Abstract
Higher plants have 2 GOGAT species, Fd-GOGAT and NADH-GOGAT. While Fd-GOGAT mainly assimilates ammonium in leaves, which is derived from photorespiration, the function of NADH-GOGAT, which is highly expressed in roots,1 needs to be elucidated. The aim of this study was to clarify the role of NADH-GOGAT in Arabidopsis roots. The supply of ammonium to the roots caused an accumulation of NADH-GOGAT, while Fd-GOGAT 1 and Fd-GOGAT 2 showed no response. A promoter–GUS fusion analysis and immunohistochemistry showed that NADH-GOGAT was located in non-green tissues like vascular bundles, shoot apical meristem, pollen, stigma, and roots. The localization of NADH-GOGAT and Fd-GOGAT was not overlapped. NADH-GOGAT T-DNA insertion lines showed a reduction of glutamate and biomass under normal CO2 conditions. These data emphasizes the importance of NADH-GOGAT in the ammonium assimilation of Arabidopsis roots.
Keywords: NADH-GOGAT, root, ammonium, glutamate, assimilation, Arabidopsis, CO2, amino acids
Glutamine oxoglutarate aminotransferase (GOGAT or glutamate synthase) catalyzes the synthesis of 2 glutamates through the transfer of a glutamine amide residue to 2-oxoglutarate.1 GOGAT uses ferredoxin or NADH as electron donor.2,3 One glutamate serves as substrate for the glutamine synthetase (GS), and the other glutamate is used for amino acid metabolism.4 The coupled reaction of the GS/GOGAT cycle is the major ammonium assimilation pathway in higher plants.4-7
Reverse genetic studies showed that Fd-GOGAT participated in assimilation of ammonium derived from photorespiration.8-10 Besides photorespiration, ammonium is generated by the protein catabolism, nitrate reduction, and the phenyl propanoid metabolism.11 Furthermore, ammonium is primarily assimilated in roots, while it is imported from the environment.12-14 NADH-GOGAT was involved in assimilation of ammonium derived from the non-photorespiratory pathway.15,16
However, it is not clear whether root NADH-GOGAT mediated ammonium assimilation is inhibited in the function under ambient air conditions or the function is only given under high CO2 conditions. The tissue and cell-type specific expression of NADH-GOGAT is not fully elucidated yet. Therefore, the article compared the growth of mutants and wild-type plants grown in medium with ammonium as a major nitrogen source under ambient air condition. In addition, the temporal and spatial distribution of NADH-GOGAT was considered.
Quantitative real-time PCR showed that ammonium supply leads to a linear increase of root NADH-GOGAT. The ratio of all root GOGATs was compared at 6 hours after ammonium supply, while NADH-GOGAT and Fd-GOGAT2 contributed 67 and 33% respectively, the content of Fd-GOGAT1 reached less than 1% of all GOGATs. Nitrate supply did not change the GOGAT composition in roots. It is likely that NADH-GOGAT is directly engaged in the root ammonium assimilation.
The contribution of shoot NADH-GOGAT to the plant growth is limited, because of 2 reasons. First, Fd-GOGAT1 is the major GOGAT isoform in shoots.17 Almost 90% of all shoot GOGATs are Fd dependent under the tested nitrogen conditions. Second, only limited organ accumulated NADH-GOGAT in shoot. Protein gel blot analysis, promoter analysis, and immunohistochemistry showed that shoot NADH-GOGAT is localized in vascular bundles of immature organs.
Furthermore, the function of NADH-GOGAT was approached with reverse genetics. The wild-type and 2 T-DNA insertion lines were grown under different nitrogen conditions. Although the ammonium supply of less than 1 mM did not show a difference between wild-type and mutants lines, the supply of 5 mM ammonium reduced the mutant biomass. The result suggests that the ammonium assimilation is NADH-GOGAT dependent under high ammonium concentrations.
Contradictory, wild-type and mutants lines did not show differences in the biomass under nitrate supplied conditions. The largest amount of incorporated nitrate is transported to the shoot, where it is reduced in the leaves to ammonium through nitrate reductase (NR) and nitrite reductase (NiR).18 Since NiR is mainly localized in mesophyll chloroplasts,19 most of the ammonium is produced there. The foliar GS/GOGAT cycle consists of GS2 and Fd-GOGAT1, thus Fd-GOGAT1 should assimilate ammonium derived from the nitrate reduction.
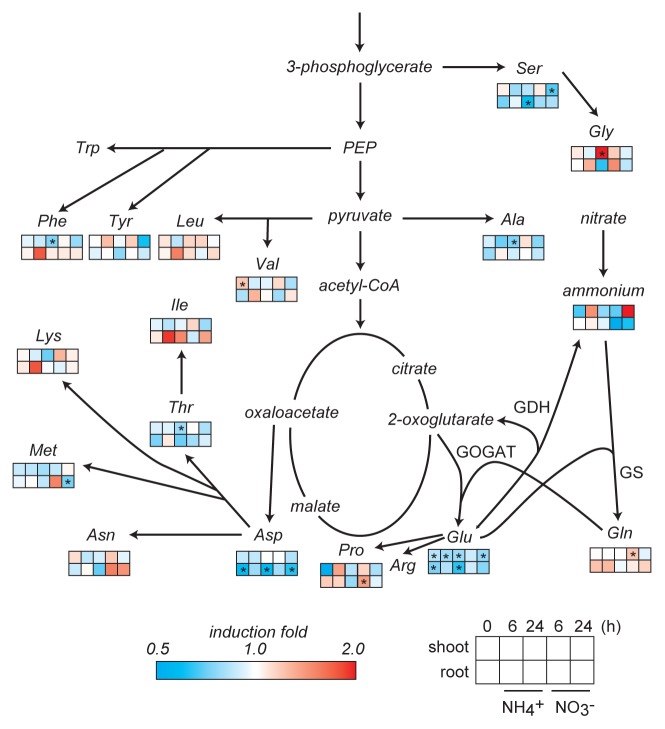
The shoot and root glutamate concentrations in NADH-GOGAT T-DNA insertion lines were lower in comparison to the wild-type at 24 h after ammonium supply (Fig. 1), what suggests a dominant contribution of NADH-GOGAT to the glutamate synthesis. Conversely, the mutant lines tended to accumulate glutamine, one of the substrates for GOGAT reactions. However, neither glutamate was depleted nor mutant lines were lethal under ammonium conditions, suggesting an alternative metabolic pathway that provides glutamate in the mutants. Fd-GOGAT2 represents the second largest accumulation of GOGAT in roots; therefore it may provide glutamate in the mutants. Beside Fd-GOGAT2, glutamate dehydrogenase (GDH) could be another candidate for root ammonium assimilation.16,21 GDH catalyzes the reversible amination of 2-oxoglutarate (2OG) for the synthesis of Glu using ammonium as a substrate.22 Recent reverse-genetic research revealed that the main physiological function of NADH-GDH is to provide 2OG for the tricarboxylic acid cycle.23 It would make sense to compare the Glu concentrations of a quadruple knock out (nadh-gogat, gdh1–2-3) with that of single knock out mutant (nadh-gogat) under high ammonium conditions in order to study the conjunction of NADH-GOGAT with GDH.23
Figure 1. Schematic representation of the free amino acid composition in NADH-GOGAT mutant lines. Plants of the wild-type and 2 T-DNA insertion lines were grown on vertical MGRL agar plates20 containing 7 mM nitrate as nitrogen source for 14 d before they were transferred to MGRL medium without nitrogen for 3 d. Afterwards plants were transferred to the medium supplemented with either 10 mM KNO3 or NH4Cl. The plants were harvested at 6 or 24 h after nitrogen supply. The concentration of amino acids in wild-type plants was compared with the 2 T-DNA insertion lines, whereas the increase or decrease is represented by different color. One-way ANOVA followed by Dunnett tests were used to identify significant differences between wild-type and T-DNA insertion lines (P < 0.05).
Since glutamate plays a pivotal role in the plant amino acid metabolism,3,24 their composition was compared between wild-type and T-DNA insertion lines (Fig. 1). The aspartate (Asp) concentration of the NADH-GOGAT T-DNA insertion line was reduced compared with the wild-type. This decrease may reflect the conversion of aspartate to glutamate as a compensation for a low glutamate concentration. Asp aminotransferase catalyzes the conversion of Asp and 2OG to oxaloacetate and Glu.25
NADH-GOGAT T-DNA insertion lines showed not only a reduction of glutamate but also changes of other amino acids. Mutants showed an increase of branched-chain amino acids, Phe, and Lys in roots, but a decrease in shoots, at 6 h after ammonium supply. These dramatic differences were weakened at 24 h after ammonium supply, suggesting 2 points. First, those amino acid concentrations seem to be dependent on the pool size of Glu, because their biosynthesis cycles are not closer in pathway distance from GS/GOGAT cycle. Second, NADH-GOGAT plays a key role in the regulation of Glu concentrations in response to ammonium in roots.
Therefore, a loss of functional NADH-GOGAT leads to a decreased biomass under high ammonium condition. This article showed that ammonium supply leads to an increase of NADH-GOGAT, which assimilates ammonium in the root.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
JSPS KAKENHI Grant Numbers 21688006 and 26450073 to S.K., and 22119003 to T.Y., supported this work. Japan Advanced Plant Research Network supported by JSPS was also acknowledged for the use of Elemental Analyzer.
Glossary
Abbreviations:
- GOGAT
glutamate synthase
- GS
glutamine synthetase
- GUS
GUS β-glucuronidase
- NADH
nicotinamide adenine dinucleotide
- PCR
polymerase chain reaction
- SAM
shoot apical meristem
- T-DNA
transfer DNA
References
- 1.Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM. MeloOliveira R, Coruzzi GM. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:569–93. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- 2.Temple SJ, Vance CP, Gantt JS. Glutamate synthase and nitrogen assimilation. Trends Plant Sci. 1998;3:51–6. doi: 10.1016/S1360-1385(97)01159-X. [DOI] [Google Scholar]
- 3.Forde BG, Lea PJ. Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot. 2007;58:2339–58. doi: 10.1093/jxb/erm121. [DOI] [PubMed] [Google Scholar]
- 4.Lea PJ, Miflin BJ. Alternative route for nitrogen assimilation in higher plants. Nature. 1974;251:614–6. doi: 10.1038/251614a0. [DOI] [PubMed] [Google Scholar]
- 5.Lea PJ, Ireland RJ. Nitrogen metabolism in higher plants. In: Plant Amino Acids: Biochemistry and Biotechnology. Singh BK, ed. New York: Marcel Dekker, Inc, 1999:1-47. [Google Scholar]
- 6.Funayama K, Kojima S, Tabuchi-Kobayashi M, Sawa Y, Nakayama Y, Hayakawa T, Yamaya T. Cytosolic glutamine synthetase1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 2013;54:934–43. doi: 10.1093/pcp/pct046. [DOI] [PubMed] [Google Scholar]
- 7.Goodall AJ, Kumar P, Tobin AK. Identification and expression analyses of cytosolic glutamine synthetase genes in barley (Hordeum vulgare L.) Plant Cell Physiol. 2013;54:492–505. doi: 10.1093/pcp/pct006. [DOI] [PubMed] [Google Scholar]
- 8.Somerville CR, Ogren WL. Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature. 1980;286:257–9. doi: 10.1038/286257a0. [DOI] [Google Scholar]
- 9.Morris PF, Layzell DB, Canvin DT. Ammonia production and assimilation in glutamate synthase mutants of Arabidopsis-thaliana. Plant Physiol. 1988;87:148–54. doi: 10.1104/pp.87.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kendall AC, Wallsgrove RM, Hall NP, Turner JC, Lea PJ. Carbon and nitrogen metabolism in barley (Hordeum vulgare L.) mutants lacking ferredoxin-dependent glutamate synthase. Planta. 1986;168:316–23. doi: 10.1007/BF00392355. [DOI] [PubMed] [Google Scholar]
- 11.Schjoerring JK, Husted S, Mäck G, Mattsson M. The regulation of ammonium translocation in plants. J Exp Bot. 2002;53:883–90. doi: 10.1093/jexbot/53.370.883. [DOI] [PubMed] [Google Scholar]
- 12.Britto DT, Kronzucker HJ. NH4+ toxicity in higher plants: a critical review. J Plant Physiol. 2002;159:567–84. doi: 10.1078/0176-1617-0774. [DOI] [Google Scholar]
- 13.Marschner H. Mineral Nutrition of Higher Plants. London: Academic Press, 1995. [Google Scholar]
- 14.Andrews M, Raven JA, Lea PJ. Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Ann Appl Biol. 2013;163:174–99. doi: 10.1111/aab.12045. [DOI] [Google Scholar]
- 15.Lancien M, Martin M, Hsieh MH, Leustek T, Goodman H, Coruzzi GM. Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J. 2002;29:347–58. doi: 10.1046/j.1365-313X.2002.01218.x. [DOI] [PubMed] [Google Scholar]
- 16.Potel F, Valadier MH, Ferrario-Méry S, Grandjean O, Morin H, Gaufichon L, Boutet-Mercey S, Lothier J, Rothstein SJ, Hirose N, et al. Assimilation of excess ammonium into amino acids and nitrogen translocation in Arabidopsis thaliana--roles of glutamate synthases and carbamoylphosphate synthetase in leaves. FEBS J. 2009;276:4061–76. doi: 10.1111/j.1742-4658.2009.07114.x. [DOI] [PubMed] [Google Scholar]
- 17.Lam HM, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh MH, Coruzzi G. Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell. 1995;7:887–98. doi: 10.1105/tpc.7.7.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sechley KA, Yamaya T, Oaks A. Compartmentation of nitrogen assimilation in higher-plants. Int Rev Cytol. 1992;134:85–163. doi: 10.1016/S0074-7696(08)62028-8. [DOI] [Google Scholar]
- 19.Crawford NM. Nitrate: nutrient and signal for plant growth. Plant Cell. 1995;7:859–68. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S. Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 1992;99:263–8. doi: 10.1104/pp.99.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benstein RM, Ludewig K, Wulfert S, Wittek S, Gigolashvili T, Frerigmann H, Gierth M, Flügge UI, Krueger S. Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell. 2013;25:5011–29. doi: 10.1105/tpc.113.118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tercé-Laforgue T, Dubois F, Ferrario-Méry S, de Crecenzo MAP, Sangwan R, Hirel B. Glutamate dehydrogenase of tobacco is mainly induced in the cytosol of phloem companion cells when ammonia is provided either externally or released during photorespiration. Plant Physiol. 2004;136:4308–17. doi: 10.1104/pp.104.047548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontaine JX, Tercé-Laforgue T, Armengaud P, Clément G, Renou JP, Pelletier S, Catterou M, Azzopardi M, Gibon Y, Lea PJ, et al. Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell. 2012;24:4044–65. doi: 10.1105/tpc.112.103689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Torre F, El-Azaz J, Avila C, Cánovas FM. Deciphering the role of aspartate and prephenate aminotransferase activities in plastid nitrogen metabolism. Plant Physiol. 2014;164:92–104. doi: 10.1104/pp.113.232462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azevedo RA, Lancien M, Lea PJ. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids. 2006;30:143–62. doi: 10.1007/s00726-005-0245-2. [DOI] [PubMed] [Google Scholar]