGC-3′, reverse 5′-AGTCCTAGTC TTAAAGAGG-3′; OCT4: forward 5′-GCTCACCCTG GGCGTTCTC-3′, reverse 5′-GGCCGCAGCT TACACATGTTC-3′; NANOG: forward 5′-CCTCCAGCAG ATGCAAGAAC TC-3′, reverse 5′-CTTCAACCAC TGGTTTTTCT GCC-3′; CTNNB1: forward 5′-GAAACGGCTT TCAGTTGAGC-3′, reverse 5′-CTGGCCATAT CCACCAGAGT-3′; GAPDH: forward 5′-CATGAGAAGT ATGACAACAG CCT-3′, reverse 5′-AGTCCTTCCA CGATACCAAA GT-3′.
The identification of resistance to PDT
The PROM1/CD133+ and PROM1/CD133− cells were incubated in the dark for 3 h with 1 µg/mL PpIX. Then, the cells were washed twice with phosphate-buffered saline (PBS) and exposed to various doses (0 J/cm2, 1 J/cm2, and 5 J/cm2) of red light generated by a light-emitting diode (LED; 633 nm) for photodynamic treatment. The cells were then incubated in fresh medium under standard cell culture conditions for 24 h. PDT-induced cell death was analyzed using the Dead Cell Apoptosis Kit with ANXA5-FITC and PI (Molecular Probes®, V13242). The percentage of apoptotic cells was measured by flow cytometry. The results are expressed as the mean ± SE of 3 different experiments.
Immunofluorescence
The PROM1/CD133+ and PROM1/CD133− cells were plated on Tissue Culture Treated Glass Slide (BD Falcon, 354114) and subjected to PDT treatment (1.3 J/cm2). After 24 h, cells were washed with PBS and fixed with 3.7% paraformaldehyde for 8 min. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min, blocked with 3% BSA for 60 min, and incubated with anti-LC3 antibodies (1:200) (Cell Signaling Technology, 3868S) overnight. After washing with PBS, cells were incubated for 1 h with DyLight 488 conjugated secondary antibody (1:500) (BioLink Biotechnology, B0210). Nuclei were counterstained with DAPI (Biotium, 40043) for 5 min. Slides were mounted with a coverslip using Fluoro-Gel (Electron Microscopy Sciences, 17985-10). Cells were imaged using a confocal laser scanning microscope (TCS SP5, Leica Microsystems, Wetzlar, Germany), and the images were analyzed using a Leica Application Suite 2.02. DyLight 488 was excited using an argon laser (488 nm), and DAPI was excited using a diode laser (405 nm). The emission lights for DAPI and DyLight 488 were collected between 410 and 490 nm and between 495 and 540 nm, respectively.
Acridine orange staining
PROM1/CD133+ and PROM1/CD133− cells were treated with PDT (1.3 J/cm2) and stained with AO (1 μg/ml) (Sigma-Aldrich, A6014) at 37 °C for 30 min after 24 h post PDT treatment. After removing AO, the stained cells were immediately analyzed in the FL-3 channel using a FACSCalibur (Becton Dickinson, BD FACSCalibur™ system, CA, USA).
Cytotoxicity assay
Disaggregated PROM1/CD133+ and PROM1/CD133− cells were plated in 100 μl of medium at a concentration of 8,000 cells/well in 96-well plates and treated with PDT at various light doses in the presence or absence of CQ (10 μM), 3-MA (5 mM), shATG3, and shATG5. After 24 h, cell survival was measured using a Premixed WST-1 Cell Proliferation Reagent (Clontech, 630118), and the absorbance of each sample was measured at 450 nm against the background control using a multi-well plate reader. Cytotoxicity was expressed as absorbance percentage of the control. The data are expressed as the mean ± SE of 3 different experiments. *P < 0.05; **P < 0.01; ***P < 0.001
Colonosphere formation assay
Disaggregated PROM1/CD133+ cells were plated at 100 cells/well in a 24-well ultra-low attachment plate and treated with PDT (1.3 J/cm2) either in the presence and absence of CQ (10 μM), 3-MA (5 mM), shATG3, and shATG5. The number of the newly formed colonospheres/well was determined after 7 d. Colonospheres containing more than 20 cells were quantified for 8 different wells. The results are expressed as the mean ± SE of 3 different experiments.
Western blotting
Cell lysates were harvested from each cell subclone by resuspending the cells in 1× RIPA Lysis Buffer (Millipore, 20-188) at 24 h after PDT treatment (1.3 J/cm2). Equal amounts of protein extracts were resolved on SDS-PAGE gels and transferred to a PVDF membrane (Millipore, IPVH00010). Primary antibodies against the following molecules were used for the analysis: BECN1 (3495P), ATG3 (3415P), ATG5 (2630S), ATG7 (2631S), ATG12 (4180P) were purchased from Cell Signaling Technology. LC3B (GTX100240) and ACTB/β-ACTIN (GTX110564) were obtained from GeneTex Inc. The specific reactive bands were detected using a goat anti-rabbit IgG antibody conjugated to horse radish peroxidase (GeneTex Inc., GTX213110-01). The immune complexes were visualized using the Western Lightning® Plus-ECL (PerkinElmer Inc., NEL104001EA), and quantification was performed using Image Quant software (GE Healthcare).
Lentivirus production and transduction
The shRNA-expression vectors, ATG3 (TRCN0000147381) and ATG5 (TRCN0000151474) shRNA constructs, were purchased from the National RNAi Core Facility (Taipei, Taiwan). For virus packaging, psPAX2 (Addgene, Plasmid 12260), pMD2.G (Addgene, Plasmid 12259), and shRNA-expression vectors were cotransfected into 293T cells (form National RNAi Core Facility). The supernatant was harvested after 48 h. The viral supernatant fraction was filtered and used to infect PROM1/CD133+ cells in the presence of 8 μg/ml Polybrene® (Santa Cruz, sc-134220). Subsequently, these infected cells were selected by 2 μg/ml puromycin (InvivoGen, ant-pr-1). A pGIPZ lentiviral vector (Fisher Scientific, NC 9668406) was used as a negative control.
Apoptosis assay
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) was performed according to the manufacturer’s instructions of DeadEnd Fluorometric TUNEL system (Promega, G3250). Confocal imaging of apoptotic cells on slides was acquired using a LSM 5 confocal laser scanning microscope (Carl Zeiss MicroImaging, Jena, Germany) using LSM 5 Pascal software version 3.0. For excitation, a 488 nm argon laser was used for fluorescein-12-dUTP (emission collected with BP 520–555 nm filter), and the 405 nm diode laser was used for DAPI (emission collected with BP 415–480 nm filter).
The CASP3 activity assay was performed using the CaspACE™ Assay System, Colorimetric (Promega, G7220) according to the manufacturer’s instructions. The results are presented as the mean CASP3 activity (± SE) of 3 different experiments.
Animal experiments
NOD/SCID mice were purchased from the National Laboratory Animal Center, Taiwan. All in vivo experimental procedures were approved by the National Taiwan University College of Medicine and College of Public Health Institutional Animal Care and Use Committee (IACUC). For xenograft tumor-seeding studies, PROM1/CD133+ PCCs pretreated with PDT (1.3 J/cm2), CQ (10 μM), or shATG5, either alone or in the indicated combinations, were allowed to recover for 3 d in the absence of treatment. Then, 1 × 103 cells were injected subcutaneously into the flanks of NOD/SCID mice. Tumor incidence was observed for 12 wk after implantation. Tumor sizes were measured with calipers, and tumor volume was calculated as (L * W2)/2 where L is the length and W is the width of the tumor.
Statistical analysis
All data from the WST-1 assays are expressed as the mean ± standard deviation. The P values were determined using an unpaired Student t test. P values < 0.05 were considered statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was funded by the National Science Council, ROC (NSC 100-2218-E-002-002) and the Ministry of Health and Welfare, ROC (MOHW103-TDU-N-211-133006).
We would like to acknowledge the service provided by the Flow Cytometric Analysis and Sorting Core Facility of the First Core Laboratory, National Taiwan University College of Medicine and the Core Laboratory of the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation.
Glossary
Abbreviations:
- 3-MA
3-methyladenine
- ABC
ATP-binding cassette
- ACTB
β-ACTIN
- ANXA5
annexin V
- AO
acridine orange
- ATG
autophagy-related
- BECN1
Beclin 1, autophagy-related
- CASP
caspase
- CQ
chloroquine
- CRC
colorectal cancer
- CSCs
cancer stem-like cells
- CTNNB1
β-catenin
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- EGF
epidermal growth factor
- FACS
fluorescence-activated cell sorting
- FITC/PI
fluorescein isothiocyanate/propidium iodide
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IgG
immunoglobulin G
- MAP1LC3A/B (LC3)
microtubule-associated protein 1A/1B-light chain 3
- LED
light-emitting diode
- MTOR
mechanistic target of rapamycin
- PBS
phosphate-buffered saline
- PCCs
primary cultured cells
- PDT
photodynamic therapy
- PE
phycoerythrin
- PpIX
protoporphyrin IX
- RNA
ribonucleic acid
- ROS
reactive oxygen species
- RT-PCR
reverse transcription-polymerase chain reaction
- shRNA
short hairpin RNA
- SP
side population
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Mulsow J, Merkel S, Agaimy A, Hohenberger W. Outcomes following surgery for colorectal cancer with synchronous peritoneal metastases. Br J Surg. 2011;98:1785–91. doi: 10.1002/bjs.7653. [DOI] [PubMed] [Google Scholar]
- 3.Wang JB, Liu LX. Use of photodynamic therapy in malignant lesions of stomach, bile duct, pancreas, colon and rectum. Hepatogastroenterology. 2007;54:718–24. [PubMed] [Google Scholar]
- 4.Geltzer A, Turalba A, Vedula SS. Surgical implantation of steroids with antiangiogenic characteristics for treating neovascular age-related macular degeneration. Cochrane Database Syst Rev. 2013;1:CD005022. doi: 10.1002/14651858.CD005022.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moan J, Berg K. Photochemotherapy of cancer: experimental research. Photochem Photobiol. 1992;55:931–48. doi: 10.1111/j.1751-1097.1992.tb08541.x. [DOI] [PubMed] [Google Scholar]
- 6.Weishaupt KR, Gomer CJ, Dougherty TJ. Identification of singlet oxygen as the cytotoxic agent in photoinactivation of a murine tumor. Cancer Res. 1976;36:2326–9. [PubMed] [Google Scholar]
- 7.Peng Q, Moan J, Nesland JM. Correlation of subcellular and intratumoral photosensitizer localization with ultrastructural features after photodynamic therapy. Ultrastruct Pathol. 1996;20:109–29. doi: 10.3109/01913129609016306. [DOI] [PubMed] [Google Scholar]
- 8.Piette J, Volanti C, Vantieghem A, Matroule JY, Habraken Y, Agostinis P. Cell death and growth arrest in response to photodynamic therapy with membrane-bound photosensitizers. Biochem Pharmacol. 2003;66:1651–9. doi: 10.1016/S0006-2952(03)00539-2. [DOI] [PubMed] [Google Scholar]
- 9.Gomer CJ, Ryter SW, Ferrario A, Rucker N, Wong S, Fisher AM. Photodynamic therapy-mediated oxidative stress can induce expression of heat shock proteins. Cancer Res. 1996;56:2355–60. [PubMed] [Google Scholar]
- 10.Xue LY, Agarwal ML, Varnes ME. Elevation of GRP-78 and loss of HSP-70 following photodynamic treatment of V79 cells: sensitization by nigericin. Photochem Photobiol. 1995;62:135–43. doi: 10.1111/j.1751-1097.1995.tb05249.x. [DOI] [PubMed] [Google Scholar]
- 11.Robey RW, Steadman K, Polgar O, Bates SE. ABCG2-mediated transport of photosensitizers: potential impact on photodynamic therapy. Cancer Biol Ther. 2005;4:187–94. doi: 10.4161/cbt.4.2.1440. [DOI] [PubMed] [Google Scholar]
- 12.Gomer CJ, Luna M, Ferrario A, Rucker N. Increased transcription and translation of heme oxygenase in Chinese hamster fibroblasts following photodynamic stress or Photofrin II incubation. Photochem Photobiol. 1991;53:275–9. doi: 10.1111/j.1751-1097.1991.tb03934.x. [DOI] [PubMed] [Google Scholar]
- 13.Brackett CM, Owczarczak B, Ramsey K, Maier PG, Gollnick SO. IL-6 potentiates tumor resistance to photodynamic therapy (PDT) Lasers Surg Med. 2011;43:676–85. doi: 10.1002/lsm.21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–61. doi: 10.1056/NEJMra061808. [Review] [DOI] [PubMed] [Google Scholar]
- 15.Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–7. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 17.Shmelkov SV, St Clair R, Lyden D, Rafii S. AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 2005;37:715–9. doi: 10.1016/j.biocel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Jin F, Zhao L, Zhao HY, Guo SG, Feng J, Jiang XB, Zhang SL, Wei YJ, Fu R, Zhao JS. Comparison between cells and cancer stem-like cells isolated from glioblastoma and astrocytoma on expression of anti-apoptotic and multidrug resistance-associated protein genes. Neuroscience. 2008;154:541–50. doi: 10.1016/j.neuroscience.2008.03.054. [DOI] [PubMed] [Google Scholar]
- 19.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 20.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8:545–54. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 21.Rich JN. Cancer stem cells in radiation resistance. Cancer Res. 2007;67:8980–4. doi: 10.1158/0008-5472.CAN-07-0895. [DOI] [PubMed] [Google Scholar]
- 22.Morgan J, Jackson JD, Zheng X, Pandey SK, Pandey RK. Substrate affinity of photosensitizers derived from chlorophyll-a: the ABCG2 transporter affects the phototoxic response of side population stem cell-like cancer cells to photodynamic therapy. Mol Pharm. 2010;7:1789–804. doi: 10.1021/mp100154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Eskelinen EL. New insights into the mechanisms of macroautophagy in mammalian cells. Int Rev Cell Mol Biol. 2008;266:207–47. doi: 10.1016/S1937-6448(07)66005-5. [DOI] [PubMed] [Google Scholar]
- 26.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–18. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 27.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim EH, Sohn S, Kwon HJ, Kim SU, Kim MJ, Lee SJ, Choi KS. Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res. 2007;67:6314–24. doi: 10.1158/0008-5472.CAN-06-4217. [DOI] [PubMed] [Google Scholar]
- 29.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–94. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 30.Reiners JJ, Jr., Agostinis P, Berg K, Oleinick NL, Kessel D. Assessing autophagy in the context of photodynamic therapy. Autophagy. 2010;6:7–18. doi: 10.4161/auto.6.1.10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen S, Kepp O, Kroemer G. The end of autophagic cell death? Autophagy. 2012;8:1–3. doi: 10.4161/auto.8.1.16618. [DOI] [PubMed] [Google Scholar]
- 32.Carew JS, Kelly KR, Nawrocki ST. Autophagy as a target for cancer therapy: new developments. Cancer Manag Res. 2012;4:357–65. doi: 10.2147/CMAR.S26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo XL, Li D, Sun K, Wang J, Liu Y, Song JR, Zhao QD, Zhang SS, Deng WJ, Zhao X, et al. Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma. J Mol Med (Berl) 2013;91:473–83. doi: 10.1007/s00109-012-0966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G, Gou X, Lin Y. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy. 2012;8:1811–21. doi: 10.4161/auto.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26:1401–10. [PubMed] [Google Scholar]
- 36.Tseng HC, Liu WS, Tyan YS, Chiang HC, Kuo WH, Chou FP. Sensitizing effect of 3-methyladenine on radiation-induced cytotoxicity in radio-resistant HepG2 cells in vitro and in tumor xenografts. Chem Biol Interact. 2011;192:201–8. doi: 10.1016/j.cbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–58. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 38.Li YY, Lam SK, Mak JC, Zheng CY, Ho JC. Erlotinib-induced autophagy in epidermal growth factor receptor mutated non-small cell lung cancer. Lung Cancer. 2013;81:354–61. doi: 10.1016/j.lungcan.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Pan X, Zhang X, Sun H, Zhang J, Yan M, Zhang H. Autophagy inhibition promotes 5-fluorouraci-induced apoptosis by stimulating ROS formation in human non-small cell lung cancer A549 cells. PLoS One. 2013;8:e56679. doi: 10.1371/journal.pone.0056679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eimer S, Belaud-Rotureau MA, Airiau K, Jeanneteau M, Laharanne E, Véron N, Vital A, Loiseau H, Merlio JP, Belloc F. Autophagy inhibition cooperates with erlotinib to induce glioblastoma cell death. Cancer Biol Ther. 2011;11:1017–27. doi: 10.4161/cbt.11.12.15693. [DOI] [PubMed] [Google Scholar]
- 41.Coupienne I, Bontems S, Dewaele M, Rubio N, Habraken Y, Fulda S, Agostinis P, Piette J. NF-kappaB inhibition improves the sensitivity of human glioblastoma cells to 5-aminolevulinic acid-based photodynamic therapy. Biochem Pharmacol. 2011;81:606–16. doi: 10.1016/j.bcp.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 42.Traganos F, Darzynkiewicz Z. Lysosomal proton pump activity: supravital cell staining with acridine orange differentiates leukocyte subpopulations. Methods Cell Biol. 1994;41:185–94. doi: 10.1016/S0091-679X(08)61717-3. [DOI] [PubMed] [Google Scholar]
- 43.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717–22. doi: 10.1002/ijc.24402. [DOI] [PubMed] [Google Scholar]
- 44.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–9. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oleinick NL, Evans HH. The photobiology of photodynamic therapy: cellular targets and mechanisms. Radiat Res. 1998;150(Suppl):S146–56. doi: 10.2307/3579816. [DOI] [PubMed] [Google Scholar]
- 46.Song LL, Miele L. Cancer stem cells--an old idea that’s new again: implications for the diagnosis and treatment of breast cancer. Expert Opin Biol Ther. 2007;7:431–8. doi: 10.1517/14712598.7.4.431. [DOI] [PubMed] [Google Scholar]
- 47.Jendzelovský R, Mikes J, Koval’ J, Soucek K, Procházková J, Kello M, Sacková V, Hofmanová J, Kozubík A, Fedorocko P. Drug efflux transporters, MRP1 and BCRP, affect the outcome of hypericin-mediated photodynamic therapy in HT-29 adenocarcinoma cells. Photochem Photobiol Sci. 2009;8:1716–23. doi: 10.1039/b9pp00086k. [DOI] [PubMed] [Google Scholar]
- 48.Henderson BW, Busch TM, Vaughan LA, Frawley NP, Babich D, Sosa TA, Zollo JD, Dee AS, Cooper MT, Bellnier DA, et al. Photofrin photodynamic therapy can significantly deplete or preserve oxygenation in human basal cell carcinomas during treatment, depending on fluence rate. Cancer Res. 2000;60:525–9. [PubMed] [Google Scholar]
- 49.Chen Q, Huang Z, Chen H, Shapiro H, Beckers J, Hetzel FW. Improvement of tumor response by manipulation of tumor oxygenation during photodynamic therapy. Photochem Photobiol. 2002;76:197–203. doi: 10.1562/0031-8655(2002)076<0197:IOTRBM>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 50.Gibson SB. A matter of balance between life and death: targeting reactive oxygen species (ROS)-induced autophagy for cancer therapy. Autophagy. 2010;6:835–7. doi: 10.4161/auto.6.7.13335. [DOI] [PubMed] [Google Scholar]
- 51.Gong C, Bauvy C, Tonelli G, Yue W, Deloménie C, Nicolas V, Zhu Y, Domergue V, Marin-Esteban V, Tharinger H, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32:2261–72, 1-11. doi: 10.1038/onc.2012.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H, Luo Z, Dong L, Tan Y, Yang J, Feng G, Wu M, Li Z, Wang H. CD133/prominin-1-mediated autophagy and glucose uptake beneficial for hepatoma cell survival. PLoS One. 2013;8:e56878. doi: 10.1371/journal.pone.0056878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S, Wang X, Chen J, Chen Y. Autophagy of cancer stem cells is involved with chemoresistance of colon cancer cells. Biochem Biophys Res Commun. 2013;434:898–903. doi: 10.1016/j.bbrc.2013.04.053. [DOI] [PubMed] [Google Scholar]
- 54.Sasnauskiene A, Kadziauskas J, Vezelyte N, Jonusiene V, Kirveliene V. Damage targeted to the mitochondrial interior induces autophagy, cell cycle arrest and, only at high doses, apoptosis. Autophagy. 2009;5:743–4. doi: 10.4161/auto.5.5.8701. [DOI] [PubMed] [Google Scholar]
- 55.Dewaele M, Martinet W, Rubio N, Verfaillie T, de Witte PA, Piette J, Agostinis P. Autophagy pathways activated in response to PDT contribute to cell resistance against ROS damage. J Cell Mol Med. 2011;15:1402–14. doi: 10.1111/j.1582-4934.2010.01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bauvy C, Gane P, Arico S, Codogno P, Ogier-Denis E. Autophagy delays sulindac sulfide-induced apoptosis in the human intestinal colon cancer cell line HT-29. Exp Cell Res. 2001;268:139–49. doi: 10.1006/excr.2001.5285. [DOI] [PubMed] [Google Scholar]
- 57.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–40. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, Hosui A, Ishida H, Tatsumi T, Kanto T, et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548–57. doi: 10.1002/ijc.26374. [DOI] [PubMed] [Google Scholar]
- 59.Song J, Guo X, Xie X, Zhao X, Li D, Deng W, Song Y, Shen F, Wu M, Wei L. Autophagy in hypoxia protects cancer cells against apoptosis induced by nutrient deprivation through a Beclin1-dependent way in hepatocellular carcinoma. J Cell Biochem. 2011;112:3406–20. doi: 10.1002/jcb.23274. [DOI] [PubMed] [Google Scholar]
- 60.Yue W, Hamaï A, Tonelli G, Bauvy C, Nicolas V, Tharinger H, Codogno P, Mehrpour M. Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy. 2013;9:714–29. doi: 10.4161/auto.23997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–44. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- 62.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 63.Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis. 2012;17:810–20. doi: 10.1007/s10495-012-0735-0. [DOI] [PubMed] [Google Scholar]
- 64.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 65.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 66.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–9. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 67.Gong C, Song E, Codogno P, Mehrpour M. The roles of BECN1 and autophagy in cancer are context dependent. Autophagy. 2012;8:1853–5. doi: 10.4161/auto.21996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–11. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.