Abstract
Nijmegen breakage syndrome (NBS) is characterized by radiation hypersensitivity, chromosomal instability, and predisposition to cancer. Nbs1, the NBS protein, forms a tight complex with Mre11 and Rad50, and these interactions contribute to proper double-strand break repair. The simian virus 40 (SV40) oncoprotein, large T antigen (T), also interacts with Nbs1, and T-containing cells experience chromosomal hyperreplication in a manner dependent on T/Nbs1 complex formation. A substantial fraction of NBS-deficient fibroblasts reinitiate DNA replication in discrete regions, and wild-type Nbs1 corrects this defect. Similarly, synthesis of an N-terminal Nbs1 fragment induced DNA rereplication and tetraploidy, in NBS-deficient but not NBS-proficient cells. Moreover, SV40 origin-containing DNA hyperreplicated in T-containing NBS-deficient cells by comparison with T-containing, Nbs1-reconstituted derivatives. Thus, Nbs1 suppresses rereplication of cellular DNA and SV40 origin-containing replicons, and T targets Nbs1, thereby enhancing the yield of new SV40 genomes during viral DNA replication.
Keywords: Nbs1, SV40 T, SV40 origin, DNA replication, endoreduplication, mammalian cells
To maintain genome stability, the extent of DNA replication is tightly controlled by a mechanism that ensures that chromosomal DNA is replicated once and only once during S phase. Regulation is achieved, in part, by licensing DNA replication initiation from each origin only once in a given S phase (Diffley 1996, 2001). When SV40 infects mammalian cells, it stimulates cellular DNA synthesis while amplifying its own genome (Hatanaka and Dulbecco 1966). Replication of SV40 DNA requires a single viral protein, SV40 large T antigen (T), and the cellular DNA replication machinery (Challberg and Kelly 1989; Stillman 1989; Hurwitz et al. 1990). Although the replication initiation function of T is partially regulated by cell-cycle-dependent phosphorylation (McVey et al. 1989; Prives 1990), SV40-infected cells escape the strict cellular control of one round of DNA replication per cell cycle and massively amplify the viral genome (Tegtmeyer 1972; Chou et al. 1974; Botchan et al. 1979). Meanwhile, SV40 induces more than one round of cellular DNA replication within a given cell cycle and regularly induces endoreduplication and hyperploidy. T is necessary for host chromosome endoreduplication (Friedrich et al. 1992, 1994; Perry and Lehman 1998), suggesting that it functions to override the normal cell cycle controls that prevent reinitiation of DNA replication within a given S phase. Although this observation was made more than 10 yr ago, the mechanisms that underlie T-mediated endoreduplication have remained a mystery.
DNA replication initiation is controlled during cell cycle progression. When the accuracy of DNA replication is challenged by DNA damage or other S-phase-perturbing events, checkpoints are activated to slow down DNA replication, in part by inhibiting late replication origin firing (Paulovich and Hartwell 1995; Santocanale and Diffley 1998; Shirahige et al. 1998). Moreover, unrepaired lesions not only lead to incorporation of incorrect nucleotides, they often translate into double-strand breaks when a replication fork passes through an unrepaired area, causing its subsequent collapse (Zou and Rothstein 1997; Seigneur et al. 2000). The intra-S-phase checkpoint senses DNA damage during S phase and slows down DNA replication in an effort to coordinate cell cycle progression with DNA repair. In mammalian cells, Nbs1 plays an essential role in the enactment of this checkpoint (Shiloh 1997; Carney et al. 1998; Varon et al. 1998).
Nijmegen breakage syndrome (NBS) is characterized by radiation sensitivity, chromosomal instability, and predisposition to cancer (Shiloh 1997; Featherstone and Jackson 1998). Cells from NBS patients exhibit radioresistant DNA synthesis, suggesting a loss of intra-S-phase checkpoint control. Both the ATM gene product and Nbs1 are active in this process. Nbs1 is phosphorylated by ATM at several serine residues after S-phase ionizing radiation, and these ATM-mediated phosphorylation events are essential for activating the intra-S-phase checkpoint in response to DNA damage (Gatei et al. 2000; Lim et al. 2000; Wu et al. 2000; Zhao et al. 2000). Nbs1 forms a tight complex with two repair proteins, Mre11 and Rad50 (Carney et al. 1998). This complex forms nuclear foci at sites of DNA damage and disappears from these sites after damage is repaired, suggesting a role for this complex in sensing DNA damage and/or in its repair (Carney et al. 1998; Nelms et al. 1998). In this regard, experimental depletion of Mre11 leads to the accumulation of DSBs during DNA replication (Costanzo et al. 2001; Mirzoeva and Petrini 2001). In addition, loss of Nbs1, Mre11, or Rad50 function in mice or in chicken cells is a lethal event (Xiao and Weaver 1997; Luo et al. 1999; Yamaguchi-Iwai et al. 1999; Zhu et al. 2001). Taken together, these findings suggest that one role of Nbs1 is to coordinate DNA replication with DNA repair.
One mechanism that slows down S-phase progression in response to DNA damage is inhibition of the firing of late replication origins (Santocanale and Diffley 1998). In Saccharomyces cerevisiae, this inhibition depends on the checkpoint genes Mec1 and Rad53 (Santocanale and Diffley 1998; Shirahige et al. 1998). The mechanism whereby Nbs1 participates in the S-phase checkpoint remains unknown. However, because S-phase checkpoint action requires specific modulation of DNA replication initiation, it is conceivable that one or more cellular components dedicated to the control of replication initiation is active in the checkpoint mechanism. In this regard, we have detected a heretofore unappreciated in vivo interaction between Nbs1 and T and observed that complex formation leads both to enhancement of SV40 DNA replication initiation and to endoreduplication of nonviral chromosomal DNA.
Results
Nbs1 interacts with SV40 large T antigen
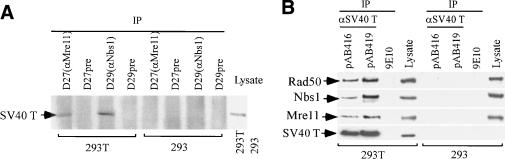
We have detected an interaction between the Nbs1/Mre11/Rad50 complex and T in multiple cell lines. Cell lysates were prepared from both 293T and 293 cells, and immunoprecipitation (IP) was performed with antibodies to Nbs1 (D29) and Mre11 (D27). Immunoblotting of these precipitates, using antibody to SV40 T (pAB419), revealed that T coimmunoprecipitated with both Nbs1 and Mre11 in 293T cell extracts (Fig. 1A). Negative results were obtained with 293 cells that lack T. Similarly, in anti-T IPs of 293T but not of 293 extracts, Nbs1, Mre11, and Rad50 all coimmunoprecipitated with T (Fig. 1B). Similar results were obtained when U20S cells that produce T were analyzed (data not shown).
Figure 1.
T interacts with the Nbs1/Mre11/Rad50 complex. (A) Cell lysates were prepared from 293T cells or 293 cells (same cell line lacking T). Polyclonal antibodies D27 (anti-Mre11) and D29 (anti-Nbs1) along with their preimmune sera were used to perform immunoprecipitation (IP) assays. Precipitates were immunoblotted with the anti-T monoclonal antibody, pAB419. We estimate that, in this protocol, ∼20%-30% of Nbs1 and Mre11 were coimmunoprecipitated with T. (B) Immunoprecipitation of 293T and 293 cell lysates with pAB416 and pAB419 (anti-T) and 9E10 (anti-myc), which served as a negative control. The Abs, EE15 (anti-Nbs1), D27 (anti-Mre11), and anti-Rad50 were used for immunoblotting.
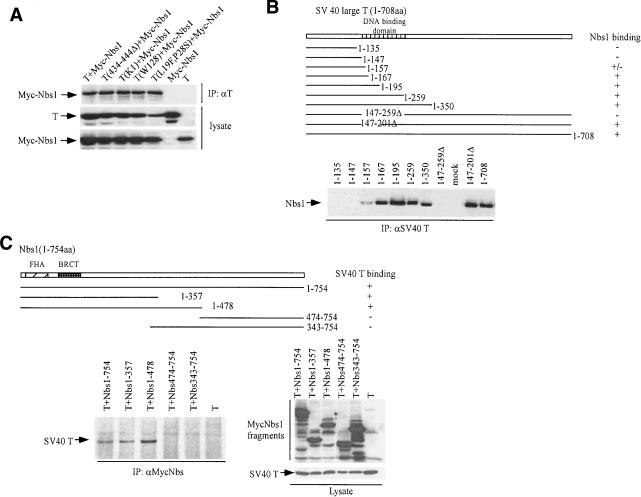
T is composed of multiple functional domains (Ali and DeCaprio 2001). It interacts with p53 in its C-terminal region and binds to Rb through its LxCxE motif. The N-terminal J domain cooperates with the LxCxE motif to inactivate the function of the Rb family members, including p107 and p130, and also possesses pocket protein binding-independent transformation cooperation function (Lin and DeCaprio 2003). The interaction of Nbs1 and SV40 T appears to proceed in the absence of the p53, the Rb binding, and the J domain. Specifically, the T mutants TΔ434-TΔ444 (a p53 binding-defective mutant), K1 (a pocket protein binding-defective mutant mapping to the LXCXE), and L19FP28S and D44N (J domain mutants; Peden et al. 1990; Kierstead and Tevethia 1993; Campbell et al. 1997), all bound to Nbs1 at the same input concentrations as wild-type T (Fig. 2A).
Figure 2.
Mutation analysis of T and Nbs1 for their interactions. (A) Analyzing the interaction of Nbs1 with wild-type and mutant T. U20S cells were transfected with myc-tagged Nbs1 and wild-type T or T mutants. Immunoprecipitation was performed with anti-T Ab (pAB419), and the precipitates were blotted with anti-myc Ab (9E10, top segment). (Bottom segment) The expression of myc-Nbs1, T, and T mutants was examined by Western blot analysis (anti-T and anti-myc) performed on transfected cell lysate. The mutants studied were T(434-444Δ), a p53-binding mutant; T(K1), an Rb-binding mutant; T(W128), an origin-binding mutant; and T(L19F,P28S), a J domain mutant. (B) T interacts with Nbs1 through a region overlapping its viral origin binding domain. Wild-type T(1-708) and a series of C-terminal and internal deletion mutants were cotransfected with myc-tagged Nbs1 into U2OS cells. Interactions of T and myc-Nbs1 were studied by immunoprecipitation of T using pAB419 and immunoblotting using 9E10 (anti-myc) as probe. The mutants, T(147-259Δ) and T(147-201Δ), have lost residues 147-259 and 147-201, respectively. In the lane labeled mock, T(1-708) was transfected in the absence of myc-Nbs1. The expression of the various T fragments is at similar levels in each transfection (data not shown). (C) The interaction of Nbs1 and T is mediated by the N-terminal region of Nbs1. U2OS cells were cotransfected with wild-type T and with each of a series of myc-tagged Nbs1 alleles, subcloned in the mammalian expression vector pCDNA3β. The sequence composition of each Nbs1 fragment is shown. T-Nbs1 complex formation was monitored by IP using Ab recognizing myc (9E10) followed by immunoblotting with pAB419 (anti-T) as probe. The expression of myc-tagged Nbs1 fragments and T was examined by Western blot analysis (anti-myc and anti-T) performed on transfected cell lysate (Lysate). (FHA) The forkhead-associated domain; (BRCT) the breast cancer C-terminal domain (Carney et al. 1998; Varon et al. 1998).
To search for a specific Nbs1-binding domain in T, a series of C-terminal dl T mutants were coexpressed with myc-tagged Nbs1 in U2OS cells (Fig. 2B). T fragments, containing as little sequence as the N-terminal 167 residues, readily bound to Nbs1, whereas the 1-157 mutant demonstrated decreased binding. No binding was evident to the 1-135 and 1-147 fragments. Thus, residues located between amino acids 147 and 167 appear to contribute to T/Nbs1 complex formation. In this regard, internal deletion mutants of T were also analyzed (Fig. 2B). Full-length T lacking residues 147-201 bound Nbs1 efficiently, unlike full-length T lacking residues 147-259. Collectively, this suggests that the T/Nbs1 core binding unit is composed of redundant sequences located between residues 147 and 259.
SV40 T binds specifically to SV40 replication origins as well as nonspecifically to both single- and double-stranded DNA. Its DNA binding function is a product of its DNA-binding domain, which extends from residues 137 to 246 (Kalderon and Smith 1984; Simmons et al. 1990; Wun-Kim et al. 1993). The Nbs1-binding unit of T, thus, overlaps part of the DNA-binding domain. Because Nbs1 also binds to DNA, we asked whether the T/Nbs1 interaction is mediated by nonspecific DNA binding. Specifically, immunoprecipitation of SV40 T or Nbs1 was performed in extracts of 293T cells in the presence of 400 μM ethidium bromide. Although DNA/protein interactions can be selectively inhibited by ethidium bromide (Lai and Herr 1992), the association of T and the Nbs1/Mre11/Rad50 complex was unaffected by this agent (data not shown). Moreover, we consistently found that the T mutant W128 (Leu156Phe; Kalderon and Smith 1984; Simmons et al. 1990), although defective in DNA binding, bound to Nbs1 like wild-type (wt) T (Fig. 2A). This implies that the T/Nbs1 interaction is not mediated by an associated DNA molecule(s).
The T-binding site on Nbs1 was also mapped and found to be located within the N-terminal region of Nbs1. Two Myc-tagged N-terminal Nbs1 fragments (amino acids 1-357 and 1-478) interacted with T when each was coexpressed with T in U2OS cells (Fig. 2C). In contrast, the Nbs1 C-terminal fragments (amino acids 474-754 and 343-754) failed to co-IP with T (Fig. 2C).
The association between SV40 T and Nbs1 does not influence Nbs1 function in response to DNA damage
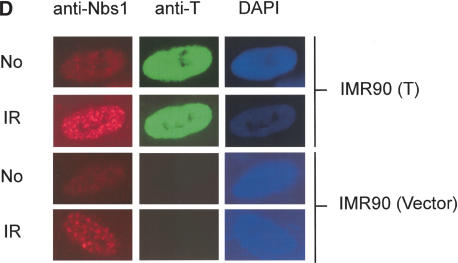
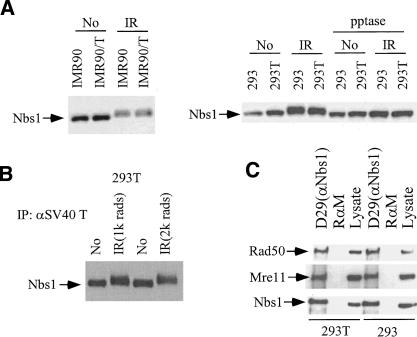
The Nbs1/Mre11/Rad50 complex plays an essential role in the response to DNA damage. Nbs1 is phosphorylated by ATM after DNA damage, and this phosphorylation is important for certain subsequent cellular responses, such as S-phase checkpoint control and cellular radiation sensitivity (Gatei et al. 2000; Lim et al. 2000; Wu et al. 2000; Zhao et al. 2000). To test whether the presence of T impairs DNA damage-driven Nbs1 phosphorylation, we compared SDS-gel mobility of Nbs1 before and after ionizing radiation (IR) of IMR 90 primary fibroblasts and a derivative that synthesizes T [IMR90(T)]. IR-induced Nbs1 phosphorylation led to a gel mobility shift (Fig. 3A) that was unaffected by T. Similar results were obtained with 293T and 293 cells. The results of exogenous phosphatase treatment confirmed that the shift was caused by IR-induced phosphorylation (Fig. 3A). We also immunoprecipitated SV40 T from 293T cells before and after IR. The same quantity of under- and hyperphosphorylated Nbs1 was coprecipitated with T before and after damage (Fig. 3B). This suggests that T/Nbs1 complex formation does not interfere with proper Nbs1 phosphorylation after DNA damage. Moreover, the formation of the Nbs1/Mre11/Rad50 complex was not perturbed by T (Fig. 3C), nor was IR-induced Nbs1, Mre11, and Rad50 nuclear focus formation (Fig. 3D). The intra-S-phase checkpoint response, represented by down-regulation of DNA synthesis in response to IR, was also unaffected in multiple T-containing cell species, including IMR90 (data not shown). These data imply that T does not negatively affect the above-noted Nbs1 functions.
Figure 3.
The interaction between T and Nbs1 does not influence post-IR Nbs1 phosphorylation and focus formation. (A) A primary human fibroblast cell line, IMR 90, and its derivative IMR 90 (T), synthesizing wild-type T were lysed before or 1 h after IR (2 krads), and lysates were fractionated by electrophoresis in 8% SDS gels. Gels were immunoblotted, using EE15 (anti-Nbs1) as probe. A similar experiment was performed in 293 and 293T cells. The lysates of 293 and 293T cells were also treated with 500 U of λ phosphastase for 30 min at 30°C before loading onto SDS gels. (B) The Nbs1 in complex with T is phosphorylated after IR. 293T cell lysates were prepared before and 1 h after IR (1 krad or 2 krads). pAB419 was used to IP T, and the gel was immunoblotted using EE15 as probe. (C) The Nbs1/Mre11/Rad50 complex formation is not disturbed by T. Nbs1 was immunoprecipitated from 293T and 293 cells using antibody D29. Mre11 and Rad50 associated with Nbs1 were detected by Western blot analysis. Rabbit anti-mouse (RαM) antibody was used as a negative immunoprecipitation control. (D) IR-induced Nbs1 nuclear focus formation is not disturbed by T synthesis. IMR90 cells infected with retroviruses encoding T or empty vector were fixed before and after IR (1 krad). Immunostaining was performed using antibodies recognizing Nbs1 (Oncogene) and T (pAB419). DAPI staining was shown. (Red) Nbs1; (green) T.
Targeting Nbs1 by SV40 large T antigen is essential for SV40 T-mediated endoreduplication
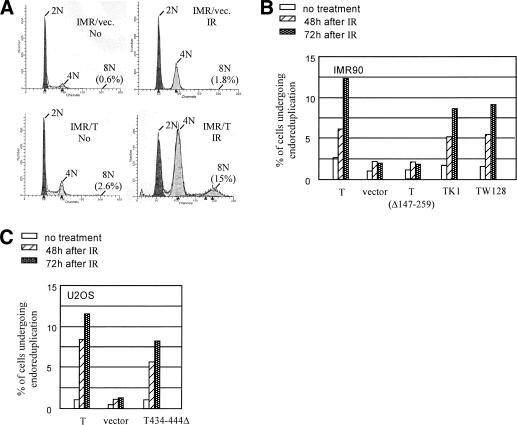
Expression of T in permissive cells, such as CV-1 induces reinitiation of DNA synthesis within a single cell cycle, resulting in the production of cells with >8N DNA content (Friedrich et al. 1992, 1994; Perry and Lehman 1998). We have found that, in semipermissive human cells, such as IMR90 primary human fibroblasts, this T-dependent cellular DNA endoreduplicative response is more apparent when cells become arrested at G2/M after ionizing radiation (IR; Fig. 4A). To test whether T/Nbs1 complex formation contributes to T-induced endoreduplication, we infected IMR 90 cells with retroviruses encoding a NeoR marker and either wild-type T or T (Δ147-259 amino acids). Cells were selected for G418 resistance. Flow cytometry was used to measure DNA content before and 48 or 72 h after IR. A significant percentage of T-producing cells (>12% at 72 h after IR) underwent a second round of DNA replication without mitosis, whereas only ∼2% of the T- (Δ147-259) or mockinfected cells revealed an 8N DNA content (Fig. 4B). Synthesis of a T mutant defective in Rb binding (K1), origin DNA binding (W128) or p53 binding (434-444Δ) also led to the accumulation of cells with an 8N DNA content (Fig. 4B,C). Similar results were obtained when another primary fibroblast cell line, BJ, was similarly analyzed (data not shown). These results are compatible with the hypothesis that T/Nbs1 complex formation allows endoreduplication of cellular DNA.
Figure 4.
T induces endoreduplication in IMR90 cells after IR. The experiment was repeated at least three times with similar results, and the results of a representative experiment are shown. (A) IMR90 were infected with a backbone retroviral vector or the vector encoding T. Cell cycle profile was monitored by flow cytometry before and 72 h after IR (2 krads). The percentage of cells with 8N DNA content is indicated. Similar results were obtained when cells were irradiated with 0.8 krad. (B) IMR90 cells were infected with retroviruses encoding wild-type T and certain T mutants, including the internal deletion mutant (Δ147-259), a pocket protein-binding-defective species, K1, and a missense species defective in SV40 replication origin binding, W128. Infected cells were selected in G418. Expression levels of T and the T mutants were approximately equal in the various cultures (data not shown). The empty vector, pBabe Neo, was used as a negative control (Vector). FACS analysis was performed before and 48 h or 72 h after IR (2 krads), and the percentage of cells that had experienced endoreduplication (i.e., displayed a >8N DNA content as defined by FACS) was plotted. (C) U2OS cells were infected with retroviruses encoding wild-type T, a T mutant defective in p53 binding (T434-444Δ), and a pBabe-Neo vector. After G418 selection, stable transformants were examined for endoreduplication 48 or 72 h after IR (2 krads). The percentage of cells that had experienced endoreduplication is depicted.
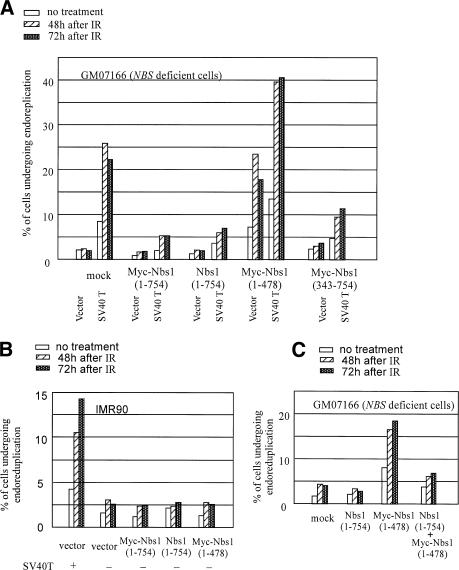
To test the T/Nbs1-endoreduplication hypothesis more stringently, we analyzed the effect of T synthesis in NBS-deficient cells before and after reconstitution with Nbs1. Indeed, T did lead to endoreduplication in NBS-deficient cells, and wild-type Nbs1 coexpression led to dramatic suppression of this effect (Fig. 5A). This result strongly suggests that Nbs1 can antagonize SV40 T to suppress the development of multiple rounds of DNA replication within one cell cycle. On the other hand, as shown earlier, there is also evidence suggesting that an excess of SV40 T can bind to Nbs1 to inactivate its endoreduplication suppression function.
Figure 5.
Ectopic Nbs1 expression suppresses T-mediated endoreduplication. Each experiment was repeated at least three times with similar results, and the results of a representative experiment are shown in each case. (A) The NBS-deficient cell line, GM07166, immortalized by expression of hTERT, was infected with retroviruses encoding wild-type Nbs1 (Nbs1), myc-tagged wild-type Nbs1 (myc-Nbs1), 1-478 and 343-754 Nbs1 fragments, and the empty pBabe-puro IRES-GFP vector (mock). After drug selection, the infected cells were superinfected with a wild-type T-encoding retrovirus (SV40 T) or empty vector (Vector). Standardized Western blotting revealed similar levels of T expression in all lines infected with T-encoding virus, and there was equal expression of all transfected Nbs1 species in this experiment (data not shown). The percentage of cells that underwent endoreduplication is shown. (B) Aliquots of IMR90 were infected with retroviruses encoding wild-type Nbs1 (Nbs1), myc-tagged wild-type Nbs1 (myc-Nbs1), the 1-478 Nbs1 fragment, and the pBabe puro IRES-GFP vector (vector). All cultures were assayed for endoreduplication as noted above. IMR90 infected by a T-encoding virus underwent endoreduplication, and they represented a positive control here. Expression of the 1-478 Nbs1 N-terminal fragment did not induce endoreduplication in IMR90 cells. (C) The myc-Nbs1 (1-478) fragment was introduced into NBS-deficient cells GM07166 and its derivative reconstituted with wild-type Nbs1 by retroviral infection. When the 1-478 Nbs1 fragment was expressed in the GM07166 cells, endoreduplication was detected. This phenotype was suppressed when the same fragment was expressed in GM07166 cells reconstituted with wild-type Nbs1.
Expression of an N-terminal Nbs1 fragment can induce endoreduplication in NBS-deficient cells in the absence of SV40 T expression
Our finding that Nbs1 can suppress SV40 T-mediated endoreduplication suggests that Nbs1 participates in restricting cellular DNA to one round of replication in each cell cycle. In this regard, NBS-deficient GM07166 cells revealed low, endogenous levels of endoreduplication (Fig. 5A). Surprisingly, when the (1-478 amino acids) Nbs1 fragment was expressed in these cells, endoreduplication was markedly enhanced (Fig. 5A, Nbs1[1-478]/vector). The enhancement was similar to that observed when these same NBS-deficient cells were engineered to express SV40 T (Fig. 5A, mock/SV40 T). Synthesis of a C-terminal (343-754) Nbs1 fragment did not result in this effect (Fig. 5A). This suggests that the Nbs1 (1-478 amino acids) fragment can, with relative specificity, disrupt cellular DNA replication control. Conceivably, it disrupts the control(s) that prevent(s) hyperreplication by interacting with and sequestering/perturbing the function of critical proteins that participate in the relevant replication control pathway(s). For example, the Nbs1 N-terminal fragment interacts with certain E2F family members known to participate in the regulation of DNA replication (Maser et al. 2001).
The endoreduplication effect mediated by the 1-478 Nbs1 fragment was detected only when cells were Nbs1-deficient. When this same fragment was expressed in endogenous wild-type Nbs1-producing cells, such as IMR90, no increment over the low, basal level of endoreduplication was observed (Fig. 5B). This was the case, even though the expression level of the 1-478 Nbs1 fragment was similar in IMR90 and NBS-deficient GM07166 cells. Notably, when the fragment was expressed in wtNbs1-reconstituted GM07166 cells, its endoreduplication-inducing effect was largely suppressed (Fig. 5C). This suggests that the Nbs1 fragment can disrupt cellular DNA replication control only when endogenous, wild-type Nbs1 function is deficient. This implies that NBS deficiency is a prerequisite for the Nbs1 N-terminal fragment to induce endoreduplication and suggests that intact Nbs1 may establish a safeguard mechanism that prevents DNA hyperreplication, even when certain replication control pathways are disturbed.
NBS deficiency leads to reinitiation of DNA replication at regions close to putative replication initiation sites
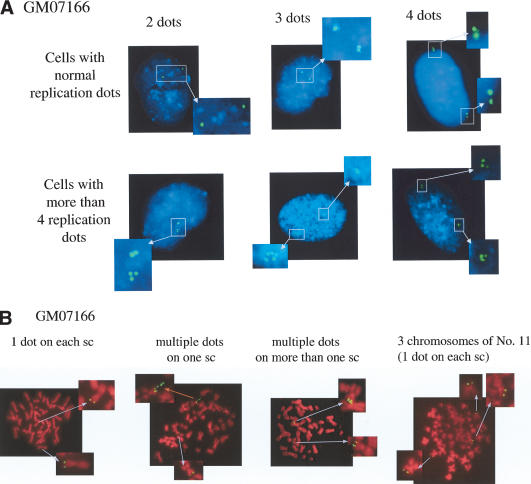
To more directly test the Nbs1 replication control hypothesis, we performed Fluorescence In Situ Hybridization (FISH) analysis on interphase cells using a DIG-labeled probe for sequences at a specific site near the β-globin locus where DNA replication is initiated (Kitsberg et al. 1993; Aladjem et al. 1995; Avni et al. 2003). FITC-conjugated secondary antibody against DIG was used to detect replication initiation foci at this site (Fig. 6A; Table 1A). Diploid cells in which the specific segment to be probed is unreplicated should reveal two FISH dots, three dots if one of the two relevant replication origins has fired, or four dots if both have fired. If there are more than four dots overlying two chromosomes, at least one segment of the probed region has rereplicated in the relevant cell cycle.
Figure 6.
NBS deficiency leads to rereplication of DNA sequences near replication initiation sites. (A) Representative FISH analysis performed on interphase NBS-deficient cells (GM07166). Fixed cells were hybridized with a BAC probe corresponding to a segment covering the β-globin locus (clone RP11-645I8), and the hybridization images appear as green spots. This cloned genomic segment contains a putative replication origin (Kitsberg et al. 1993; Aladjem et al. 1995). Chromosomal DNA was stained with DAPI (blue). Examples of GM07166 cells that contain two, three, four, or more dots at the β-globin locus are shown. (B) FISH of metaphase nuclei of GM07166 cells. Cells were treated with colchicine to select for metaphase cells, and metaphase spreads were prepared. The fixed cells were hybridized with the above-noted β-globin probe (green), and chromosomes were stained with PI (red). FISH-positive segments of chromatids are identified with arrows in magnified copies of the relevant segments of each spread. In the various images are depicted GM07166 nuclei containing two pairs of β-globin dots (normal; one on each sister chromatid [sc]); multiple dots on one sc; multiple dots on more than one sc; and three pairs of dots each on a chromosome (indicating the presence of three No. 11 chromosomes). In some cases, the highly packed metaphase chromosomes likely “opened up” in certain locations during preparation, possibly allowing DNA to loop out, which, in turn, led to FISH signals extending out from the main body of the chromosome. An example is shown in the “multiple dots on one sc” and is marked with a red arrow. Here, the amplification of the β-globin DNA sequence of interest in the indicated chromatid is reflected by the presence of multiple FISH dots in a linear array extending out from the relevant chromatid.
Table 1.
β-Globin locus FISH analysis of NBS-deficient cells (GM07166)
| A) FISH of unsynchronized cells a
| ||||
|---|---|---|---|---|
| Cell lines | Total number | 2 dots | 3 or 4 dots | >4 dots |
| IMR90 | 807 | 423 (52.4%) | 365 (45.2%) | 19 (2.4%) |
| NBS−/− | 219 | 67 (30.6%) | 102 (46.6%) | 50 (22.8%) |
| NBS−/− (Nbslwt) | 432 | 232 (53.7%) | 177 (41.0%) | 23 (5.3%) |
| B) FISH of metaphase nuclei b
| ||||
|---|---|---|---|---|
| Cell lines | Total number | 1 on each sc | 2 or more on one sc | 2 or more on more than one sc |
| IMR90 | 129 | 125 (96.9%) | 4 (3.1%) | 0 (0.0%) |
| NBS−/− | 110 | 70 (63.6%) | 29 (26.4%) | 11 (10.0%) |
A BAC clone containing a large DNA segment (clone RP11-64518) of the β-globin locus, which includes a DNA replication initiation site (Kitsberg et al. 1993; Aladjem et al. 1995), was used as a FISH probe.
Interphase nuclei from primary IMR90 cells, NBS-deficient GM07166 cells (NBS), and wild-type (wt) Nbs1-reconstituted GM07166 cells [NBS−/− (Nbs1wt)] were analyzed by FISH using this probe. The number of cells containing two, three, four, or more than four dots at the β-globin locus is indicated.
Metaphase nuclei of IMR90 and GM07166 cells were analyzed by FISH, using the above-noted probe. The number of IMR90 and GM07166 cells with two pairs of β-globin dots, one on each sister chromatid (sc one on each sc), with multiple dots on one sc (two or more on one sc), and with multiple dots on more than one sc (two or more on more than one sc), are shown. The percentage of cells in each category is shown in parentheses.
In a culture of unperturbed IMR90, most cells contained two, three, or four dots, and only ∼2% of cells contained more than four dots (Table 1A). In contrast, >20% of the aforementioned NBS-deficient cell line contained more than four dots—even in the unperturbed state (Table 1A). Examples of NBS-deficient cells with two, three, four, or more than four replication dots are illustrated in Figure 6A. Expression of wild-type Nbs1 in NBS-deficient cells reduced the percentage of cells with more than four dots by more than fourfold (Table 1A). Probes that recognize DNA regions containing known sites of DNA synthesis initiation near the HSP70 and Lamin B2 genes (Taira et al. 1994; Abdurashidova et al. 2000) yielded similar results (data not shown). Almost all NBS-deficient cells (GM07166) were characterized by normal chromosomal numbers.
We also performed FISH analysis on metaphase chromosomes. In >97% of IMR90 cells (wtNbs1), we detected two pairs of dots at the β-globin replication initiation region per cell, one on each chromatid, implying that, in G2-arrested cells, the adjacent DNA has replicated once (Table 1B). In contrast, ∼26% of NBS-deficient cells (GM07166) contained two or more β-globin replication dots on one chromatid, and 10% contained two or more dots on more than one chromatid (Fig. 6B; Table 1B). Occasionally, we observed that certain GM07166 cells carry an additional Chromosome 11 (three copies of Chromosome 11), where the β-globin gene is located (<1%; Fig. 6B). These findings strongly suggest that NBS deficiency permits more than one round of DNA replication at certain loci close to replication origins. The additional round of DNA replication appears regional and does not cover the entire chromosome, in keeping with the prior observation that only low/background levels of gross chromosomal endoreduplication and hyperploidy were detected in NBS-deficient cells (see Fig. 5A).
Nbs1 suppresses SV40 T-mediated viral DNA replication
Our study suggests that SV40 T binds to Nbs1 and, thereby, perturbs the normal control of cellular DNA replication initiation. One outcome appears to be a state of cellular endoreduplication. This being the case, it seemed reasonable to ask whether this process is relevant to the phenomenon of autonomous replication of the viral genome, an event that depends on repetitive firing of the viral origin in a single S phase.
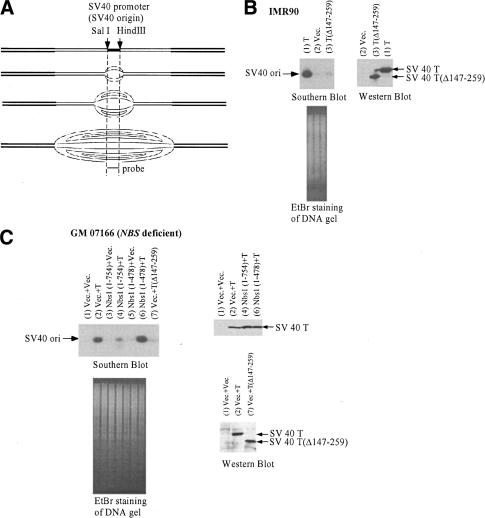
In an effort to address this question, we introduced an integrated SV40 replication origin into IMR90 cells by infecting with a retroviral vector. The vector encodes the murine ecotropic retroviral receptor (ER) and contains an SV40 replication origin. Subsequently, these cells were infected with another retrovirus encoding wild-type T or with an empty vector (pBaba-Neo-oric-) that carries a neo-resistance gene but lacks an SV40 replication origin. After several passages, genomic DNA from these cultures was purified and digested with SalI and HindIII, a step that results in the generation of an intact and unique SV40 origin-containing fragment detectable by Southern blotting (Fig. 7A). The integrated SV40 origin-containing fragment became amplified when T was expressed (Fig. 7B). In contrast, cells infected with empty vector or T(Δ147-259), which encodes a protein defective in replication origin binding, did not amplify this fragment (Fig. 7B).
Figure 7.
Nbs1 suppresses replication initiation from the SV40 replication origin. (A) The SV40 replication origin present in the pBabe vector used in this experiment is flanked by SalI and HindIII restriction sites. The indicated probe was used for Southern blot analysis. Genomic DNA surrounded the viral replication origin may be amplified when the viral origin fires multiple times in a given cell cycle. (B) IMR90 cells were infected by a retrovirus (pBabe-hygro-ER) encoding a murine ecotropic retroviral receptor (ER). The vector also contained an SV40 replication origin. Cells were subsequently infected with retroviruses encoding wild-type T, the T(Δ147-259) mutant, or empty vector pBabe-Neooric- (Vec.). Standardized quantities of purified genomic DNA from the indicated cultures were digested with SalI and HindIII. Southern blot analysis was performed using the SalI to HindIII probe identified in A. An EtBr-stained agarose gel depicts the quantities of SalI + HindIII-digested genomic DNA analyzed. The expression of T and the deletion mutant T(Δ147-259) was assessed by Western blot analysis. (C) NBS-deficient cells (GM07166) immortalized by hTERT were infected with pBabe-hygro-ER containing an SV40 origin. They were then reconstituted by infection with myc-tagged wild-type Nbs1 retrovirus, myc-Nbs1 fragment (1-478) virus, or empty vector virus. T or empty vector (Vec.) virus was subsequently used to superinfect these cultures. Southern blot analysis of standardized amounts of SalI- and HindIII-digested genomic DNA from each of these cultures was then performed in search of SV40 origin-containing DNA, using the above-noted SalI-HindIII fragment as probe (see A). An EtBr-stained gel revealed the quantity of DNA from each culture that was loaded onto the relevant gel. The expression level of T in each culture is shown by Western blotting.
We also asked whether wild-type Nbs1 affects this process. Forced expression of an ectopic, wild-type Nbs1 allele (amino acids 1-754) in the NBS-deficient cells suppressed T-mediated viral DNA replication in these cells (Fig. 7C, cf. lanes 2 and 4). In contrast, the Nbs1(1-478) fragment led to enhanced T-mediated viral DNA replication (Fig. 7C, cf. lanes 6 and 2), implying that the processes that elicit Nbs1(1-478)-dependent cellular endoreduplication and autonomous firing of the SV40 replication origin are, at least in part, related.
Thus, we propose that T targets Nbs1, suppressing its ability to down-regulate the autonomous firing of both cellular and viral replication origins. The data also imply that Nbs1 is involved in suppressing the development of more than one round of replication in a given S phase and suggest that Nbs1 regulates cellular and viral replication by a related mechanism(s).
Discussion
SV40 induces cellular endoreduplication when it infects host cells, and SV40 T is required for this process (Tegtmeyer 1972; Chou et al. 1974; Botchan et al. 1979). The mechanisms by which SV40 T drives more than one round of cellular DNA replication in a given S phase have been largely unclear. In this report, we have identified an association of the Nbs1/Mre11/Rad50 complex with SV40 T. The data suggest that this association underlies, at least in part, the ability of SV40 T to inactivate the processes that guarantee one round of DNA replication of cellular replicons in a given cell cycle. One outcome of this T-mediated perturbation is endoreduplication of chromosomal DNA. For example, the T deletion mutant (Δ147-259) was both impaired in Nbs1 binding and unable to induce host chromosomal endoreduplication. This is consistent with the previous observation that the N-terminal 1-259 region of T induced a tetraploid DNA content in CV-1 cells—a line that is permissive for viral DNA replication (Perry and Lehman 1998). Expression of SV40 T also induced much higher levels of cellular endoreduplication in NBS-deficient cells than in wild-type Nbs1-reconstituted isogenic cell lines. These findings are compatible with a hypothesis in which SV40 T targets Nbs1, thereby inactivating its rereplication suppression function and inducing multiple rounds of cellular DNA replication in a given cell cycle.
Given the finding that excess Nbs1 antagonized T-mediated cellular endoreduplication, one might predict that NBS deficiency would result in signs of rereplication. Results of FISH analysis support this hypothesis. We observed that DNA at positions close to certain replication origins was hyperreplicated in >22% of NBS-deficient fibroblast cells (GM07166), although the data do not explicitly demonstrate that rereplication initiated from the relevant origins. Where analyzed, ectopic expression of wild-type Nbs1 efficiently suppressed this regional hyperreplication phenomenon.
Although a large fraction of NBS-deficient cells spontaneously rereplicated their DNA in regions close to replication initiation sites, FACS analysis did not reveal significant numbers of cells with DNA contents of >8N. However, when the N-terminal fragment (amino acids 1-478) of Nbs1 was expressed in NBS-deficient cells, profound endoreduplication was observed independent of SV40 T function. This phenomenon was observed only in NBS-deficient cells, and not in NBS-proficient cells such as IMR90. Taken together, these findings, in part, suggest that Nbs1 may contribute to the activation of a checkpoint that prevents additional refiring of replication origins that have already fired.
However, florid DNA rereplication requires additional molecular disruptions beyond simple loss of NBS function. It appears likely that the rereplication suppression function of Nbs1 is inhibited following SV40 T/Nbs1 complex formation. Yet, because T was required to maximize endoreduplication in NBS-deficient cells, florid, T-mediated endoreduplication likely requires at least one as-yet-undefined function of this viral protein. Whatever this step(s) may be, like T, the 1-478 fragment of Nbs1 also induced marked endoreduplication. Notably, it was only detected in NBS-deficient cells. Conceivably, unopposed by wild-type Nbs1, this mutant polypeptide interacts in an abnormal manner with certain cell protein(s) (which might include one or more polypeptides with which intact Nbs1 normally interacts), an outcome of which is a gross override of rereplication control. It will eventually be interesting to determine whether T and this particular Nbs1 mutant operate by perturbing the same pathway(s).
Marked amplification of the viral genome is essential for SV40 propagation, but the factors that optimize this process have, in the past, been incompletely defined. In this regard, we observed that a chromosomally integrated SV40 origin-containing segment was more extensively amplified in NBS-deficient than in wild-type Nbs1-reconstituted GM07166. This suggests that, by targeting Nbs1, SV40 T creates an environment that contributes to maximal amplification of its own genome.
Results suggesting that Nbs1 regulates DNA replication control—both cellular and viral—are consistent with the observation that Nbs1 interacts with E2F1, a protein known to associate with certain replication initiation sites, and locates near these regions in S-phase cells (Maser et al. 1997). Because T can also perturb E2F1 function by displacing it from the Rb protein, one wonders whether it can also displace it and its partner Nbs1 from certain replication origins. This is, however, not likely the case, because the association of Nbs1 with the β-globin and B2 lamin replication initiation sites was not affected by T, as revealed by the results of chromatin immunoprecipitation (ChIP) assays (X. Wu and D. Livingston, unpubl.).
Nbs1 also operates in S-phase checkpoint control. In response to IR delivered during S phase, the protein becomes phosphorylated by ATM at several sites, and these phosphorylation events are important for Nbs1 function in the ensuing intrareplication checkpoint (Lim et al. 2000). Although it remains unclear how Nbs1 contributes to the slowdown of DNA replication following DNA damage, it is conceivable that, when S-phase DNA damage is sensed or after replicons have fired during a normal cell cycle, Nbs1 communicates with the DNA replication machinery to ensure that no further initiation events occur.
However, it should also be noted that the signals that activate Nbs1 checkpoint function and its hyperreplication suppression function might be different. For example, T-Nbs1 complex formation did not interfere with DNA damage-induced Nbs1 phosphorylation (Gatei et al. 2000; Lim et al. 2000; Wu et al. 2000; Zhao et al. 2000), and one or more of these phosphorylation events participates in the activation of the intrareplication checkpoint (Lim et al. 2000). Consistently, this checkpoint operated normally in the presence of T. In contrast, the interaction of T and Nbs1 did interfere with the function of Nbs1 in preventing hyperreplication. How signals are generated and transduced to Nbs1 when the duplication of a given replicon has been initiated and/or is complete remains unclear.
Nbs1 forms a tight complex with Mre11 and Rad50 (Carney et al. 1998). Both of these proteins are essential for DSB repair (Johzuka and Ogawa 1995; Moore and Haber 1996). In mammalian cells, damage-induced foci containing Mre11 are detected during S phase, and focus formation depends on Nbs1 function (Maser et al. 1997; Carney et al. 1998; Dong et al. 1999). Depletion of Mre11 from Xenopus extracts leads to dramatic accumulation of DSBs during DNA replication (Costanzo et al. 2001). The Nbs1 S-phase checkpoint function suppresses DNA replication initiation once damage is sensed, and Nbs1 activates Mre11 and Rad50 through a direct association, leading to repair of the relevant DNA damage. The studies described in this report reveal a new function for Nbs1 in preventing DNA hyperreplication during the cell cycle. This finding reinforces the notion that Nbs1 is a multifunctional protein that contributes to proper DNA replication and the maintenance of genome stability.
Materials and methods
Cell culture
GM07166 was obtained from the Coriell Human Mutant Cell Repository. IMR90, 293T, 293, and U2OS cells were from ATCC. Murine ecotropic retroviral receptor (ER; Albritton et al. 1989) was first introduced into IMR90 and GM07166 cells by retroviral infection. These viruses were generated using the Phoenix amphotropic packaging cell line from ATCC. Subsequently wild-type Nbs1, Nbs1 mutants, wild-type T, and T mutants were introduced into these ER+ cells with suitable recombinant murine ecotropic retroviruses generated in Phoenix ecotropic packaging cells (ATCC). GM07166 cells were immortalized by infection with a retrovirus (pBabe/hygro) encoding hTERT (Meyerson et al. 1997). U2OS cells that stably express SV40 T [U2OS(T)] were generated by transfecting U2OS cells with CMV T neo (Campbell et al. 1997) and selecting for G418-resistant cells.
Antibodies
Monoclonal antibodies against SV40 T (pAB419 and pAB416) and against Nbs1 (EE15 and D29) have been described previously (Harlow et al. 1981; Wu et al. 2000). Polyclonal Ab (D27) against Mre11 was raised against the C-terminal fragment of human Mre11 (Mre11360-709). Monoclonal antibody against Rad50 was purchased from Novus Biologicals.
Plasmids
Full-length Nbs1 and Nbs1 fragments were subcloned into a mammalian expression vector, pCDNA3β, that carries the sequence encoding the myc epitope (Chen et al. 1998). Full-length T and its derivative mutants were subcloned into either pCDNA3β (Scully et al. 1997) or pSG5 (Stratagene). Alleles encoding full-length Nbs1 with or without a myc-tag and myc-tagged Nbs1 fragments (1-478, 343-754) were inserted into a retroviral vector, pBabe-puro, that was modified by adding an IRES-EGFP sequence from pIRES-EGFP (Clontech). The retroviral vector, pBabe/neo/oric-, was generated by replacing the SV40 promoter of pBabe/neo with the comparable DNA region of pBaba/puro/oric-, a gift from Kathy Rundell (Northwestern University, Chicago, IL). The retroviral vector, pBabe-Blasticidine, encoding ER, was constructed by replacing the Hygro marker of pBabe-hygro-ER with a Blasticidine marker, a gift from Peiqing Sun (The Scripps Research Institute, LaJolla, CA). Detailed information on plasmid construction will be provided upon request.
Immunoassays
Cells were lysed in NETN (150 mM NaCl, 1 mM EDTA, 20 mM Tris-Cl at pH 8.0, 0.5% NP-40). Primary antibodies were incubated with cell lysates at 4°C for 3 h, followed by adding protein A- or G-Sepharose beads for another hour's incubation. Beads were washed four times with NETN before SDS-gel sample buffer was added. Primary antibodies were routinely rocked with blots overnight at 4°C. HRP-conjugated secondary antibodies were used at 1:3000 dilution (Amersham).
For immunostaining, cells were fixed in 70% methanol and 30% acetone for 15 min at -20°C. After 1 h of drying at room temperature and three subsequent PBS washes, cells were stained with antibody recognizing Nbs1 (rabbit polyclonal; Oncogene) and/or T (monoclonal pAB419) for 4 h at room temperature. Subsequently, cells were stained with FITC-conjugated anti-mouse and Rhodamine-conjugated anti-rabbit antibodies (Jackson ImmunoResearch) for 1 h at room temperature.
Analysis of DNA content by flow cytometry
At each time point, cells were trypsinized, washed once with PBS, and fixed with prechilled 70% ethanol. After incubating for several hours or overnight at 4°C, cells were washed with PBS and resuspended in a buffer containing 38 mM Na Citrate, 70 μM propidium iodide (PI), and 20 μg/mL RNase A, incubated for 30 min at 37°C in the dark, and subjected to flow cytometry analysis.
Fluorescence In Situ Hybridization (FISH) analysis
FISH was performed as described (Heng et al. 1992; Heng and Tsui 1993; Boggs and Chinault 1997). Human BAC clones that were used as probes for DNA sequences at the B2-Lamin, HSP70, and β-globin loci were obtained through the Children's Hospital Oakland Research Institute (CHORI). The BAC clone numbers are: B2-Lamin, RP11-211I3; HSP70, RP11-425A7; and β-globin, RP11-645I8.
Acknowledgments
We thank James DeCaprio and Ole Gjoerup for sharing SV40 T DNA-encoding plasmids and for many helpful discussions. We also thank William Hahn and Robert Weinberg for providing pBabe-hygro-hTERT, Peiqing Sun for pBabe-hygro-ER, and Kathy Rundell for pBabe-puro-oric-. William Hahn graciously helped us in the immortalization of GM07166 with hTERT.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1182804.
Corresponding author.
References
- Abdurashidova, G., Deganuto, M., Klima, R., Riva, S., Biamonti, G., Giacca, M., and Falaschi, A. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287: 2023-2026. [DOI] [PubMed] [Google Scholar]
- Aladjem, M.I., Groudine, M., Brody, L.L., Dieken, E.S., Fournier, R.E., Wahl, G.M., and Epner, E.M. 1995. Participation of the human β-globin locus control region in initiation of DNA replication. Science 270: 815-819. [DOI] [PubMed] [Google Scholar]
- Albritton, L.M., Tseng, L., Scadden, D., and Cunningham, J.M. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57: 659-666. [DOI] [PubMed] [Google Scholar]
- Ali, S.H. and DeCaprio, J.A. 2001. Cellular transformation by SV40 large T antigen: Interaction with host proteins. Semin. Cancer Biol. 11: 15-23. [DOI] [PubMed] [Google Scholar]
- Avni, D., Yang, H., Martelli, F., Hofmann, F., ElShamy, W.M., Ganesan, S., Scully, R., and Livingston, D.M. 2003. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 12: 735-746. [DOI] [PubMed] [Google Scholar]
- Boggs, B.A. and Chinault, A.C. 1997. Analysis of DNA replication by fluorescence in situ hybridization. Methods 13: 259-270. [DOI] [PubMed] [Google Scholar]
- Botchan, M., Topp, W., and Sambrook, J. 1979. Studies on simian virus 40 excision from cellular chromosomes. Cold Spring Harb. Symp. Quant. Biol. 43: 709-719. [DOI] [PubMed] [Google Scholar]
- Campbell, K.S., Mullane, K.P., Aksoy, I.A., Stubdal, H., Zalvide, J., Pipas, J.M., Silver, P.A., Roberts, T.M., Schaffhausen, B.S., and DeCaprio, J.A. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes & Dev. 11: 1098-1110. [DOI] [PubMed] [Google Scholar]
- Carney, J.P., Maser, R.S., Olivares, H., Davis, E.M., Le Beau, M., Yates III, J.R., Hays, L., Morgan, W.F., and Petrini, J.H. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477-486. [DOI] [PubMed] [Google Scholar]
- Challberg, M.D. and Kelly, T.J. 1989. Animal virus DNA replication. Annu. Rev. Biochem. 58: 671-717. [DOI] [PubMed] [Google Scholar]
- Chen, J., Silver, D.P., Walpita, D., Cantor, S.B., Gazdar, A.F., Tomlinson, G., Couch, F.J., Weber, B.L., Ashley, T., Livingston, D.M., et al. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2: 317-328. [DOI] [PubMed] [Google Scholar]
- Chou, J.Y., Avila, J., and Martin, R.G. 1974. Viral DNA synthesis in cells infected by temperature-sensitive mutants of simian virus 40. J. Virol. 14: 116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo, V., Robertson, K., Bibikova, M., Kim, E., Grieco, D., Gottesman, M., Carroll, D., and Gautier, J. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8: 137-147. [DOI] [PubMed] [Google Scholar]
- Diffley, J.F. 1996. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 10: 2819-2830. [DOI] [PubMed] [Google Scholar]
- ———. 2001. DNA replication: Building the perfect switch. Curr. Biol. 11: R367-R370. [DOI] [PubMed] [Google Scholar]
- Dong, Z., Zhong, Q., and Chen, P.L. 1999. The Nijmegen breakage syndrome protein is essential for Mre11 phosphorylation upon DNA damage. J. Biol. Chem. 274: 19513-19516. [DOI] [PubMed] [Google Scholar]
- Featherstone, C. and Jackson, S.P. 1998. DNA repair: The Nijmegen breakage syndrome protein. Curr. Biol. 8: R622-R625. [DOI] [PubMed] [Google Scholar]
- Friedrich, T.D., Laffin, J., and Lehman, J.M. 1992. Simian virus 40 large T-antigen function is required for induction of tetraploid DNA content during lytic infection. J. Virol. 66: 4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1994. Induction of tetraploid DNA content by simian virus 40 is dependent on T-antigen function in the G2 phase of the cell cycle. J. Virol. 68: 4028-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatei, M., Young, D., Cerosaletti, K.M., Desai-Mehta, A., Spring, K., Kozlov, S., Lavin, M.F., Gatti, R.A., Concannon, P., and Khanna, K. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25: 115-119. [DOI] [PubMed] [Google Scholar]
- Harlow, E., Crawford, L.V., Pim, D.C., and Williamson, N.M. 1981. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 39: 861-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka, M. and Dulbecco, R. 1966. Induction of DNA synthesis by SV40. Proc. Natl. Acad. Sci. 56: 736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, H.H. and Tsui, L.C. 1993. Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma 102: 325-332. [DOI] [PubMed] [Google Scholar]
- Heng, H.H., Squire, J., and Tsui, L.C. 1992. High-resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc. Natl. Acad. Sci. 89: 9509-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, J., Dean, F.B., Kwong, A.D., and Lee, S.H. 1990. The in vitro replication of DNA containing the SV40 origin. J. Biol. Chem. 265: 18043-18046. [PubMed] [Google Scholar]
- Johzuka, K. and Ogawa, H. 1995. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139: 1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D. and Smith, A.E. 1984. In vitro mutagenesis of a putative DNA binding domain of SV40 large-T. Virology 139: 109-137. [DOI] [PubMed] [Google Scholar]
- Kierstead, T.D. and Tevethia, M.J. 1993. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J. Virol. 67: 1817-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg, D., Selig, S., Keshet, I., and Cedar, H. 1993. Replication structure of the human β-globin gene domain. Nature 366: 588-590. [DOI] [PubMed] [Google Scholar]
- Lai, J.S. and Herr, W. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. 89: 6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, D.S., Kim, S.T., Xu, B., Maser, R.S., Lin, J., Petrini, J.H., and Kastan, M.B. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404: 613-617. [DOI] [PubMed] [Google Scholar]
- Lin, J.Y. and DeCaprio, J.A. 2003. SV40 large T antigen promotes dephosphorylation of p130. J. Biol. Chem. 278: 46482-46487. [DOI] [PubMed] [Google Scholar]
- Luo, G., Yao, M.S., Bender, C.F., Mills, M., Bladl, A.R., Bradley, A., and Petrini, J.H. 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. 96: 7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R.S., Monsen, K.J., Nelms, B.E., and Petrini, J.H. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17: 6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R.S., Mirzoeva, O.K., Wells, J., Olivares, H., Williams, B.R., Zinkel, R.A., Farnham, P.J., and Petrini, J.H. 2001. Mre11 complex and DNA replication: Linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21: 6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey, D., Brizuela, L., Mohr, I., Marshak, D.R., Gluzman, Y., and Beach, D. 1989. Phosphorylation of large tumour antigen by cdc2 stimulates SV40 DNA replication. Nature 341: 503-507. [DOI] [PubMed] [Google Scholar]
- Meyerson, M., Counter, C.M., Eaton, E.N., Ellisen, L.W., Steiner, P., Caddle, S.D., Ziaugra, L., Beijersbergen, R.L., Davidoff, M.J., Liu, Q., et al. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90: 785-795. [DOI] [PubMed] [Google Scholar]
- Mirzoeva, O.K. and Petrini, J.H. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21: 281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J.K. and Haber, J.E. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms, B.E., Maser, R.S., MacKay, J.F., Lagally, M.G., and Petrini, J.H. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280: 590-592. [DOI] [PubMed] [Google Scholar]
- Paulovich, A.G. and Hartwell, L.H. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82: 841-847. [DOI] [PubMed] [Google Scholar]
- Peden, K.W., Spence, S.L., Tack, L.C., Cartwright, C.A., Srinivasan, A., and Pipas, J.M. 1990. A DNA replication-positive mutant of simian virus 40 that is defective for transformation and the production of infectious virions. J. Virol. 64: 2912-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, M.B. and Lehman, J.M. 1998. Activities of SV40 T antigen necessary for the induction of tetraploid DNA content in permissive CV-1 cells. Cytometry 31: 251-259. [PubMed] [Google Scholar]
- Prives, C. 1990. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell 61: 735-738. [DOI] [PubMed] [Google Scholar]
- Santocanale, C. and Diffley, J.F. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615-618. [DOI] [PubMed] [Google Scholar]
- Scully, R., Chen, J., Plug, A., Xiao, Y., Weaver, D., Feunteun, J., Ashley, T., and Livingston, D.M. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265-275. [DOI] [PubMed] [Google Scholar]
- Seigneur, M., Ehrlich, S.D., and Michel, B. 2000. RuvABC-dependent double-strand breaks in dnaBts mutants require recA. Mol. Microbiol. 38: 565-574. [DOI] [PubMed] [Google Scholar]
- Shiloh, Y. 1997. Ataxia-telangiectasia and the Nijmegen breakage syndrome: Related disorders but genes apart. Annu. Rev. Genet. 31: 635-662. [DOI] [PubMed] [Google Scholar]
- Shirahige, K., Hori, Y., Shiraishi, K., Yamashita, M., Takahashi, K., Obuse, C., Tsurimoto, T., and Yoshikawa, H. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618-621. [DOI] [PubMed] [Google Scholar]
- Simmons, D.T., Wun-Kim, K., and Young, W. 1990. Identification of simian virus 40 T-antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J. Virol. 64: 4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman, B. 1989. Initiation of eukaryotic DNA replication in vitro. Annu. Rev. Cell Biol. 5: 197-245. [DOI] [PubMed] [Google Scholar]
- Taira, T., Iguchi-Ariga, S.M., and Ariga, H. 1994. A novel DNA replication origin identified in the human heat shock protein 70 gene promoter. Mol. Cell. Biol. 14: 6386-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer, P. 1972. Simian virus 40 deoxyribonucleic acid synthesis: The viral replicon. J. Virol. 10: 591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon, R., Vissinga, C., Platzer, M., Cerosaletti, K.M., Chrzanowska, K.H., Saar, K., Beckmann, G., Seemanova, E., Cooper, P.R., Nowak, N.J., et al. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93: 467-476. [DOI] [PubMed] [Google Scholar]
- Wu, X., Ranganathan, V., Weisman, D.S., Heine, W.F., Ciccone, D.N., O'Neill, T.B., Crick, K.E., Pierce, K.A., Lane, W.S., Rathbun, G., et al. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response [see comments]. Nature 405: 477-482. [DOI] [PubMed] [Google Scholar]
- Wun-Kim, K., Upson, R., Young, W., Melendy, T., Stillman, B., and Simmons, D.T. 1993. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J. Virol. 67: 7608-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. and Weaver, D.T. 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25: 2985-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., Sonoda, E., Sasaki, M.S., Morrison, C., Haraguchi, T., Hiraoka, Y., Yamashita, Y.M., Yagi, T., Takata, M., Price, C., et al. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18: 6619-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S., Weng, Y.C., Yuan, S.S., Lin, Y.T., Hsu, H.C., Lin, S.C., Gerbino, E., Song, M.H., Zdzienicka, M.Z., Gatti, R.A., et al. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products [see comments]. Nature 405: 473-477. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Petersen, S., Tessarollo, L., and Nussenzweig, A. 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11: 105-109. [DOI] [PubMed] [Google Scholar]
- Zou, H. and Rothstein, R. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90: 87-96. [DOI] [PubMed] [Google Scholar]