Abstract
Plants have developed mechanisms to protect themselves against both biotic and abiotic environmental stress. Specialized/secondary metabolism is one of the stress response mechanisms. Recently, we reported that flavonoids, a class of specialized metabolites, including flavonols and anthocyanins with strong radical scavenging activity contributed to the mitigation of oxidative and drought stress in Arabidopsis thaliana (Arabidopsis). However, the behavior of flavonoids during drought stress is still not well-documented. Herein we investigated the time-series alternation of flavonoids in the aerial part of Arabidopsis (wild type, Col-0) during drought stress by LC-QTOF-MS. The drastic alternation of 5 flavonols and 5 anthocyanins was revealed together with changes in marker metabolites of drought stress, e.g., proline, raffinose, and galactinol. These findings indicate that flavonols and anthocyanins can mitigate drought stress.
Keywords: metabolomics, flavonol, anthocyanin, specialized/secondary metabolite, Arabidopsis
Flavonoids are a major component of specialized/secondary metabolites in plants.1 These metabolites are thought to be defense metabolites against environmental stress such as biotic and abiotic stress.2 Recently, we identified the role of flavonoids as a mitigator of oxidative and drought stress in Arabidopsis thaliana (Arabidopsis). We further showed that the overaccumulation of anthocyanins enhanced tolerance against both oxidative and drought stress.3 This result suggested that identification of flavonoid alternation behavior during environmental stress will lead to a better understanding of the flavonoid biosynthetic pathway at both the metabolite and gene levels in plants. A previous extensive study using several mass spectrometry methods including GC-MS and CE-MS showed drought stress responsive metabolites in Arabidopsis.4 The profiling of polar specialized metabolites including flavonoids during drought stress was not performed with LC-MS, which can widely detect such specialized metabolites.5 Hence, we investigated the time-series alternation of flavonoids in the aerial parts of Arabidopsis using LC-QTOF-MS.
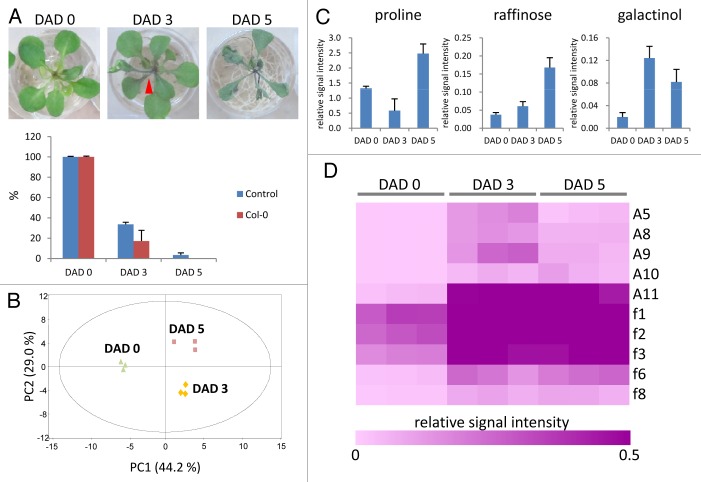
To investigate flavonoid alternation during drought stress in Arabidopsis (wild type, Col-0), 3-week-old plants were exposed to drought stress using 24-well plates for up to 5 d. During drought stress, anthocyanin accumulation and wilting were visually confirmed (Fig. 1A, upper). The extent of water loss (drought stress) was evaluated as the remaining water measured from the comparison of the control (without plants) with the experiment (with plants) (Fig. 1A, lower).
Figure 1. Flavonoid alternation during drought stress in Arabidopsis. (A) Upper: the phenotype of Arabidopsis (Col-0) plants. The red arrow indicates the site of anthocyanin accumulation. Lower: the extent of water loss during drought stress. The water remaining in wells was indicated as the percentage of water in day 0. Control indicates that no plants were in the wells. Differences and standard deviations (error bars) were calculated from the results of 12 wells. (B) Principal component analysis (PCA) of untargeted profiling data obtained by LC-QTOF-MS. The plot of PC1 (44.2%) vs. PC2 (29.0%) is presented. Three biological replicates were used for this analysis. (C) Relative changes of 3 positive marker metabolites of drought stress. These metabolite peaks were assigned by comparison with standard compounds. Means and standard deviations (error bars) were calculated from the results of 3 replicates. (D) Hierarchical cluster analysis (HCA) of flavonoid peak intensities using the processed data. “A” and “f” represent anthocyanin and flavonol, respectively, as previously described.6,7
Untargeted metabolomic analysis by LC-QTOF-MS was performed with the aerial samples harvested at 0, 3, and 5 d after drought (DAD). Principal component analysis (PCA) using the processed data, which consisted of 4093 metabolite peaks, exhibited clear separation between the groups with different periods of exposure to drought stress (Fig. 1B). An increase of the levels of known positive markers of drought stress (proline, raffinose, and galactinol) was confirmed; the peak identity of these metabolites was unequivocally confirmed by comparison with standard chemicals (level 1: identification according to the guide line of MSI8,9) (Fig. 1C). These data indicated that the marker metabolites of drought response were also altered under this experimental condition. The glycosides of kaempferol (f1, f2, and f3), quercetin (f6 and f8), and cyanidin (A5, A8, A9, A10, and A11) were then chemically assigned by comparison to reference MS/MS spectra6,7 (level 2: annotation). Given that the flavonoids were main flavonoids in the aerial parts of seedlings in Arabidopsis,7 they were assigned for identifying their behavior during drought stress. Hierarchical clustering analysis (HCA) using the data of the assigned flavonoids showed that all flavonols and anthocyanins were clustered together, suggesting a metabolic response to the drought stress (Fig. 1D).
In this study, we profiled flavonoids of the Arabidopsis plants in the time-series experiments of drought stress by untargeted metabolomic analysis using LC-QTOF-MS. The data demonstrated that 10 flavonoids increased from DAD 3 to DAD 5, indicating that all flavonoids are drought stress responsive metabolites that can be used as positive markers and potential mitigators for drought stress. The experiments revealed the alternation of 3 types of flavonoids, glycosides of kaempferol, quercetin, and cyanidin, during drought stress; however, the signaling/regulation mechanisms of flavonoids or the individual role of each molecule in the stress mitigation mechanism is still unclear. The experiment using the well plates can easily mimic drought stress conditions using pods or pads with soils in a laboratory. Therefore, integrated metabolome and transcriptome analysis using mutant lines lacking or overaccumulating each glycoside of kaempferol, quercetin, and cyanidin with the plate experiments will lead to the identification of the signaling/regulation mechanisms and the specific function of each molecule under drought stress. Finally, the experiment using the plates will significantly promote studies related to drought stress due to its simplicity.
Methods
Plant materials and growth conditions
Arabidopsis (Col-0) plants were used in this study. All plates were placed in a growth chamber at 22 °C with a relative humidity of approximately 15% for a 14 h light (approximately 40 μmol sec−1 m−2)/10 h dark cycle. Harvested samples were immediately frozen and freeze-dried. The samples were stored at −80 °C until use.
Drought stress
Three-week-old plants vertically grown on GM plates3 were incubated in 24-well plates (TPP, http://www.tpp.ch/index.php). Each plant was put in a well with 3 ml water. Three-week-old plants were exposed to drought stress by stopping watering. The aerial samples were harvested at the time points of DAD 0, 3, and 5.
Untargeted metabolomic analysis of LC-QTOF-MS
The untargeted metabolomic analysis using LC-QTOF-MS and the metabolome data processing of alignment, de-isotoping, noise cutting, and normalization was conducted as previously described.3
Statistical analysis
Principal component analysis (PCA) using the processed data consisting of 4093 metabolite peaks was performed with the SIMCA-P 11.5 software, and HCA using the data of the assigned flavonoids was performed by MeV 4.8 (http://mev.tm4.org/).3
Data upload
Metabolomic data are available on the website DROP Met < http://prime.psc.riken.jp/?action=drop_index>.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Yutaka Yamada and Tetsuya Sakurai (RIKEN CSRS) for technical help. This work was supported by Strategic International Collaborative Research Program (SICORP) and Japan Science and Technology Agency (JST).
Glossary
Abbreviations:
- Col-0
Columbia-0
- LC-QTOF-MS
liquid chromatography-quadrupole time-of-flight-mass spectrometry
- DAD
day after drought
- PCA
principal component analysis
- MSI
metabolomics standard initiatives
- HCA
hierarchical cluster analysis
- GC-MS
gas chromatography‒mass spectrometry
- CE-MS
capillary electrophoresis-mass spectrometry
References
- 1.Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Dixon RA, Paiva NL. Stress-Induced Phenylpropanoid Metabolism. Plant Cell. 1995;7:1085–97. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–79. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009;57:1065–78. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- 5.Matsuda F, Hirai MY, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart NJ, Sakurai T, Shimada Y, Saito K. AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol. 2010;152:566–78. doi: 10.1104/pp.109.148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell. 2008;20:2160–76. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–35. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- 8.Neumann S, Böcker S. Computational mass spectrometry for metabolomics: identification of metabolites and small molecules. Anal Bioanal Chem. 2010;398:2779–88. doi: 10.1007/s00216-010-4142-5. [DOI] [PubMed] [Google Scholar]
- 9.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–21. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]