Abstract
Recently we reported that the Arabidopsis thaliana PHD-finger protein EDM2 (enhanced downy mildew 2) impacts disease resistance by affecting levels of di-methylated lysine 9 of histone H3 (H3K9me2) at an alternative polyadenylation site in the immune receptor gene RPP7. EDM2-dependent modulation of this post-translational histone modification (PHM) shifts the balance between full-length RPP7 transcripts and prematurely polyadenylated transcripts, which do not encode the RPP7 protein. Our previous work genetically linked, for the first time, PHMs to alternative polyadenylation and established EDM2 as a critical component mediating PHM-dependent polyadenylation control. However, how EDM2 is recruited to its genomic target sites and how it affects H3K9me2 levels is unknown. Here we show the PHD-finger module of EDM2 to recognize histone H3 bearing certain combinations of 3 distinct PHMs. Our results suggest that targeting of EDM2 to specific genomic regions is mediated by the histone-binding selectivity of its PHD-finger domain.
Keywords: PHD-finger, histone methylation, EDM2, Arabidopsis thaliana, RPP7
The Arabidopsis thaliana gene EDM2 (enhanced downy mildew 2) is required for race-specific immunity mediated by the disease resistance gene RPP7.1-3 The EDM2 protein is a nuclear-localized and chromatin-associated epigenetic regulator that physically interacts with putative nucleosome remodelers related to the human oncoprotein EMSY.3,4 EDM2 affects silencing states of several transposons by modulating levels of di-methylated lysine 9 of histone H3 (H3K9me2), a repressive epigenetic mark.5 H3K9me2 is one of several types of post-translational histone modifications (PHMs) that have been recognized as key components of the histone-code, a set of chemical tags covalently linked to specific residues of N-terminal histone tails correlated with defined local chromatin configurations and transcriptional states.6 Particularly well characterized PHMs are ε-N-acetylated lysines (Kac) and ε-N-mono-, -di– or –tri-methylated lysines (Kme1, Kme2 or Kme3) as well as mono-methylated, or symmetrically or asymmetrically di-methylated arginine residues (Rme1; Rme2s; Rme2a).7,8Our previous work revealed that, besides affecting transcriptional activity, PHMs can also control alternative polyadenylation.9-11 EDM2 impacts immunity mediated by the immune receptor gene RPP7 by controlling levels of H3K9me2 at the COPIA-R7 retrotransposon present in the 1st RPP7 intron. EDM2-dependent modulation of H3K9me2 at an alternative polyadenylation site in this RPP7 intron shifts the balance between full-length RPP7 transcripts and prematurely polyadenylated transcripts, which do not encode the RPP7 immune receptor.
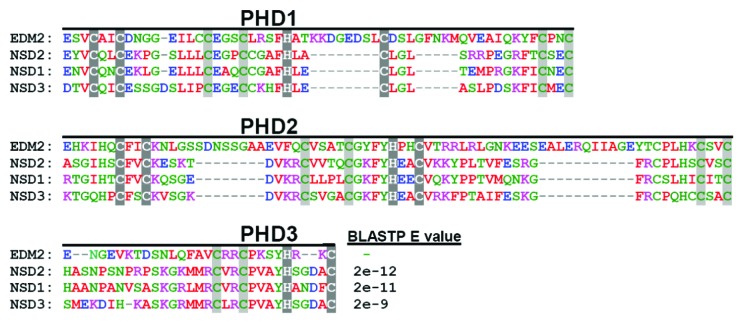
EDM2 contains a module of 2 ½ adjacent units of atypical PHD (Plant Homeodomain)-finger motifs (PHD221–381).1 The PHD-finger is an epigenetic reader domain defined by a characteristic cysteine/histidine (C4HC3) pattern of 8 zinc-ligands which stabilizes (by binding 2 zinc ions) the PHD-finger fold, a conserved structural arrangement of 2 anti-parallel β sheets.12 Distinct variants of the PHD-finger fold can harbor pockets that allow amino acid sequence-specific docking to several forms of methylated or unmethylated lysine or arginine. Particularly well characterized are PHD-finger units containing an aromatic cage that docks to H3K4me2 or H3K4me3.13,14 Although parts of EDM2 PHD221–381 are recognized by standard domain prediction algorithms as PHD-fingers,15 these motifs do not perfectly match the consensus C4HC3 pattern of canonical PHD-fingers.1 The spacings between some of the putative zinc-ligands in PHD221–381 are longer than usual. In addition, its third unit is truncated at both ends and appears to consist only of the central 4 (C3, C4, H and C5) of the 8 zinc ligands of standard PHD-fingers (Fig. 1). A recent study by Lee et al. predicted the PHD-finger region of EDM2 unlikely to form aromatic cages, which are critical for H3K4me2 or H3K4me3-binding.15 The sequences and spacings between potential zinc-ligands of PHD221–381 are highly conserved among the plant specific family of EDM2-like proteins (ELPs).1 In addition, EDM2-related PHD-finger domains are also present other eukaryotes such as algae, oomycetes, protozoa and metazoans, including humans; but they appear to be absent in fungi and prokaryotes.1
Figure 1. The EDM2 PHD-finger module is conserved between Arabidopsis and humans. BlastP searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with the PHD221–381 sequence against human non-redundant protein sequences revealed high similarity with stretches of the human SET-domain proteins NSD1, NSD2 and NSD3. Shown is a ClustalW-generated (http://www.ebi.ac.uk/Tools/msa/clustalw2/) alignment of EDM2-PHD221–381 with the respective regions of NSD1, NSD2 and NSD3. For each protein the 3 shown sequence stretches constitute one contiguous and uninterrupted sequence. Highlighted in dark gray or light gray are ligands of the characteristic C4HC3 zinc finger motifs likely to bind the first, or second zinc ion, respectively, of each hypothetical PHD-finger unit.
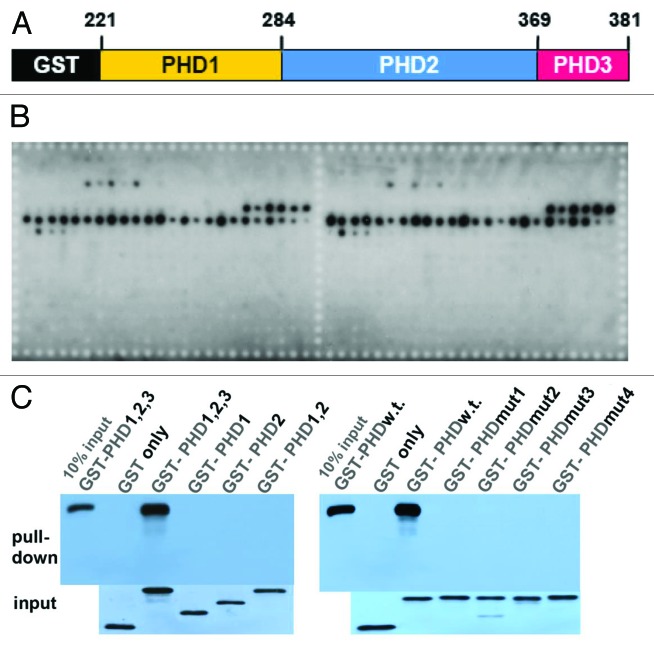
To test if PHD221–381 can bind to histones, we expressed this part of EDM2 in E.coli and used it for in vitro binding assays with the Modified Histone Peptide Array (Active Motif; Figs. 2A and B). This array features peptides (19mers) representing 59 different PHMs of the N-terminal tails of the nucleosome core histones H2A, H2B, H3 and H4. It contains 384 unique histone modification combinations as well as unmodified histones in duplicate. PHD221–381 bound exclusively to modified versions of H3 peptides and no other histone peptides. Intriguingly, it only bound strongly to H3 peptides with dimethylation marks on either R2 or R8 combined with methylation or acetylation marks on K4 and K9 (Table 1). Thus, EDM2 may be exclusively recruited by PHD221–381 to chromatin regions harboring H3 with certain triple PHM marks. One of the H3 peptides most strongly bound by PHD221–381 contains the triple PHM K4me1/R8me2a/K9me1 (Table 1). In additional in vitro-pull down assays, binding to the H3-K4me1/R8me2a/K9me1 peptide was only detectable with the full-length wild-type PHD221–381 region, but not mutated or truncated derivatives not containing all 2 ½ wild-type PHD-finger units (Fig. 2C). Therefore strong cooperativity between PHM-binding pockets present in all 2 ½ PHD-finger units of the PHD221–381 region seems to be required for binding to triple PHMs on H3.
Figure 2. The EDM2 PHD-finger module binds in vitro preferentially to H3 harboring triple PHMs.(A) GST-tagged PHD221–381 used for in vitro binding assays. PHD3 consists only of ½ of a full PHD-finger unit. (B) Both duplicate regions of the Modified Histone Peptide Array (Active Motif) show nearly identical binding represented by dark spots resulting from western blot-detection with GST antibodies. Software provided by Active Motif was used to quantify binding intensities and correlated them with defined PHMs (see Table 1). GST-PHD221–381 was expressed in the E.coli strain Rosetta-gami B (DE3) (Millipore) using the expression vector pGGWA.17 After blocking with the blocking buffer (2% ECL Blocking Agent (GE healthcare) in TBST), the peptide array was incubated with 100nM GST-PHD221–381 in binding/wash buffer (50mM TRIS-HCl pH7.5, 300mM NaCl, 1mM ZnCl2, 0.1% NP-40, complete proteinase inhibitor, EDTA-free (Roche)) overnight at 4 °C. The array was washed 3 times with binding/wash buffer, and binding signals was detected using ECL Advance Western Blotting Detection Kit (GE healthcare) and anti-GST antibody (SantaCruz, GST (Z-5): sc-459 HRP). (C) In vitro histone peptide pull-down assays. Different GST-fused EDM2-PHD-finger versions were separately incubated with biotinylated H3-K4me1-R8me2a-K9me1 peptides and pulled down using streptavidin agarose beads. GST-fusions were visualized by western blotting. Only the full-length wild type PHD221–281 (labeled either as “GST-PHD1,2,3” or “GST-PHDw.t.”) exhibited strong binding to the H3-K4me1-R8me2a-K9me1 peptide. GST-PHD1, -PHD2, and -PHD1,2 contain only of the 1st, 2nd or 1st and 2nd PHD-finger unit, respectively. In GST-PHDmut1, -PHDmut2, -PHMmut3 or -PHDmut4 the 1st Cys/Cys pair of the 1st, 2nd, 3rd or 1st and 2nd PHD-finger unit, respectively, is changed to Gly/Gly. Binding to the H3 peptide is only observed when all 3 PHD-finger units are present and of wild type sequence.
Table 1. Quantified intensities of interactions of recombinant PHD221–381 with modified and unmodified histone peptides.
| H3-R2/R8 | H3-K4 | H3-K9 | Intensity av. | Error | H3-R2/R8 | H3-K4 | H3-K9 | Intensity av. | Error | |
|---|---|---|---|---|---|---|---|---|---|---|
| R2me2s | K4me3 | K9me3 | 0.9962 | 0.0038 | R8me2s | - | - | 0.2821 | 0.0423 | |
| R2me2s | K4me2 | K9me3 | 0.9903 | 0.0045 | R8me2a | - | - | 0.3156 | 0.0235 | |
| R8me2s | K4me1 | K9me2 | 0.9845 | 0.0080 | - | - | K9me1 | 0.2868 | 0.0325 | |
| R8me2a | K4me1 | K9me1 | 0.9819 | 0.0030 | - | - | K9me2 | 0.2911 | 0.0310 | |
| R8me2a | K4ac | K9me2 | 0.9771 | 0.0102 | - | - | K9me3 | 0.3338 | 0.0819 | |
| R8me2s | K4me2 | K9me3 | 0.9762 | 0.0042 | - | - | K9ac | 0.2386 | 0.0053 | |
| R8me2a | K4me2 | K9me2 | 0.9738 | 0.0059 | unmodifyed H3 | 0.2327 | 0.0226 | |||
| R2me2a | K4me1 | K9ac | 0.9610 | 0.0131 | ||||||
| . | . | . | . | . | ||||||
| . | . | . | . | . | ||||||
| . | . | . | . | . | ||||||
| R8me2a | - | K9me1 | 0.1300 | 0.0205 | non-H3 peptides | Intensity av. | Error | |||
| R8me2a | - | K9me2 | 0.5966 | 0.0452 | unmodifyed H4 | 0.0926 | 0.0248 | |||
| R8me2a | - | K9me3 | 0.2383 | 0.1386 | unmodified H2A | 0.2180 | 0.0061 | |||
| R8me2a | - | K9ac | 0.5753 | 0.1233 | unmodified H2B | 0.2916 | 0.0352 | |||
| R2me2s | - | - | 0.2603 | 0.0211 | biotin | 0.2958 | 0.0618 | |||
| R2me2a | - | - | 0.3166 | 0.0061 | c-myc tag | 0.2757 | 0.0475 | |||
| - | K4me1 | - | 0.3884 | 0.0758 | neg. contol | 0.2229 | 0.0229 | |||
| - | K4me2 | - | 0.3689 | 0.0287 | background 01 | 0.3200 | 0.0994 | |||
| - | K4me3 | - | 0.3365 | 0.0312 | background 02 | 0.2726 | 0.0812 | |||
| - | K4ac | - | 0.2487 | 0.0116 | ||||||
Selected data from Histone Peptide Array analysis for H3 with single and selected double or triple PHMs as well as unmodified versions of additional peptides. Signal intensities up to 0.32 are background. Except for 2 H3 peptides with double PHMs (R8me2a/K9me2 and R8me2a/K9ac) signal intensities above 0.4 were only observed for triple PHMs on K4 and K9 as well as R2 or R8, of H3; of these, only the 8 examples with the highest binding intensity are listed in the table. None of the single or quadruple PHMs yielded signals with intensities clearly above background. Average intensity values (Intensity av.) clearly above background levels (> 0.4) are printed in bold
Taken together, our data show that the EDM2 PHD-finger module is able in vitro to bind specifically to modified N-terminal tails of H3. Furthermore, PHMs at 3 of 4 defined H3 residues (K4 and K9 as well as R2 or R8) are necessary for docking of PHM221–381. However, the EDM2 PHD-finger module seems not to be able to discriminate between certain types of PHMs at these positions. At K4 and K9 me1, me2, me3 or ac are recognized, while the third type of PHM can be either R2me2a, R2me2s, R8me2a or R8me2s. Strong cooperativity between individual PHD/PHM interactions seems to enable detectable levels of H3 binding of PHD221–381 only when defined triple PHM combinations are present at H3. Such an “all-or-nothing” mode of binding to combinatorial PHMs has so far not been reported for any PHD-finger module and seems to be rare among other types of epigenetic readers. Thus, EDM2 appears to be a unique PHD-finger-based epigenetic reader likely capable to simultaneously integrate multiple epigenetic signals and to decode combinatorial PHM marks.
Consistent with our findings, another group also reported that the EDM2 PHD-finger module is able to bind to H3.16 However, the binding specificity of the PHD-finger module seemed significantly broader in their study as compared with ours. This discrepancy may have been caused by the use of a binding buffer containing Zn2+, which preserves the PHD-finger structure, as well as higher stringency washing conditions (2x higher concentrations of NaCl and Nonidet P-40) in our assays. Furthermore, the recombinant PHD-finger module used by the other group included in addition of PHD221–381 an extra sequence representing a part of the adjacent PGR domain, which we previously showed to be a protein-protein interaction domain.3 The existence of the partial PGR domain may have distorted the structure and binding specificity of the EDM2 PHD-finger module.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Mercedes Schroeder (University of California at Riverside) for critical reading of the manuscript. This work was supported by National Science Foundation Grant IOS 1052556 to T. E.
References
- 1.Eulgem T, Tsuchiya T, Wang XJ, Beasley B, Cuzick A, Tör M, Zhu T, McDowell JM, Holub E, Dangl JL. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 2007;49:829–39. doi: 10.1111/j.1365-313X.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- 2.Tsuchiya T, Eulgem T. Co-option of EDM2 to distinct regulatory modules in Arabidopsis thaliana development. BMC Plant Biol. 2010;10:203. doi: 10.1186/1471-2229-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsuchiya T, Eulgem T. The Arabidopsis defense component EDM2 affects the floral transition in an FLC-dependent manner. Plant J. 2010;62:518–28. doi: 10.1111/j.1365-313X.2010.04169.x. [DOI] [PubMed] [Google Scholar]
- 4.Tsuchiya T, Eulgem T. EMSY-like genes are required for full RPP7-mediated race-specific immunity and basal defense in Arabidopsis. Mol Plant Microbe Interact. 2011;24:1573–81. doi: 10.1094/MPMI-05-11-0123. [DOI] [PubMed] [Google Scholar]
- 5.Tsuchiya T, Eulgem T. Mutations in EDM2 selectively affect silencing states of transposons and induce plant developmental plasticity. Sci Rep. 2013;1701 doi: 10.1038/srep01701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Roudier F, Teixeira FK, Colot V. Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet. 2009;25:511–7. doi: 10.1016/j.tig.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–5. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuchiya T, Eulgem T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc Natl Acad Sci U S A. 2013;110:E3535–43. doi: 10.1073/pnas.1312545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L, Guo C, Li QQ. Role of alternative polyadenylation in epigenetic silencing and antisilencing. Proc Natl Acad Sci U S A. 2014;111:9–10. doi: 10.1073/pnas.1321025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDowell JM, Meyers BC. A transposable element is domesticated for service in the plant immune system. Proc Natl Acad Sci U S A. 2013;110:14821–2. doi: 10.1073/pnas.1314089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36:364–72. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee WY, Lee D, Chung WI, Kwon CS. Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 2009;58:511–24. doi: 10.1111/j.1365-313X.2009.03795.x. [DOI] [PubMed] [Google Scholar]
- 16.Lei M, La H, Lu K, Wang P, Miki D, Ren Z, Duan CG, Wang X, Tang K, Zeng L, et al. Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc Natl Acad Sci U S A. 2014;111:527–32. doi: 10.1073/pnas.1320106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busso D, Delagoutte-Busso B, Moras D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal Biochem. 2005;343:313–21. doi: 10.1016/j.ab.2005.05.015. [DOI] [PubMed] [Google Scholar]