Abstract
SN1-type alkylating agents represent an important class of chemotherapeutics, but the molecular mechanisms underlying their cytotoxicity are unknown. Thus, although these substances modify predominantly purine nitrogen atoms, their toxicity appears to result from the processing of O6-methylguanine (6MeG)-containing mispairs by the mismatch repair (MMR) system, because cells with defective MMR are highly resistant to killing by these agents. In an attempt to understand the role of the MMR system in the molecular transactions underlying the toxicity of alkylating agents, we studied the response of human MMR-proficient and MMR-deficient cells to low concentrations of the prototypic methylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). We now show that MNNG treatment induced a cell cycle arrest that was absolutely dependent on functional MMR. Unusually, the cells arrested only in the second G2 phase after treatment. Downstream targets of both ATM (Ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) kinases were modified, but only the ablation of ATR, or the inhibition of CHK1, attenuated the arrest. The checkpoint activation was accompanied by the formation of nuclear foci containing the signaling and repair proteins ATR, the S*/T*Q substrate, γ-H2AX, and replication protein A (RPA). The persistence of these foci implied that they may represent sites of irreparable damage.
Keywords: ATM/ATR, cell cycle arrest, DNA damage signaling, G2 check point, methylating agents, mismatch repair
Treatment of cells with clastogenic DNA damaging agents such as ionizing radiation (IR) generally results in the rapid activation of damage signaling pathways, cell cycle arrest and, depending on the extent of damage, either recovery or cell death. IR causes predominantly DNA base modifications (Cooke et al. 2003), which are rapidly and efficiently processed by the base excision repair (BER) system. Interestingly, this metabolic pathway does not appear to trigger DNA damage checkpoints. Instead, IR-induced signaling events are believed to be associated exclusively with the detection or processing of single- and double-strand breaks (DSBs), which rapidly activate the ATM (Ataxia telangiectasia mutated) kinase (Bakkenist and Kastan 2003) and, later, also ATR (ATM and Rad3-related; Brown and Baltimore 2003). DNA damage-induced signaling cascades can be activated also by DNA replication forks stalled by DNA damage (e.g., ultraviolet-induced photodimers or cross-links), nucleotide depletion (e.g., on hydroxyurea treatment), or polymerase arrest (e.g., by aphidicolin). In all the latter cases, the signaling events are triggered in the first S phase after treatment and involve primarily the activation of ATR kinase and its downstream targets (Abraham 2001; Osborn et al. 2002; Shiloh 2003).
DNA damage signaling induced by SN1-type methylating agents has to date not been studied in detail. Treatment of cells with N-methyl-N-nitrosourea (MNU) and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) gives rise predominantly to N7-methylguanine (7MeG), N3-methyladenine (3MeA), O4-methylthymine (4MeT), O6-methylguanine (6MeG), and methyl-phosphotriesters in their DNA. The major adducts, 7MeG and 3MeA, represent ∼70% of the damage. However, both these methylated bases are efficiently removed from DNA by alkyl-adenine DNA-glycosylase (Scharer and Jiricny 2001), and the resulting abasic sites are repaired by the BER pathway (Seeberg et al. 1995), without causing undue cytotoxicity at low concentrations. Interestingly, the cytotoxicity of the above methylating agents is ascribed to 6MeG, detoxified by methylguanine methyl transferase (MGMT), which reverts it back to guanine (Sedgwick and Lindahl 2002). 6MeG residues were implicated in cell killing when cells expressing high levels of MGMT were shown to be highly resistant to killing by MNU (Karran 2001), but how can persistent 6MeG residues in DNA lead to cell death?
In 1993, the presence of 6MeG in plasmid DNA was shown to inhibit DNA replication, but also to stimulate DNA repair synthesis (Ceccotti et al. 1993). This evidence was extended to show that 6MeG residues did not inhibit DNA polymerases per se, but that DNA replication was arrested through a trans-acting signal generated during the processing of 6MeG residues in DNA (Zhukovskaya et al. 1994). The discovery that cells defective in both mismatch repair (MMR) and MGMT were resistant to killing by methylating agents implicated the MMR system in this processing. The MMR substrates are thought to be 6MeG/T mispairs, which arise during replication of methylated DNA because of the propensity of 6MeG to preferentially base pair with thymine. The recognition of the 6MeG/T mispairs by the mismatch binding factor hMutSα (Duckett et al. 1996) is believed to activate a signal transduction pathway that results in a G2/M arrest (Aquilina et al. 1999; Cejka et al. 2003). However, how this arrest is activated is currently unclear. One hypothesis proposes that the repeated loading of the mismatch binding proteins at the mismatch site may be sufficient to activate a DNA damage-signaling cascade (Fishel 1998). The other suggests that the cell cycle arrest is activated by nonproductive, repetitive processing of 6MeG/T mispairs by the MMR system, or by intermediates arising as a result of this processing (for review, see Bellacosa 2001; Karran 2001). We set out to gain more insights into the molecular transactions underlying the G2/M cell cycle arrest induced by methylating agents of SN1 type. To this end, we studied the behavior of proteins involved in DNA damage signaling and processing in a cell line in which MMR status can be tightly controlled (Cejka et al. 2003).
Results
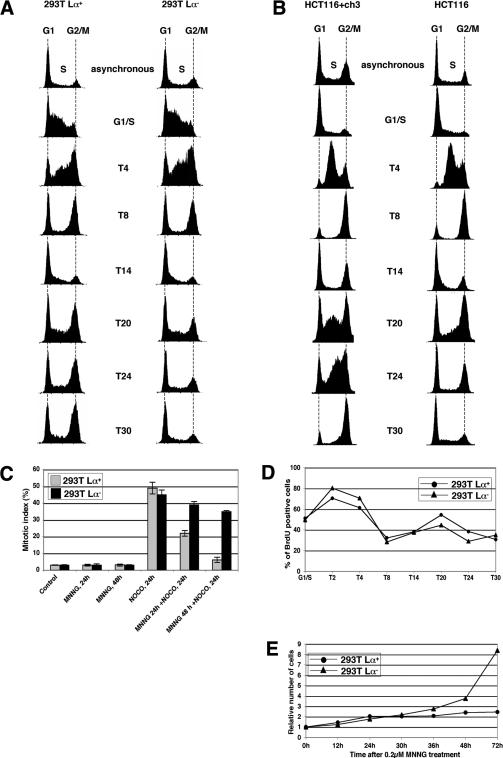
MNNG-induced MMR-dependent G2 arrest occurs in the second cell cycle
The human embryonic kidney cell line 293T is MMR-deficient and does not convert MeG in its DNA back to G, as the promoters of the hMLH1 (Trojan et al. 2002) and MGMT (Cejka et al. 2003) genes are epigenetically silenced. We used these cells to generate the 293T Lα cell line, which carries a stably integrated hMLH1 cDNA minigene controlled by the TetOff expression system. In the absence of doxycycline (Dox), these cells, referred to as 293T Lα+, express hMLH1, are MMR-proficient, and are sensitive to killing by MNNG (Cejka et al. 2003; Di Pietro et al. 2003). In contrast, when the same cells are grown in the presence of 50 ng/mL Dox (293T Lα- cells), they shut off hMLH1 expression, display a MMR defect, and are 125-fold more resistant to MNNG than 293T Lα+ cells. Flow cytometric analysis showed that on treatment with 0.2 μM MNNG, the 293T Lα+ cells arrested with a DNA content of 4n (Cejka et al. 2003). Interestingly, the arrest did not take place in the first cell cycle, as synchronized 293T Lα cells treated with MNNG at the G1/S-transition progressed through the first G2/M boundary and mitosis irrespective of their MMR status. The arrest was activated after the second S phase, and only in the MMR-proficient 293T Lα+ cells (Fig. 1A). However, 293T cells express the SV40 large T antigen, as well as the adenoviral E1A and E1B proteins, which inhibit the functions of the retinoblastoma (Rb) and p53 tumor suppressor proteins in regulating the G1/S transition on DNA damage (Bartek and Lukas 2001). To ensure that the proper functioning of DNA damage response in 293T Lα+ cells was not affected, and to show that the observed arrest in the second cell cycle was not limited to this cell line, we repeated this experiment with synchronized HCT116 (hMLH1-deficient) and HCT116 + ch3 (hMLH1-proficient) cells that carry both functional p53 and pRb. As shown in Figure 1B, both these latter cell lines proceeded through the first cell cycle in a similar manner. However, 20 h posttreatment, the MMR-proficient HCT116 + chr3 cells began to accumulate in the second S phase and then proceeded to arrest with a DNA content of 4n (T30), whereas the MMR-deficient HCT116 cells exited the second S phase normally and continued to cycle.
Figure 1.
Kinetics of the G2/M arrest in cells treated with 0.2 μM MNNG. (A) FACS analysis of 293T Lα cell cultures synchronized in G1/S with a double thymidine block and treated with 0.2 μM MNNG. (T4-T30) FACS analyses carried out 4-30 h posttreatment. (B) FACS analysis of MNNG-treated HCT116 and HCT116 + ch3 cell cultures synchronized in G1/S with HU. (T4-T30) FACS analyses carried out 4-30 h posttreatment. The 4n peak in the unsynchronized and HU-synchronized HCT116 + chr3 cells is larger than in the HCT116 cells. This is not the result of a larger fraction of diploid cells in the G2 phase of the cell cycle, but to a subpopulation of tetraploid cells, which arrest after MNNG treatment with a content of 8n (not shown). (C) Mitotic index of 293T Lα cells after MNNG treatment. The cells were treated with MNNG, and nocodazole was added 24 or 48 h later. The cells were microscopically examined after an additional 24 h. As shown, substantially more MMR-deficient 293T Lα- cells were arrested in mitosis than were MMR-proficient 293T Lα+ cells, which indicates that the latter cells were more frequently arrested in G2. (D) Synchronized 293T Lα cells (as in A) were pulse-labeled with BrdU, and the number of cells in S phase was estimated by CELLQuest software. Both 293T Lα+ (•) and 293T Lα- (▴) cells entered the second S phase between T14 and T24. (E) Growth curves of unsynchronized MNNG-treated 293T Lα cells. Although the treated 293T Lα+ cells doubled their number 24 h after treatment and then ceased to proliferate, the 293T Lα- cells continued to grow.
We next had to confirm that the cells indeed arrested in the G2 phase of the cell cycle, rather than stopping because of a mitotic catastrophe. To this end, we added nocodazole, an inhibitor of mitotic spindle formation, to the MNNG-treated cell cultures 24 h before cytological analysis. If the treated cells were arrested in G2, they could not traverse to mitosis. Thus, nocodazole should block only cells that failed to arrest and continued to cycle. As shown in Figure 1C, the MMR-deficient 293T Lα- cells treated first with MNNG and then with nocodazole were frequently arrested in mitosis. This indicates that they did not arrest before this phase. In contrast, when nocodazole was added to the MNNG-treated MMR-proficient 293T Lα+ cells, the number of cells reaching mitosis was substantially lower, which shows that more MMR-proficient cells preferentially arrested in G2 after MNNG treatment. The finding that arrest took place after the second S phase was further confirmed by bromodeoxyuridine (BrdU) labeling experiments, in which synchronized, MNNG-treated 293T Lα cells were shown to enter the second S phase between 14 and 24 h after treatment, irrespective of their MMR status (Fig. 1D). As shown in the graph, the number of cells in the second S phase appeared lower than in the first. To see whether some cells died during the course of this experiment and were therefore lost, we followed the proliferation of the unsynchronized, MNNG-treated cell populations. As shown in Figure 1E, no appreciable cell loss occurred: The MNNG-treated MMR-proficient cells doubled in number during the first 24 h and then arrested, whereas the treated MMR-deficient cells continued to proliferate. This showed that the decrease in cell number in the second S phase (Fig. 1D) was only apparent and was most likely the result of the gradual loss of synchronization. In summary, the MNNG-induced checkpoint in MMR-proficient cells is activated after the second S phase and is absolutely dependent on a functional MMR system.
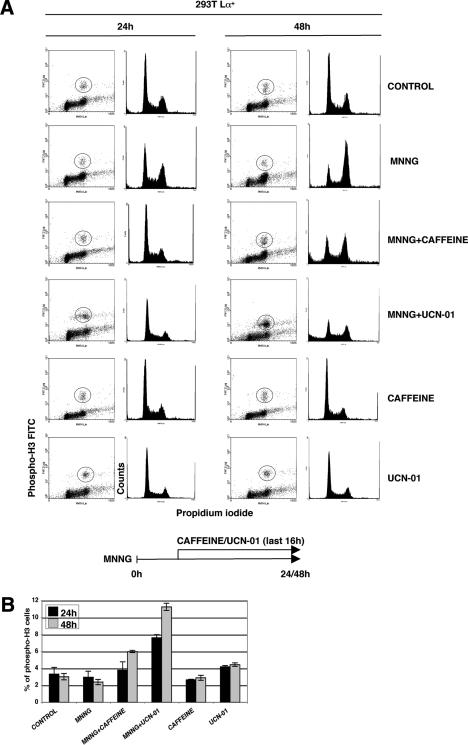
Caffeine and UCN-01 abrogate the MMR-dependent G2 arrest
We wanted to check whether the MNNG-induced cell cycle arrest observed in the 293T Lα+ cells was brought about by a physical block to DNA synthesis (e.g., collapsed replication forks, aberrant recombination intermediates) or whether it was caused by the activation of a DNA damage checkpoint. As the latter process involves the major DNA damage-signaling protein kinases ATM and ATR, which are inhibited by caffeine (Sarkaria et al. 1999; Zhou et al. 2000), we decided to test whether the MNNG-induced arrest was sensitive to this drug. As shown in Figure 2, this was indeed the case. Fluorescence-activated cell sorting (FACS) analysis of cell populations doubly stained with propidium iodide and an antibody against the phosphorylated form of histone H3 (Xu et al. 2001) allowed us to distinguish between G2-arrested and mitotic cells, as H3 is phosphorylated on Ser 10 only during mitosis (Crosio et al. 2002). In the initial set of experiments (data not shown), we pretreated the cells with caffeine 30 min before adding MNNG and then incubated the cells for a further 24 or 48 h. Using this protocol, we failed to observe any differences between caffeine-treated and untreated cells, as measured by Western blotting with the phospho-H3 antibody, probably because the half-life of caffeine is only 4.5 h. We therefore added the kinase inhibitor some hours after the MNNG treatment. Using this protocol, MNNG-treated cells with a DNA content of 4n accumulated as observed previously (Fig. 1B), but the addition of caffeine to the treated cells 16 h before harvesting reduced the number of arrested cells by a substantial amount at the 24- and 48-h time points, as well as causing substantial cell death (Fig. 2A). That the latter effect was linked to an increased fraction of cells arriving in mitosis with unrepaired DNA is witnessed by a greater number of mitotic cells with phosphorylated histone H3 (Fig. 2B).
Figure 2.
The G2 arrest in MMR-proficient 293T Lα+ cells is caffeine- and UCN-01-sensitive. (A) 293T Lα+ cells were treated with MNNG (0.2 μM) for the indicated times, and caffeine (2 mM) or UCN-01 (100 nM) was added 16 h before harvesting. The cells were stained with propidium iodide (PI) and phospho-histone H3 antibody (cells in circle) to distinguish mitotic cells from those in G2. The results show that both inhibitors attenuated the G2 arrest in MNNG-treated cells. (B) Quantification of phospho-H3-positive cells from A. The number of cells entering mitosis in samples treated with MNNG and caffeine or UCN-01 was higher than in the controls, which shows that these kinase inhibitors abrogated the G2 arrest and allowed more cells to enter mitosis.
The initiation of G2 arrest requires CHK1, the major transducer of ATR-dependent DNA damage signaling (Liu et al. 2000). As CHK1 kinase activity can be preferentially inhibited by the staurosporine analog UCN-01 (Busby et al. 2000; Graves et al. 2000), we studied the response of MNNG-treated cells to this drug in a way analogous to that deployed for caffeine. FACS analysis (Fig. 2A) showed that UCN-01 treatment abrogated the MNNG-induced G2 checkpoint in 293T Lα+ cells to an even greater extent than caffeine. Correspondingly, the fraction of cells arriving in mitosis, seen in FACS analysis as phospho-H3-positive cells, was higher in the MNNG- and UCN-01-treated samples than in cells treated with MNNG and caffeine (Fig. 2B).
Taken together, these results demonstrate that the MNNG-induced cell cycle arrest was indeed induced by a DNA damage-signaling cascade. Our data thus help explain the nature of the in trans inhibition of DNA replications in cells treated with methylating agents that was described more than a decade ago (Zhukovskaya et al. 1994).
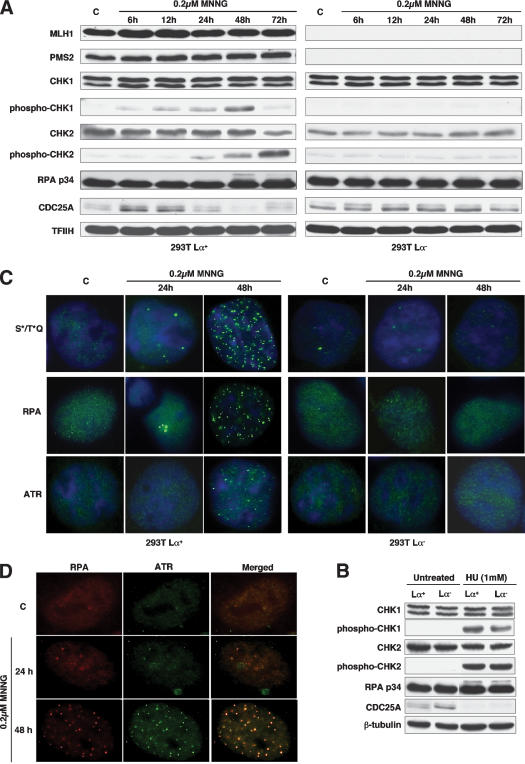
Low-dose MNNG treatment brings about MMR-dependent phosphorylation of downstream targets of both ATM and ATR
ATM and ATR are both activated by DNA damage. However, whereas ATM responds rapidly to clastogenic damage such as that induced by IR (Bakkenist and Kastan 2003), ATR responds slower and cooperates with ATM in the later phases of the response (Brown and Baltimore 2003). ATR is also known to be preferentially activated on replication fork arrest induced by ultraviolet (UV) light, hydroxyurea (HU), or DNA polymerase inhibitors such as aphidicolin (Abraham 2001; Osborn et al. 2002). As MNNG treatment is thought to exert its cytotoxicity through the processing of 6MeG residues during DNA synthesis (Karran and Bignami 1992), it might be anticipated that the damage-induced signaling cascade would initiate in S phase and involve ATR rather than ATM. Indeed, when the 293T Lα+ cells were treated with 0.2 μM MNNG, phosphorylation of the ATR-activated checkpoint kinase CHK1 on Ser 345 became detectable after 12 h and peaked at 48 h, whereas phosphorylation of Thr 68 of CHK2, a preferred target for ATM, lagged by 12 h and increased steadily until 72 h (Fig. 3A).
Figure 3.
MMR-dependent DNA damage signaling in 293T Lα cells. (A) The 293T Lα+ cells express hMLH1 and hPMS2 and are MMR proficient. Treatment with 0.2 μM MNNG brought about the phosphorylation of CHK1 and CHK2, as well as that of the single-strand DNA-binding protein RPA (p34), while CDC25A was gradually degraded. None of these modifications was observed in the MMR-deficient 293T Lα- cells. The phosphorylation status of RPA is indicated by the slower migration of the modified polypeptides through polyacrylamide gels (48-h time point). TFIIH was used as loading control. (B) Treatment of 293T Lα cells with 1 mM HU brought about a MMR-independent phosphorylation of CHK1, CHK2, and RPA (p34) and the degradation of CDC25A within 24 h. β-tubulin was used as loading control. (C) Indirect immunofluorescence imaging of nuclear foci formed by protein targets of the ATM/ATR kinases phosphorylated on serine and threonine residues in the SQ or TQ motifs, RPA (p34) and ATR. As shown, the foci formed only in the MMR-proficient 293T Lα+ cells and were most numerous 48 h after treatment. (D) Indirect immunofluorescence imaging of nuclear foci formed by RPA (p34) and ATR in HeLa cells treated with 0.2 μM MNNG. The images were superimposed using Adobe Photoshop software. (C) Control, untreated cells.
We also examined the posttranslational modification of the single-stranded DNA-binding protein RPA, reported to redirect its function from replication to repair (Wang et al. 2001) through recruitment of the ATR/ATRIP (ATR-interacting protein) complex onto sites of DNA damage, which leads to an ATR-mediated activation of CHK1 (Zou and Elledge 2003). The p34 subunit of RPA was indeed phosphorylated after MNNG treatment, and the timing of its posttranslational modification coincided with the appearance of the highest levels of phosphorylated CHK1 (Fig. 3A).
The steady-state levels of CDC25A, a cell cycle phosphatase that is degraded on phosphorylation by CHK1 or CHK2 (Falck et al. 2001; Zhao et al. 2002), began to decline 24 h after treatment, at which time point only CHK1 kinase appeared to be activated. CDC25A controls the activation of CDK1 and CDK2 kinases and is known to regulate the G1 (Hoffmann et al. 1994), intra-S (Falck et al. 2001), and G2/M (Mailand et al. 2002) checkpoints. Its phosphorylation by CHK1/CHK2 leads to its destruction by the proteasome and thus also to cell cycle arrest. Indeed, 24 h after treatment, CDC25A levels were substantially lower than at the earlier time points. Taken together, this evidence suggests that ATR downstream targets were posttranslationally modified during the first cell cycle and that CHK2, a target of ATM, became activated later, after the second S phase. Importantly, none of these phenomena were apparent in the MMR-deficient 293T Lα- cells (Fig. 3A), which failed to arrest following MNNG treatment (Fig. 1A,E).
In a control experiment, we treated the 293T Lα cells with HU, which is known to bring about a cell cycle arrest in the first S phase after treatment. As shown in Figure 3B, CHK1, CHK2, and RPA-p34 phosphorylation was detectable already at the 24 h time point and, as anticipated, no differences were observed between the MMR-proficient and the MMR-deficient cells. CDC25A was undetectable in the treated cells at this time point, again irrespective of the cells' MMR status. We failed to observe MMR-dependent differences in phosphorylation patterns and CDC25A degradation also after 6 and 48 h (data not shown). These results both confirm that the 293T Lα- cells do not have defective checkpoint activating pathways and show that the signals triggering the HU- and MNNG-dependent G2 checkpoints are different.
MNNG treatment induces ATM/ATR activation in vivo
We set out to seek evidence of the activation of ATM and ATR protein kinases in living cells. To this end, we employed the phospho-(Ser/Thr) ATM/ATR substrate (S*/T*Q) antibody that was raised against peptides carrying SQ or TQ amino acid motifs known to be posttranslationally modified by these kinases in several different substrates, and that is an accepted marker of ATM/ATR-dependent phosphorylation events (DiTullio et al. 2002). As shown in Figure 3C, foci of phosphorylated polypeptides began to appear after 24 h, but they were most numerous 48 h posttreatment, where they were visible in 67% of the cells. A similar phenomenon was observed also for RPA (78% of cells with foci) and ATR (75% of cells with foci). Again, these changes were observed exclusively in the MMR-proficient 293T Lα+ cells. In MNNG-treated MMR-proficient HeLa cells, the ATR and RPA foci colocalized (Fig. 3D; Zou and Elledge 2003). Notably, the initial signs of checkpoint activation in the form of phosphorylation of CHK1 and degradation of CDC25A in 293T Lα+ cells (Fig. 3A) preceded the appearance of the foci in both cell types by ∼40 h. This implied that the ATM/ATR kinases were activated long before the ATR, RPA, and S*/T*Q proteins formed the foci. As the appearance of the foci coincides with the formation of chromosomal aberrations (N. Mojas, L. Stojic, and J. Jirivny, in prep.), it is possible that the nuclear foci represent recombination intermediates arising during the second S phase.
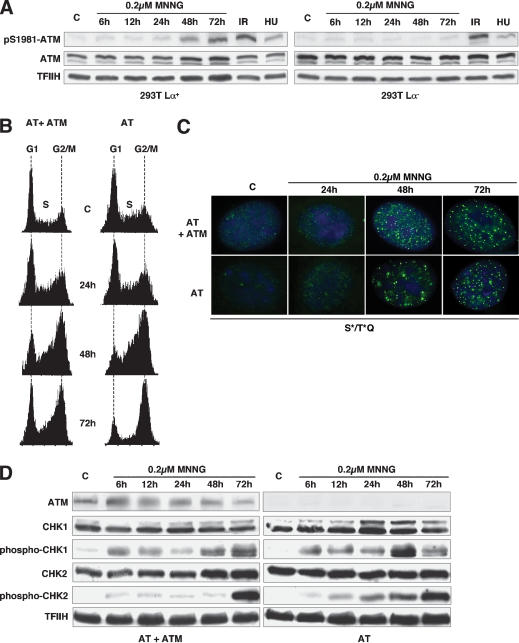
ATM is dispensable for cell cycle arrest induced by low dose MNNG treatment
As shown above, the MNNG-induced G2 checkpoint was released by UCN-01, which inhibits preferentially the CHK1 kinase, the preferred target of ATR. Given that the latter kinase has been implicated in the control of S-phase checkpoints triggered by arrested replication forks (Abraham 2001; Osborn et al. 2002) and that the nonproductive processing of 6MeG/T mispairs by the MMR system should also signal during the S phase, it seemed logical that ATR also should be involved in the control of the MNNG-induced checkpoint described above. However, as ATR and ATM display a certain degree of functional redundancy, we wanted to exclude the involvement of the latter kinase in checkpoint activation. Under normal conditions, ATM is present in the nucleus in an inactive, dimeric form, but it can be rapidly activated by stress stimuli. This process involves disruption of the dimer and is accompanied by ATM autophoshorylation of Ser 1981 (Bakkenist and Kastan 2003). Using a specific antibody against this phosphorylated isoform, we were able to follow activation of the ATM kinase in the 293T Lα cells following treatment with 0.2 μM MNNG. In a control experiment, ATM was efficiently activated by IR treatment, irrespective of the MMR status of the cells, whereas HU treatment was significantly less effective in activating ATM, as anticipated (Fig. 4A). Treatment with MNNG resulted in ATM activation, although only at the 48- and 72-h time points, which coincided with the peak of phosphorylation of CHK2, a known downstream target of ATM (Fig. 3A). Notably, both these events were observed exclusively in the MMR-proficient 293T Lα+ cells.
Figure 4.
ATM is activated but dispensable for the MNNG-induced G2 arrest in MMR-proficient cells. (A) ATM was activated early in both 293T Lα+ and 293T Lα- cells on ionizing radiation (10 Gy, 1 h) and, to a lesser degree, after HU treatment (1 mM, 6 h). In contrast, on treatment with MNNG, ATM was activated late, and only in MMR-proficient (293T Lα+) cells. ATM activation was assessed using an antibody against phosphorylated Ser 1981. (B) FACS analysis of unsynchronized cultures of AT and AT + ATM fibroblasts following treatment with 0.2 μM MNNG. Both ATM-proficient (AT + ATM) and ATM-deficient (AT) cells proceeded through the cell cycle with similar kinetics and began to accumulate in G2/M after 2 d. (C) Indirect immunofluorescence imaging of nuclear foci formed by protein targets of the ATM/ATR kinases phosphorylated on serine and/or threonine residues in the SQ or TQ motifs. As shown, the foci began to form in both ATM-proficient and ATM-deficient cells after the 24 h time point. At 48 h, both cell types contained foci, even though they were less numerous in the AT cells. However, at 72 h, no significant differences in focus number or intensity were observed in the two cell types. (D) MNNG treatment leads to ATM-independent CHK1 and CHK2 activation, albeit with different kinetics.
Although the above experiment demonstrated that ATM was activated in a MMR-dependent manner by MNNG treatment in 293T Lα+ cells, it failed to show whether this kinase was indispensable for activation of the cell cycle arrest. This question was answered with the help of a matched pair of ATM-positive and ATM-negative fibroblast lines (Ziv et al. 1997), which displayed no major differences in G2 arrest efficiency on MNNG treatment, as assessed by FACS analysis (Fig. 4B).
Consistent with the above evidence, the number and kinetics of appearance of S*/T*Q foci on treatment with 0.2 μM MNNG was similar in the AT and AT + ATM cells (Fig. 4C). These foci were not detected in the AT cells on IR treatment (DiTullio et al. 2002), which strongly suggested that the lesions generated by the MMR system during processing of MNNG-induced damage are distinct from IR-induced strand breaks.
Analysis of protein phosphorylation cascades by Western blotting revealed that CHK1 and CHK2 were posttranslationally modified in both cell lines, albeit with different kinetics (Fig. 4D). In a recent report, Wang and colleagues (2003) showed that in IR-treated AT-deficient cells, the ATR kinase compensated for the lack of ATM through overactivation of CHK1. We now extend these findings to MNNG treatment, as the phosphorylation of CHK1 at the 48-h time point was substantially stronger in the AT cells than in the corrected AT + ATM line. Taken together, the results presented in Figure 4 demonstrate that although MNNG treatment led to activation of ATM, this kinase was dispensable for triggering the protein phosphorylation cascade and the G2 cell cycle arrest.
The MNNG-induced G2 arrest and DNA damage-dependent signaling requires ATR
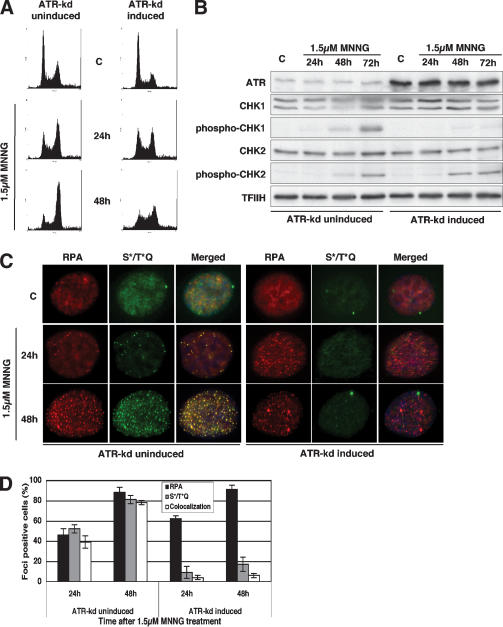
As ATM was not required for the MNNG-induced checkpoint activation (Fig. 4), we set out to confirm the involvement of ATR. Unlike in the case of ATM, there are no stable ATR-defective cell lines, as the loss of this kinase is lethal, and we therefore had to resort to using U2OS cells, which overexpress a kinase-dead variant of ATR (ATR-kd) under the control of the Dox-regulated TetOn expression system (Nghiem et al. 2002). ATR-kd overexpression was shown to abrogate the G2 arrest (Cliby et al. 2002) and to sensitize cells to several DNA damaging agents (Nghiem et al. 2002), and we wanted to see how it affected the cellular response to MNNG. The U2OS cells were substantially more resistant to MNNG than the 293T Lα+, AT, and ATM + AT lines (even though the MGMT activity of all the lines was inhibited with O6-benzylguanine), and we therefore had to use a 1.5-μM concentration of the drug to obtain cytotoxicity comparable to that exerted on the latter cells by 0.2 μM MNNG. Under these experimental conditions, the uninduced U2OS cells were largely arrested in the G2 phase of the cell cycle 48 h after treatment (Fig. 5A, left panel), similar to the 293T Lα+ cells (Fig. 1B). However, this arrest was substantially attenuated when the cells were induced to overexpress ATR-kd (Fig. 5A, right panel). Phosphorylation of CHK1, seen in the uninduced cells, was totally abrogated by overexpression of ATR-kd, whereas CHK2 phosphorylation remained largely unchanged (Fig. 5B). Moreover, overexpression of ATR-kd had a dramatic effect on the formation of S*/T*Q foci (Fig. 5C,D). The uninduced cells displayed no defect in focus formation: Both RPA and S*/T*Q foci were abundant 48 h after MNNG treatment, and the fact that they largely colocalized substantiated recent reports that demonstrated the requirement for RPA-bound stretches of single-stranded DNA for the recruitment of ATR and for focus formation (Zou and Elledge 2003). In ATR-kd overexpressing cells, the RPA foci formed earlier, but we failed to observe foci of S*/T*Q. This demonstrated that the kinase activity of ATR is required for the formation of the latter foci. This experimental evidence also demonstrated that the formation of RPA foci is ATR independent (i.e., that RPA is recruited to sites of damage before ATR, as discussed by others [Zou and Elledge 2003]).
Figure 5.
The G2 checkpoint induced by low MNNG doses is ATR-dependent. (A) FACS analysis of U2OS cells that overexpress the kinase-dead ATR variant under doxycycline control. The figure shows that the G2 arrest activated by MNNG treatment in these cells was attenuated by overexpression of the ATR-kd protein. (B) CHK2 phosphorylation was largely unaffected by overexpression of ATR-kd in the treated U2OS cells. In contrast, activation of CHK1 was dependent on the presence of functional ATR. (C) Indirect immunofluorescence of ATR-kd inducible U2OS cells showing that formation of S*/T*Q foci and their colocalization with RPA (p34) after MNNG treatment is ATR-dependent. The fraction of foci-displaying cells on each microscope slide is shown in panel D.
MNNG treatment induces formation of γ-H2AX foci that are not associated with double-strand breaks, but colocalize with the foci of RPA
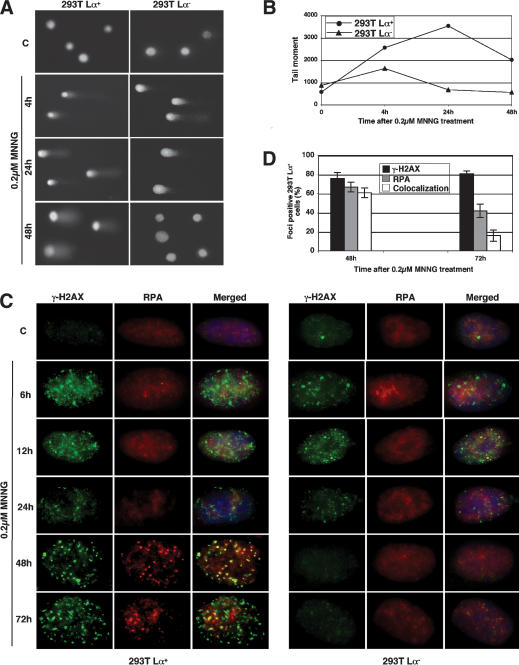
We wanted to gain information about the type of damage generated by the MMR-dependent processing of MNNG-induced DNA damage. Using alkaline comet assays, we found evidence of extensive single-stranded DNA degradation in both MMR-deficient and MMR-proficient cells already 4 h posttreatment. Under these conditions, apurinic sites generated by removal of methylated bases are cleaved and the strand breaks become detectable as the double-stranded DNA is denatured. Importantly, the single-strand breaks completely disappeared with time in the MMR-deficient cells, whereas in the MMR-proficient cells, a substantial proportion persisted even 48 h after treatment (Fig. 6A,B). On the basis of this evidence, we expected to observe no nuclear foci of the phosphorylated form of histone H2AX (γ-H2AX), which was reported to associate with DSBs (Rogakou et al. 1999) and to recruit repair factors to these sites (Paull et al. 2000). Surprisingly, numerous γ-H2AX foci appeared soon after MNNG treatment (Fig. 6C). The lesions associated with these early foci were apparently not responsible for triggering the signaling cascade, as they appeared in similar numbers and with similar kinetics in both MMR-proficient and MMR-deficient cells and as no signaling that could be ascribed to DSBs was detected. Moreover, when the treated cells were examined by pulse field gel electrophoresis, TUNEL, and neutral pH comet assays, no DSBs could be detected (data not shown). It is therefore highly unlikely that the early-appearing γ-H2AX foci seen in this study represent sites of DSB formation. A more likely scenario is that they represent regions in which chromatin structure is disrupted because of the processing of modified purines by the BER system.
Figure 6.
MMR-dependent processing of methylation damage. (A) Alkaline comet assays showing the appearance and repair of DNA single-strand breaks in 293T Lα cells on 0.2 μM MNNG treatment. The panel shows representative cells. (B) Quantification of the tail moment of 50 randomly selected cells per slide. As shown, the single-strand breaks (or gaps) were repaired in the MMR-deficient cells but persisted in the MMR-proficient ones. (C) Kinetics of histone H2AX phosphorylation and the colocalization of γ-H2AX foci with those of RPA in MNNG-treated 293T Lα cells. The γ-H2AX foci appeared soon after treatment, independent of the MMR status of the cells. They then diminished in number in both cell types but began to reappear in the MMR-proficient 293T Lα+ cells after 24 h. At the 48-h time point, they largely colocalized with the foci of RPA, but this overlap diminished by 72 h. In contrast, in the MMR-deficient 293T Lα- cells, the γ-H2AX foci disappeared completely. The panel shows representative cells. The fraction of cells on each microscope slide displaying a similar phenotype is shown in D.
At later time points, the γ-H2AX foci gradually disappeared from the nuclei of MMR-deficient cells, while in the MMR-proficient cells they appeared to change morphology and increased in number and intensity. We wanted to test whether these foci colocalized with those formed by RPA, given that the single-stranded DNA-binding protein was seen to colocalize with ATR and the S*/T*Q substrate (Figs. 3D, 5C, respectively). As shown in Figure 6C and D, the foci of γ-H2AX and RPA were seen to colocalize in ∼60% of the treated cells at the 48-h time point. At the 72-h time point, the intensity of the RPA foci diminished and very little colocalization with the γ-H2AX foci could be seen.
Discussion
A functional MMR system has been postulated to be required for the activation of a G2/M cell cycle arrest (Hawn et al. 1995; Claij and Te Riele 2002; Cejka et al. 2003) and for apoptosis (D'Atri et al. 1998) induced in mammalian cells by SN1 type methylating agents and 6-thioguanine. Using an isogenic system developed in our laboratory (Cejka et al. 2003), in which the MMR status of the 293T Lα cells can be controlled by Dox, we show that the MMR-proficient cells treated with 0.2 μM MNNG arrested in the G2 phase of the second cell cycle (Fig. 1A,B), rather than undergoing a mitotic catastrophe (Fig. 1C). This was highly unusual, because cells generally arrest a few hours after DNA damage. We therefore set out to identify the molecular mechanisms underlying this phenomenon. In the first series of experiments, we showed that the accumulation of MNNG-treated cells in G2 was attenuated by caffeine and UCN-01, which are known to inhibit preferentially the ATM/ATR and CHK1 kinases, respectively (Fig. 2). This evidence further confirmed that the increase in the number of cells with a 4n DNA content, as observed by FACS, was the result of activation of a checkpoint, rather than of a physical block imposed by the DNA damage. Correspondingly, we could show that MNNG treatment of the MMR-proficient cells activated a protein phosphorylation cascade that modified a number of downstream targets of the ATM/ATR kinases (Fig. 3). It appeared most likely that these phosphorylation events actually triggered the arrest, as the posttranslational modification of these targets temporally coincided with its activation. We were able to rule out the requirement for ATM in the activation of the MNNG-induced checkpoint: Although the kinase appeared to be activated at late time points in the MNNG-treated 293T Lα+ cells (Fig. 4A), the AT fibroblasts lacking this kinase arrested similarly to ATM-proficient ones (Fig. 4B). This hypothesis is further supported by earlier findings, which showed that HCT15 cells that lack CHK2, one of the downstream targets of ATM, arrested normally on treatment with methylating agents when their MMR defect was corrected (Umar et al. 1997). In contrast, ATR kinase and its downstream target CHK1 were shown to be required for the efficient activation of the MNNG-induced checkpoint, as the number of cells with a 4n DNA content was dramatically decreased in MNNG-treated U2OS cells overexpressing the kinase-dead ATR variant (Fig. 5), or in the 293T Lα+ cells when the CHK1 activity was inhibited by UCN-01 (Fig. 2).
Our finding that the ATM kinase is activated only very late after MNNG treatment seemingly contrasts with the data of Adamson and colleagues (2002), who reported that MNNG treatment rapidly activates this enzyme. These differences are probably the result of the 120-fold higher dose of the reagent used in the latter study. High concentrations of DNA methylating agents bring about levels of base damage that are too high to be effectively processed by the BER system. As a result, strand breaks arising through incomplete BER appear soon after treatment and activate damage-signaling pathways that are more reminiscent of those induced by other clastogenic DNA damaging agents. Importantly, this processing is largely independent of the MMR system (L. Stojic, N. Mojas, P. Cejka, and J. Jiricny, in prep.).
The involvement of damage-specific kinases other than ATM and ATR in the MNNG-induced cell cycle arrest has not been ruled out. However, it is unlikely that DNA-dependent protein kinase (DNA-PK) plays a key role, as it generally does not appear to be required for DNA damage signaling (Durocher and Jackson 2001). Moreover, cells mutated in its Ku80 subunit are hyper-sensitive to IR but appear to respond normally to MNNG (Jeggo and Kemp 1983).
If ATR is the most upstream DNA damage-signaling kinase, what is the nature of the MNNG-induced lesions that trigger its activation? Our results show that the kinase cascade is not activated directly by 6MeG/T mispairs (e.g., through interaction with the mismatch binding heterodimer hMSH2/hMSH6; Duckett et al. 1996; Fishel 1999). First, these mispairs should have already arisen during the first S phase, and even if they were to activate the signaling cascade directly, there is no reason why cells should be arrested in the second cell cycle, when the number of these mispairs is reduced by 50% because of the semiconservative nature of DNA replication. (The half-life of MNNG in culture medium is ∼1 h; it has thus been inactivated long before the onset of the second cell division.) Second, cells lacking hMLH1/hPMS2 (e.g., HCT116, 293T, 293T Lα-) express normal levels of the hMSH2/hMSH6 heterodimer, yet are also highly resistant to killing by MNNG and do not arrest in G2. This would require that the DNA damage signaling be mediated by the hMSH2/hMSH6/hMLH1/hPMS2 heterotetramer. Although formally possible, the appearance of RPA foci suggests that the signaling was initiated through processing rather than just detection of the damage—but what is the nature of this processing?
More than 30 years ago, Plant and Roberts (1971) suggested that replication past 6MeG in the template strand during the first S phase may give rise to single-stranded gaps, which are converted into DSBs during the second replication cycle. This was long thought to be unlikely, as DNA polymerases were expected to repair gaps remaining from incomplete replication during the G2 phase. However, it is conceivable that such gaps do indeed arise in DNA methylated by SN1-type agents. During DNA replication, the polymerases may incorporate a T or a C into the newly synthesized strand opposite the 6MeG residues in the template strand, and it has been suggested that the MMR system will detect these non-Watson-Crick structures (Duckett et al. 1996) and attempt to repair them. The repair process would exonucleolytically degrade a short stretch of the newly replicated DNA (i.e., the strand containing the pyrimidines). However, as the 6MeG residues persist in the template strand, resynthesis of this region would again generate 6MeG/T or 6MeG/C mispairs. The repeated processing of these mispairs by the MMR system (Karran and Bignami 1996) will likely lead to stalling of the replication fork. One might pose the question of why these structures would fail to activate the S-phase checkpoint, when other polymerase-arresting agents such as HU or aphidicolin do so extremely efficiently (Abraham 2001; Osborn et al. 2002; Shiloh 2003). One possible explanation is that HU and aphidicolin inhibit all active replicons, whereas the number of 6MeG residues generated by MNNG treatment may be too low to trigger an S-phase arrest. An alternative explanation is that unlike HU, which brings about a depletion of the purine nucleotide pool, or aphidicolin, which directly inhibits the replicative polymerases, 6MeG residues in the template strand do not prevent a replication restart downstream from the modified base. Indirect support for the replication restart hypothesis comes from in vitro experiments carried out with MNU-modified plasmid DNA; the observed DNA repair synthesis required nucleoside triphosphates (NTPs), which would be required by a primase (Ceccotti et al. 1993, 1996). Furthermore, these experiments demonstrated that the repair synthesis triggered by the presence of 6MeG residues in the DNA is RPA independent and that it gives rise to open circular DNA, in contrast to the in vitro replication reaction of the same, but unmodified, plasmid, which yielded almost exclusively super-coiled DNA molecules. These results could be taken as further evidence of the persistence of unligatable single-stranded regions or breaks in the plasmid DNA incubated with MMR-proficient cell extracts (Ceccotti et al. 1996). Direct support for the persistence of incompletely replicated DNA comes from our experiments, in which the genomic DNA of MNNG-treated MMR-proficient cells after the first S phase was shown to contain numerous single-strand breaks, as witnessed by the appearance of DNA tails in alkaline comet assays (Fig. 6A,B). Our findings are further supported by recent evidence showing that treatment of cells with 6-thioguanine, which is believed to exert its cytotoxicity via a mechanism analogous to MNNG (Swann et al. 1996), also results in the accumulation of MMR-dependent single-strand DNA breaks (Yan et al. 2003).
The latter hypothesis raises two important questions. First, if genomic DNA containing 6MeG residues does indeed contain single-strand gaps after the first S phase, why do such gaps not activate the checkpoint already at this time? One possibility is that they are too few in number. Alternatively, the gaps might not stall the replisome, or they might be too short to be bound by RPA. Indeed, the RPA foci began to appear only at 24 h, and their number peaked at the 48-h time point (Figs. 3C,D, 5C,D). As RPA has been shown to be required for the efficient recruitment of the ATR/ATRIP complex to the sites of damage, single-strand gaps that are not RPA bound would fail to efficiently activate the CHK1 kinase, which has been identified in complexes that associate with strand breaks and with single-stranded DNA (Goudelock et al. 2003) and which was shown to be involved in the MNNG-induced G2 checkpoint (Fig. 2).
Second, assuming that the single-stranded gaps do indeed form, how could they persist until the subsequent S phase as suggested (Plant and Roberts 1971; Kaina et al. 1997)? This could be the result of a combination of factors. As discussed above, it is possible that, in the absence of bound RPA, the damage sites may signal too weakly to effectively activate the checkpoint. The other reason might be that the filling of a gap opposite a 6MeG residue may not be trivial. Thus, proliferating cell nuclear antigen (PCNA)-dependent polymerases tested to date would generate 6MeG/C or 6MeG/T mispairs (Reha-Krantz et al. 1996), which would be again addressed by the MMR system, because of its ability to interact with the processivity factor (Kleczkowska et al. 2001). Other polymerases might have problems extending from the non-Watson-Crick 6MeG/C or 6MeG/T structures, in which case the DNA synthesis would stall at the mispairs because of the activation of the 3′ → 5′ proofreading activity (Khare and Eckert 2001).
Persistent single-strand gaps opposite the 6MeG residues would become DSBs during the second S phase, and the affected replication forks would collapse unless rescued by recombination events such as sister chromatid exhanges (SCEs). That events of this type indeed arise in cells treated with methylating agents was suggested by an increase in SCE frequency in the treated MMR-proficient 293T Lα+ cells (data not shown; N. Mojas, L.Stojic, and J. Jiricny, in prep.; see also Galloway et al. 1995; Kaina et al. 1997). The timing of these events broadly coincided with the formation of foci containing RPA, ATR (Fig. 3C,D), and γ-H2AX (Fig. 6C), which may represent the sites in chromatin at which processing is taking place and also from which the signaling events may be originating.
In conclusion, treatment of mammalian cells with a low dose of MNNG was shown to bring about a G2 cell cycle arrest that was absolutely dependent on a functional MMR system and that, to a substantial degree, was also dependent on the ATR and CHK1 kinases. This checkpoint was highly atypical, inasmuch as it came into effect only in the second cell cycle after treatment. Its activation was accompanied by a number of changes in the nuclei of the treated cells, possibly indicative of recombination events. Our present findings thus suggest that SN1 type methylating agents such as the chemotherapeutics procarbazine and temozolomide, which act similarly to MNNG, kill cells with the help of MMR, which generates intermediates that cannot be effectively processed by the cellular repair machinery. The persistence of these lesions into the second cell cycle may kill cells through the generation of aberrant recombination intermediates. We are currently attempting to elucidate the structures of these lesions by biophysical techniques.
Materials and methods
Cell lines
The 293T Lα cell line was established in our laboratory and propagated as described (Cejka et al. 2003). HeLa cells were maintained in DMEM (OmniLab) supplemented with 10% fetal calf serum (FCS; Life Technologies), penicillin (100 U/mL), and streptomycin (100 μg/mL). The ATM-deficient (AT) fibroblasts AT22IJE-T and the matched line complemented with ATM minigene (AT + ATM) were kindly provided by Yosef Shiloh (Tel Aviv University, Israel) and were maintained as described (Ziv et al. 1997). The U2OS cell line conditionally expressing ATR kinase-dead protein (Paul Nghiem, Harvard University, Cambridge, MA) was maintained in DMEM supplemented with 10% FCS, 200 μg/mL G418, and 200 μg/mL Hygromycin B. Induction of ATR-kd was accomplished by supplementing the growth medium with Dox (1 μg/mL) for 48 h, as described (Nghiem et al. 2002). The hMLH1-deficient human colon cancer cell line HCT116 and its MMR-proficient subline HCT116 + ch3 were maintained in McCoy's 5A medium (OmniLab) with 10% FCS. The chromosome-complemented cell line was maintained in medium containing 400 μg/mL G418. Expression of all MMR proteins was confirmed in both AT fibroblasts and ATR-inducible cells by immunobloting (data not shown). To inhibit MGMT activity, HeLa cells, HCT116 and HCT116 + ch3 cells, AT and AT + ATM fibroblasts, and the ATR-kd-inducible cells were pretreated with 10 μM O6-benzylguanine 2 h before MNNG treatment. All cell lines were cultured at 37°C in a 5% CO2 humidified atmosphere.
Chemicals and irradiations
MNNG (Sigma) was dissolved in DMSO and stored at -20°C in the dark. O6-benzylguanine (Sigma) was dissolved in ethanol and stored at -80°C. HU (Sigma) and Dox (Clontech) were dissolved in water and stored at -20°C. UCN-01 (Sally Hausman, Cancer Therapy Evaluation Program, National Cancer Institute, National Institutes of Health, Rockville, MD) was dissolved in DMSO and stored at -20°C. Caffeine (Calbiochem) was dissolved in water and always prepared fresh. Irradiations were carried out at the doses indicated, using a Philips PW2184/00-Monitor SN4.
Mitotic index assays
The 293T Lα cells were treated with 0.2 μM MNNG and incubated for 24 or 48 h. Nocodazole (0.3 μg/mL, Sigma) was then added and the cells were incubated for a further 24 h. The floating and attached cells were then harvested and centrifuged at 400g. The pellet was suspended in 3 mL of 75 mM KCl for 10 min, centrifuged again as above, and resuspended in Carnoy's fixative (1:3 v/v acetic acid:methanol). This latter step was repeated three times. Twenty microliters of the cell suspension were spotted onto a microscope slide and allowed to air dry. Once dry, the cells were stained with 0.1 μg/mL 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI; Sigma) for 10 min, washed with water, and mounted in SlowFade Antifade (Molecular Probes). Using a fluorescence microscope, cells with broken nuclei and condensed chromatin were counted as mitotic. Five hundred cells were counted per sample.
Cell synchronizations
The 293T Lα cells were grown to 50% confluency in a serum-rich medium. Thymidine (2 mM, SynGen Inc.) was added, and the cells were incubated for 18 h, washed three times with PBS, and released into thymidine-free medium for 9 h. Thymidine (2 mM) was then added for a further 15 h. The cells were then washed three times with PBS. At this point (G1/S, Fig. 1A), the cells were treated with 0.2 μM MNNG in a serum-rich medium without thymidine, and time points were collected 4 (T 4), 8 (T 8), 14 (T 14), 20 (T 20), 24 (T 24), and 30 (T 30) h later. HCT116 and HCT116 + ch3 cells were synchronized in a medium containing 2 mM HU for14h. O6-benzylguanine (10 μM) was added for the last 2 h, when the cells were washed with PBS. The cells were then incubated in fresh medium containing 0.2 μM MNNG (G1/S, Fig. 1B) and O6-benzylguanine. Cells were harvested and analyzed by propidium iodide-flow cytometric analysis as described (Cejka et al. 2003).
Cell cycle analyses
For BrdU labeling, cells were pulse-labeled with 10 μM BrdU (Sigma) for 30 min before harvesting and fixation in 70% ethanol at 4°C. The cells were then processed as described (Cliby et al. 2002). BrdU incorporation studies and cell cycle distributions were analyzed by Becton Dickinson CELLQuest software. For immunofluorescence-based detection of phosphorylated histone H3, the cells were treated with 0.2 μM MNNG. Sixteen hours before harvesting, the growth medium was supplemented with caffeine (2 mM) or UCN-01 (100 nM). The cells were collected 24 or 48 h after MNNG treatment. The subsequent steps were carried out as described (Xu et al. 2001).
Cell doubling assays
Cell doubling assays were carried out as described previously (Cejka et al. 2003).
Alkaline comet assays
Alkaline comet assays were carried out using Trevigen Comet-Slides according to the manufacturer's recommendations. DNA was stained with ethidium bromide (10 μg/mL) and visualized using a fluorescence microscope. Fifty comets were analyzed per slide, using National Institutes of Health images with Comet macro (Helma and Uhl 2000).
Antibodies and immunoblotting
Anti-MLH1 (554072) and anti-PMS2 (556415) monoclonal antibodies were from BD Pharmingen; anti-CHK1 (611152) was from BD Transduction Laboratories; and anti-β-tubulin (D-10), anti-TFIIH p89 (S-19), anti-CDC25A (F-6), and anti-ATR (FRP1, N-19) were from Santa Cruz Biotechnology. Anti-RPA p34 (Ab-3) was from Oncogene. Anti-phospho-CHK1 (Ser 345), anti-phospho-CHK2 (Thr 68), and anti-phospho-Ser/Thr (S*/T*Q) ATM/ATR substrate antibodies were from Cell Signaling. Anti-CHK2 (07-126) and anti-γ-H2AX (Ser 139) antibodies were from Upstate Biotechnology. The anti-ATM phospho-Ser 1981 antibody was obtained from Rockland. The anti-ATM antibody was kindly provided by Stephen P. Jackson (Wellcome/CRC Institute, Cambridge, UK). Immunoblotting and total protein extractions were performed as described previously (Cejka et al. 2003).
Immunofluorescence studies
Cells grown on glass cover slips were treated or mock-treated with MNNG and incubated for the indicated time periods. Fixation was done in 3.7% formaldehyde/PBS (15 min, 4°C), followed by permeabilization in 0.2% Triton X-100/PBS (5 min, 4°C). In the case of anti-γ-H2AX, the cells were fixed in ice-cold methanol (20 min, -20°C). The coverslips were blocked with 3% low-fat milk/PBS and incubated with anti-phospho-(Ser/Thr) ATM/ATR substrate, anti-γ-H2AX (Ser 139), anti-ATR, and anti-RPA p34 antibodies, all at 1:100 dilution. After washing, the cells were incubated with FITC-conjugated anti-rabbit or anti-goat antibodies (Sigma) and TR-conjugated anti-mouse antibodies (Abcam) for 1 h at 37°C. The nuclei were counter-stained with DAPI (0.1 μg/mL, Sigma). Images were captured on a Leica DC 200 fluorescence microscope.
Acknowledgments
We thank Stephen P. Jackson for the anti-ATM antibody, Yosef Shiloh for the AT and AT + ATM cell lines, Sally Hausman (Cancer Therapy Evaluation Program, National Cancer Institute, National Institutes of Health) for providing UCN-01, and Paul Nghiem for the ATR-inducible U2OS cells. We also thank Jiri Bartek, Stefania D'Atri, Stephen P. Jackson, and Primo Schär for helpful discussions; Primo Schär for help with the irradiations; Conny Marty for help with the BrdU analyses; and Eva Niederer in the Flow Cytometry Laboratory of the Institute of Biomedical Engineering of the Swiss Federal Institute of Technology (ETH) and the University of Zurich for help with the FACS analyses. The contributions of the remaining members of our laboratory are also gratefully acknowledged. L.S. was supported by European Community grant QLG1-CT-2000-001230 and P.C. by a grant from UBS AG, and N.M. and M. D. were supported by the Swiss National Science Foundation grants 3168182 and 3238-064650, awarded to J.J.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.294404.
References
- Abraham, R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 15: 2177-2196. [DOI] [PubMed] [Google Scholar]
- Adamson, A.W., Kim,W.J., Shangary, S., Baskaran, R., and Brown, K.D. 2002. ATM is activated in response to N-methyl-N′-nitro-N-nitrosoguanidine-induced DNA alkylation. J. Biol. Chem. 277: 38222-38229. [DOI] [PubMed] [Google Scholar]
- Aquilina, G., Crescenzi, M., and Bignami, M. 1999. Mismatch repair, G(2)/M cell cycle arrest and lethality after DNA damage. Carcinogenesis 20: 2317-2326. [DOI] [PubMed] [Google Scholar]
- Bakkenist, C.J. and Kastan, M.B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499-506. [DOI] [PubMed] [Google Scholar]
- Bartek, J. and Lukas, J. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13: 738-747. [DOI] [PubMed] [Google Scholar]
- Bellacosa, A. 2001. Functional interactions and signaling properties of mammalian DNA MMR proteins. Cell Death Differ. 8: 1076-1092. [DOI] [PubMed] [Google Scholar]
- Brown, E.J. and Baltimore, D. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes & Dev. 17: 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby, E.C., Leistritz, D.F., Abraham, R.T., Karnitz, L.M., and Sarkaria, J.N. 2000. The radiosensitizing agent 7-hydroxy-staurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 60: 2108-2112. [PubMed] [Google Scholar]
- Ceccotti, S., Dogliotti, E., Gannon, J., Karran, P., and Bignami, M. 1993. O6-methylguanine in DNA inhibits replication in vitro by human cell extracts. Biochemistry 32: 13664-13672. [DOI] [PubMed] [Google Scholar]
- Ceccotti, S., Aquilina, G., Macpherson, P., Yamada, M., Karran, P., and Bignami, M. 1996. Processing of O6-methylguanine by mismatch correction in human cell extracts. Curr. Biol. 6: 1528-1531. [DOI] [PubMed] [Google Scholar]
- Cejka, P., Stojic, L., Mojas, N., Russell, A.M., Heinimann, K., Cannavo, E., di Pietro, M., Marra, G., and Jiricny, J. 2003. Methylation-induced G(2)/M arrest requires a full complement of the MMR protein hMLH1. EMBO J. 22: 2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claij, N. and Te Riele, H. 2002. Methylation tolerance in MMR proficient cells with low MSH2 protein level. Oncogene 21: 2873-2879. [DOI] [PubMed] [Google Scholar]
- Cliby, W.A., Lewis, K.A., Lilly, K.K., and Kaufmann, S.H. 2002. S phase and G2 arrests induced by topoisomerase I poisons are dependent on ATR kinase function. J. Biol. Chem. 277: 1599-1606. [DOI] [PubMed] [Google Scholar]
- Cooke, M.S., Evans, M.D., Dizdaroglu, M., and Lunec, J. 2003. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 17: 1195-1214. [DOI] [PubMed] [Google Scholar]
- Crosio, C., Fimia, G.M., Loury, R., Kimura, M., Okano, Y., Zhou, H., Sen, S., Allis, C.D., and Sassone-Corsiet, P. 2002. Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell Biol. 22: 874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Atri, S., Tentori, L., Lacal, P.M., Graziani, G., Pagani, E., Benincasa, E., Zambruno, G., Bonmassar, E., and Jiricny, J. 1998. Involvement of the MMR system in temozolomide-induced apoptosis. Mol. Pharmacol. 54: 334-341. [DOI] [PubMed] [Google Scholar]
- Di Pietro, M., Marra, G., Cejka, P., Stojic, L., Menigatti, M., Cattaruzza, M.S., and Jiricny, J. 2003. Mismatch repair-dependent transcriptome changes in human cells treated with the methylating agent N-methyl-n′-nitro-N-nitrosoguanidine. Cancer Res. 63: 8158-8166. [PubMed] [Google Scholar]
- DiTullio Jr., R.A., Mochan, T.A., Venere, M., Bartkova, J., Sehested, M., Bartek, J., and Halazonetis, T.D. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4: 998-1002. [DOI] [PubMed] [Google Scholar]
- Duckett, D.R., Drummond, J.T., Murchie, A.I., Reardon, J.T., Sancar, A., Lilley, D.M., and Modrich, P. 1996. Human Mut-Salpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. 93: 6443-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher, D. and Jackson, S.P. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: Variations on a theme? Curr. Opin. Cell Biol. 13: 225-231. [DOI] [PubMed] [Google Scholar]
- Falck, J., Mailand, N., Syljuasen, R.G., Bartek, J., and Lukas, J. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410: 842-847. [DOI] [PubMed] [Google Scholar]
- Fishel, R. 1998. Mismatch repair, molecular switches, and signal transduction. Genes & Dev. 12: 2096-2101. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Signaling MMR in cancer. Nat. Med. 5: 1239-1241. [DOI] [PubMed] [Google Scholar]
- Galloway, S.M., Greenwood, S.K., Hill, R.B., Bradt, C.I., and Bean, C.L. 1995. A role for MMR in production of chromosome aberrations by methylating agents in human cells. Mutat. Res. 346: 231-245. [DOI] [PubMed] [Google Scholar]
- Goudelock, D.M., Jiang, K., Pereira, E., Russell, B., and Sanchez, Y. 2003. Regulatory interactions between the checkpoint kinase Chk1 and the proteins of the DNA-PK complex. J. Biol. Chem. 278: 29940-29947. [DOI] [PubMed] [Google Scholar]
- Graves, P.R., Yu, L., Schwarz, J.K., Gales, J., Sausville, E.A., O'Connor, P.M., Piwnica-Worms, H. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275: 5600-5605. [DOI] [PubMed] [Google Scholar]
- Hawn, M.T., Umar, A., Carethers, J.M., Marra, G., Kunkel, T.A., Boland, C.R., and Koi, M. 1995. Evidence for a connection between the MMR system and the G2 cell cycle checkpoint. Cancer Res. 55: 3721-3725. [PubMed] [Google Scholar]
- Helma, C. and Uhl, M. 2000. A public domain image-analysis program for the single-cell gel-electrophoresis (comet) assay. Mutat. Res. 466: 9-15. [DOI] [PubMed] [Google Scholar]
- Hoffmann, I., Draetta, G., and Karsenti, E. 1994. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 13: 4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo, P.A. and Kemp, L.M. 1983. X-ray-sensitive mutants of Chinese hamster ovary cell line. Isolation and cross-sensitivity to other DNA-damaging agents. Mutat. Res. 112: 313-327. [DOI] [PubMed] [Google Scholar]
- Kaina, B., Ziouta, A., Ochs, K., and Coquerelle, T. 1997. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex-, Mex+ and methylation-tolerant MMR compromised cells: facts and models. Mutat. Res. 381: 227-241. [DOI] [PubMed] [Google Scholar]
- Karran, P. 2001. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis 22: 1931-1937. [DOI] [PubMed] [Google Scholar]
- Karran, P. and Bignami, M. 1992. Self-destruction and tolerance in resistance of mammalian cells to alkylation damage. Nucleic. Acids. Res. 20: 2933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. Drug-related killings: A case of mistaken identity. Chem. Biol. 3: 875-879. [DOI] [PubMed] [Google Scholar]
- Khare, V. and Eckert, K.A. 2001. The 3′ → 5′ exonuclease of T4 DNA polymerase removes premutagenic alkyl mispairs and contributes to futile cycling at O6-methylguanine lesions. J. Biol. Chem. 276: 24286-24292. [DOI] [PubMed] [Google Scholar]
- Kleczkowska, H.E., Marra, G., Lettieri, T., and Jiricny, J. 2001. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes & Dev. 15: 724-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Guntuku, S., Cui, X.S., Matsuoka, S., Cortez, D., Tamai, K., Luo, G., Carattini-Rivera, S., DeMayo, F., Bradley, A., et al. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 14: 1448-1459. [PMC free article] [PubMed] [Google Scholar]
- Mailand, N., Podtelejnikov, A.V., Groth, A., Mann, M., Bartek, J., and Lukas, J. 2002. Regulation of G(2)/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J. 21: 5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem, P., Park, P.K., Kim, Y.S., Desai, B.N., and Schreiber, S.L. 2002. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J. Biol. Chem. 277: 4428-4434. [DOI] [PubMed] [Google Scholar]
- Osborn, A.J., Elledge, S.J., and Zou, L. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12: 509-516. [DOI] [PubMed] [Google Scholar]
- Paull, T.T., Rogakou, E.P., Yamazaki, V., Kirchgessner, C.U., Gellert, M., and Bonner, W.M. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10: 886-895. [DOI] [PubMed] [Google Scholar]
- Plant, J.E. and Roberts, J.J. 1971. A novel mechanism for the inhibition of DNA synthesis following methylation: the effect of N-methyl-N-nitrosourea on HeLa cells. Chem. Biol. Interact. 3: 337-342. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz, L.J., Nonay, R.L., Day, R.S., and Wilson, S.H. 1996. Replication of O6-methylguanine-containing DNA by repair and replicative DNA polymerases. J. Biol. Chem. 271: 20088-20095. [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., Boon, C., Redon, C., and Bonner, W.M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146: 905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria, J.N., Busby, E.C., Tibbetts, R.S., Roos, P., Taya, Y., Karnitz, L.M., and Abraham, R.T. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59: 4375-4382. [PubMed] [Google Scholar]
- Scharer, O.D. and Jiricny, J. 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays 23: 270-281. [DOI] [PubMed] [Google Scholar]
- Sedgwick, B. and Lindahl, T. 2002. Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene 21: 8886-8894. [DOI] [PubMed] [Google Scholar]
- Seeberg, E., Eide, L., and Bjoras, M. 1995. The base excision repair pathway. Trends Biochem. Sci. 20: 391-397. [DOI] [PubMed] [Google Scholar]
- Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3: 155-168. [DOI] [PubMed] [Google Scholar]
- Swann, P.F., Waters, T.R., Moulton, D.C., Xu, Y.Z., Zheng, Q., Edwards, M., and Mace, R. 1996. Role of postreplicative DNA MMR in the cytotoxic action of thioguanine. Science 273: 1109-1111. [DOI] [PubMed] [Google Scholar]
- Trojan, J., Zeuzem, S., Randolph, A., Hemmerle, C., Brieger, A., Raedle, J., Plotz, G., Jiricny, J., and Marra, G. 2002. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology 122: 211-219. [DOI] [PubMed] [Google Scholar]
- Umar, A., Koi, M., Risinger, J.I., Glaab, W.E., Tindall, K.R., Kolodner, R.D., Boland, C.R., Barrett, J.C., and Kunkel, T.A. 1997. Correction of hypermutability, N-methyl-N′-nitro-N-nitrosoguanidine resistance, and defective DNA MMR by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res. 57: 3949-3955. [PubMed] [Google Scholar]
- Wang, H., Guan, J., Perrault, A.R., Wang, Y., and Iliakis, G. 2001. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 61: 8554-8563. [PubMed] [Google Scholar]
- Wang, X., Khadpe, J., Hu, B., Iliakis, G., and Wang, Y. 2003. An over-activated ATR/CHK1 pathway is responsible for the prolonged G2 accumulation in irradiated AT cells. J. Biol. Chem. 278: 30869-30874. [DOI] [PubMed] [Google Scholar]
- Xu, B., Kim, S., and Kastan, M.B. 2001. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell Biol. 21: 3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, T., Berry, S.E., Desai, A.B., and Kinsella, T.J. 2003. DNA MMR (MMR) mediates 6-thioguanine genotoxicity by introducing single-strand breaks to signal a G(2)-M arrest in MMR-proficient RKO cells. Clin. Cancer Res. 9: 2327-2334. [PubMed] [Google Scholar]
- Zhao, H., Watkins, J.L., and Piwnica-Worms, H. 2002. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl. Acad. Sci. 99: 14795-14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B.B., Chaturvedi, P., Spring, K., Scott, S.P., Johanson, R.A., Mishra, R., Mattern, M.R., Winkler, J.D., and Khanna, K.K. 2000. Caffeine abolishes the mammalian G(2)/M DNA damage checkpoint by inhibiting ataxia-telangiectasia-mutated kinase activity. J. Biol. Chem. 275: 10342-10348. [DOI] [PubMed] [Google Scholar]
- Zhukovskaya, N., Branch, P., Aquilina, G., and Karran, P. 1994. DNA replication arrest and tolerance to DNA methylation damage. Carcinogenesis 15: 2189-2194. [DOI] [PubMed] [Google Scholar]
- Ziv, Y., Bar-Shira, A., Pecker, I., Russell, P., Jorgensen, T.J., Tsarfati, I., and Shiloh, Y. 1997. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 15: 159-167. [DOI] [PubMed] [Google Scholar]
- Zou, L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542-1548. [DOI] [PubMed] [Google Scholar]