Abstract
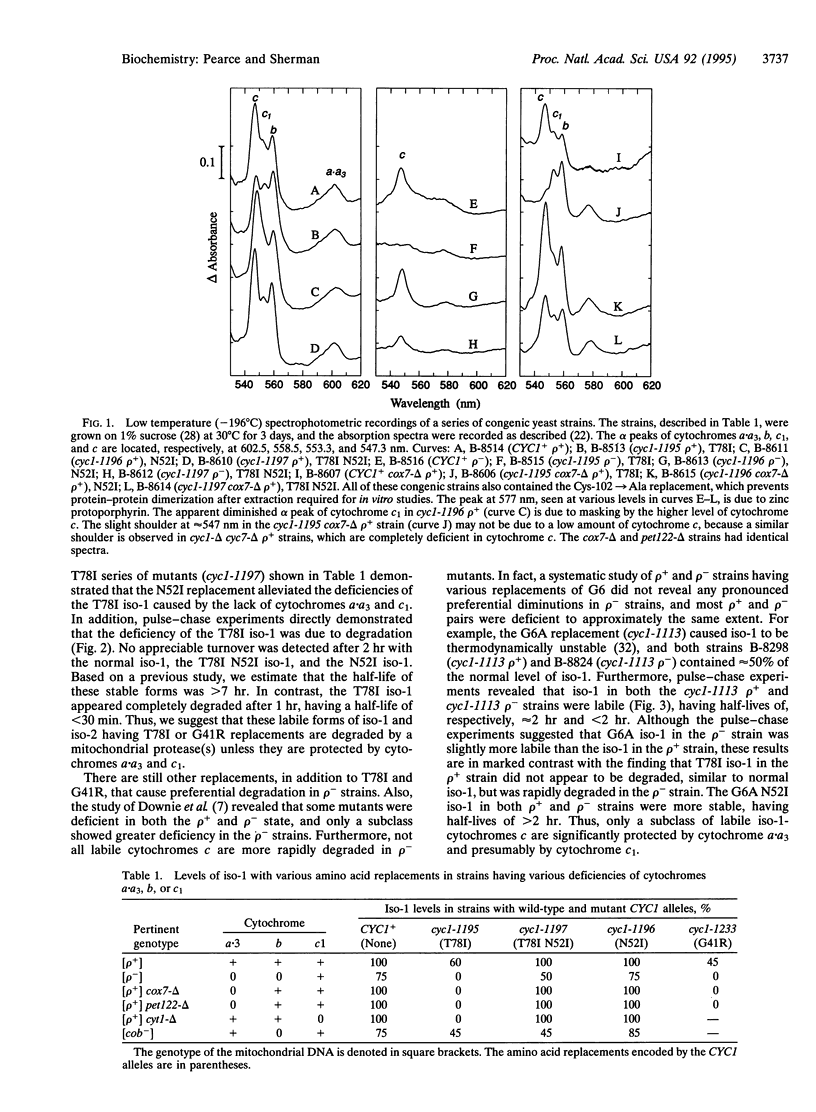
The level and structure of yeast iso-1-cytochrome c and iso-2-cytochrome c, encoded by the nuclear genes CYC1 and CYC7, respectively, are normally not altered in rho- mutants, which completely lack the cytochromes a.a3 subunits and cytochrome b that are encoded by mitochondrial DNA. In contrast, iso-cytochromes c containing the amino acid change Thr-78-->Ile (T78I) were observed at the normal or near-normal wild-type level in rho+ strains but were completely absent in rho- mutants. We have demonstrated with the "global" suppressor mutation Asn-52-->Ile and by pulse-chase labeling that the T78I iso-1-cytochrome c undergoes rapid cellular degradation in rho- mutants. Furthermore, specific mutations revealed that the deficiency of T78I iso-1 cytochrome c can be caused by the lack of cytochrome a.a3 or cytochrome c1, but not by the lack of cytochrome b. Thus, this and certain other, but not all, labile forms of cytochrome c are protected from degradation by the interaction with its physiological partners.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggeler R., Capaldi R. A. Yeast cytochrome c oxidase subunit VII is essential for assembly of an active enzyme. Cloning, sequencing, and characterization of the nuclear-encoded gene. J Biol Chem. 1990 Sep 25;265(27):16389–16393. [PubMed] [Google Scholar]
- Berroteran R. W., Hampsey M. Genetic analysis of yeast iso-1-cytochrome c structural requirements: suppression of Gly6 replacements by an Asn52----Ile replacement. Arch Biochem Biophys. 1991 Jul;288(1):261–269. doi: 10.1016/0003-9861(91)90193-m. [DOI] [PubMed] [Google Scholar]
- Das G., Hickey D. R., McLendon D., McLendon G., Sherman F. Dramatic thermostabilization of yeast iso-1-cytochrome c by an asparagine----isoleucine replacement at position 57. Proc Natl Acad Sci U S A. 1989 Jan;86(2):496–499. doi: 10.1073/pnas.86.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Stewart J. W., Brockman N., Schweingruber A. M., Sherman F. Structural gene for yeast iso-2-cytochrome c. J Mol Biol. 1977 Jun 25;113(2):369–384. doi: 10.1016/0022-2836(77)90147-4. [DOI] [PubMed] [Google Scholar]
- Downie J. A., Stewart J. W., Sherman F. Yeast mutants defective in iso-2-cytochrome c. J Mol Biol. 1977 Dec 5;117(2):369–386. doi: 10.1016/0022-2836(77)90133-4. [DOI] [PubMed] [Google Scholar]
- Dumont M. D., Mathews A. J., Nall B. T., Baim S. B., Eustice D. C., Sherman F. Differential stability of two apo-isocytochromes c in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990 Feb 15;265(5):2733–2739. [PubMed] [Google Scholar]
- Dumont M. E., Ernst J. F., Hampsey D. M., Sherman F. Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae. EMBO J. 1987 Jan;6(1):235–241. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelin E., Rep M., Grivell L. A. Sequence of the AFG3 gene encoding a new member of the FtsH/Yme1/Tma subfamily of the AAA-protein family. Yeast. 1994 Oct;10(10):1389–1394. doi: 10.1002/yea.320101016. [DOI] [PubMed] [Google Scholar]
- Hickey D. R., Berghuis A. M., Lafond G., Jaeger J. A., Cardillo T. S., McLendon D., Das G., Sherman F., Brayer G. D., McLendon G. Enhanced thermodynamic stabilities of yeast iso-1-cytochromes c with amino acid replacements at positions 52 and 102. J Biol Chem. 1991 Jun 25;266(18):11686–11694. [PubMed] [Google Scholar]
- Hickey D. R., Jayaraman K., Goodhue C. T., Shah J., Fingar S. A., Clements J. M., Hosokawa Y., Tsunasawa S., Sherman F. Synthesis and expression of genes encoding tuna, pigeon, and horse cytochromes c in the yeast Saccharomyces cerevisiae. Gene. 1991 Aug 30;105(1):73–81. doi: 10.1016/0378-1119(91)90515-d. [DOI] [PubMed] [Google Scholar]
- Lancashire W. E., Mattoon J. R. Genetic manipulation of a latent defect in yeast cytochrome biosynthesis utilizing cytoduction. Biochem Biophys Res Commun. 1979 Oct 12;90(3):801–809. doi: 10.1016/0006-291x(79)91899-0. [DOI] [PubMed] [Google Scholar]
- Maicas E., Pluthero F. G., Friesen J. D. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol Cell Biol. 1988 Jan;8(1):169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matner R. R., Sherman F. Differential accumulation of two apo-iso-cytochromes c in processing mutants of yeast. J Biol Chem. 1982 Aug 25;257(16):9811–9821. [PubMed] [Google Scholar]
- McKnight G. L., Cardillo T. S., Sherman F. An extensive deletion causing overproduction of yeast iso-2-cytochrome c. Cell. 1981 Aug;25(2):409–419. doi: 10.1016/0092-8674(81)90059-3. [DOI] [PubMed] [Google Scholar]
- Moerschell R. P., Hosokawa Y., Tsunasawa S., Sherman F. The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. Processing of altered iso-1-cytochromes c created by oligonucleotide transformation. J Biol Chem. 1990 Nov 15;265(32):19638–19643. [PubMed] [Google Scholar]
- Pajic A., Tauer R., Feldmann H., Neupert W., Langer T. Yta10p is required for the ATP-dependent degradation of polypeptides in the inner membrane of mitochondria. FEBS Lett. 1994 Oct 17;353(2):201–206. doi: 10.1016/0014-5793(94)01046-3. [DOI] [PubMed] [Google Scholar]
- REILLY C., SHERMAN R. GLUCOSE REPRESSION OF CYTOCHROME A-SYNTHESIS IN CYTOCHROME-DEFICIENT MUTANTS OF YEAST. Biochim Biophys Acta. 1965 Apr 19;95:640–651. doi: 10.1016/0005-2787(65)90518-6. [DOI] [PubMed] [Google Scholar]
- Reid G. A. Pulse labeling of yeast cells and spheroplasts. Methods Enzymol. 1983;97:324–329. doi: 10.1016/0076-6879(83)97144-6. [DOI] [PubMed] [Google Scholar]
- SHERMAN F., SLONIMSKI P. P. RESPIRATION-DEFICIENT MUTANTS OF YEAST. II. BIOCHEMISTRY. Biochim Biophys Acta. 1964 Jul 15;90:1–15. doi: 10.1016/0304-4165(64)90113-8. [DOI] [PubMed] [Google Scholar]
- Sanders H. K., Mied P. A., Briquet M., Hernandez-Rodriguez J., Gottal R. F., Mattoon J. R. Regulation of mitochondrial biogenesis: yeast mutants deficient in synthesis of delta-aminolevulinic acid. J Mol Biol. 1973 Oct 15;80(1):17–39. doi: 10.1016/0022-2836(73)90231-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall R., Mannhaupt G., Stucka R., Tauer R., Ehnle S., Schwarzlose C., Vetter I., Feldmann H. Identification of a set of yeast genes coding for a novel family of putative ATPases with high similarity to constituents of the 26S protease complex. Yeast. 1994 Sep;10(9):1141–1155. doi: 10.1002/yea.320100903. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974 Jun;77(2):255–284. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Margoliash E., Parker J., Campbell W. The structural gene for yeast cytochrome C. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1498–1504. doi: 10.1073/pnas.55.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Taber H., Campbell W. Genetic determination of iso-cytochromes c in yeast. J Mol Biol. 1965 Aug;13(1):21–39. doi: 10.1016/s0022-2836(65)80077-8. [DOI] [PubMed] [Google Scholar]
- Stueber D., Ibrahimi I., Cutler D., Dobberstein B., Bujard H. A novel in vitro transcription-translation system: accurate and efficient synthesis of single proteins from cloned DNA sequences. EMBO J. 1984 Dec 20;3(13):3143–3148. doi: 10.1002/j.1460-2075.1984.tb02271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C. K., Suda K., Wang N., Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994 Apr 8;264(5156):273–276. doi: 10.1126/science.8146662. [DOI] [PubMed] [Google Scholar]
- Tauer R., Mannhaupt G., Schnall R., Pajic A., Langer T., Feldmann H. Yta10p, a member of a novel ATPase family in yeast, is essential for mitochondrial function. FEBS Lett. 1994 Oct 17;353(2):197–200. doi: 10.1016/0014-5793(94)01045-5. [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., White K. H., Fox T. D. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Sep;13(9):5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y. F., Thompson J. R., Rotenberg M. O., Larkin J. C., Woolford J. L., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 1988 Jun;2(6):664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Yue J., Jang J., Paul M. F. A new member of a family of ATPases is essential for assembly of mitochondrial respiratory chain and ATP synthetase complexes in Saccharomyces cerevisiae. J Biol Chem. 1994 Oct 21;269(42):26144–26151. [PubMed] [Google Scholar]
- Van Dyck L., Pearce D. A., Sherman F. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1994 Jan 7;269(1):238–242. [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Moerschell R. P., Wakem L. P., Komar-Panicucci S., Sherman F. Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics. 1992 Aug;131(4):811–819. doi: 10.1093/genetics/131.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- elBaradi T. T., van der Sande C. A., Mager W. H., Raué H. A., Planta R. J. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr Genet. 1986;10(10):733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]