Abstract
Changes in DNA supercoiling are induced by a wide range of environmental stresses in Escherichia coli, but the physiological significance of these responses remains unclear. We now demonstrate that an increase in negative supercoiling is necessary for transcriptional activation of a large subset of osmotic stress-response genes. Using a microarray-based approach, we have characterized supercoiling-dependent gene transcription by expression profiling under conditions of high salt, in conjunction with the microbial antibiotics novobiocin, pefloxacin, and chloramphenicol. Algorithmic clustering and statistical measures for gauging cellular function show that this subset is enriched for genes critical in osmoprotectant transport/synthesis and rpoS-driven stationary phase adaptation. Transcription factor binding site analysis also supports regulation by the global stress ς factor rpoS. In addition, these studies implicate 60 uncharacterized genes in the osmotic stress regulon, and offer evidence for a broader role for supercoiling in the control of stress-induced transcription.
Changes in the level of DNA supercoiling coincide with a diverse spectrum of environmental events including nutritional upshift, entry into stationary phase, temperature stress, peroxide stress, and osmotic shock (Hengge-Aronis 1999; Lopez-Garcia and Forterre 2000; Weinstein-Fischer et al. 2000; Travers et al. 2001). These phenomena have been best characterized in the stress responses of Escherichia coli but have been described in a number of other bacterial species as well (Rohde et al. 1994; Jordi et al. 1995; Alice and Sanchez-Rivas 1997; Ali et al. 2002). Concurrently, expression and activity of global transcriptional regulators such as Fis (fis), cyclic AMP receptor protein (crp), and stress-induced ς factor ςS (rpoS) have been shown to be dependent on the supercoiling state of the cell (Finkel and Johnson 1992; Schneider et al. 1999). Supercoiling modulates the transcriptional effects of these regulators directly by affecting the efficiency of protein binding to their DNA targets, or indirectly by altering the transcriptional expression of the regulators themselves. These observations have raised the possibility that DNA supercoiling may play a functional role in coupling stress signals to transcriptional activity (Dorman 1996).
In this paper, we report the application of whole-genome microarrays toward understanding the significance of supercoiling in the osmotic stress response. Osmotic shock is among the most common environmental challenges faced by bacterial organisms (Kempf and Bremer 1998; Wood 1999), and serves as an ideal model system because of the rapidity of its effects. High osmolarity causes a rapid increase in negative supercoiling, with subsequent relaxation to preinduction values as the cell recovers. These supercoiling effects are accompanied by a distinct sequence of cellular events: plasmolysis, followed by active potassium influx and glutamate synthesis to restore intracellular water, and finally, replacement of potassium glutamate with osmoprotectants more compatible with cell growth. Artificially induced positive supercoiling delays recovery and retards growth. Meanwhile, increased negative supercoiling is reported to stimulate transcription of several osmoregulated genes in vitro and in vivo, including the primary active transporter for the osmoprotectants proline and glycine–betaine (proU) (Higgins et al. 1988; Jordi and Higgins 2000) and the lipoprotein osmE (Conter et al. 1997), as well as to repress transcription of the outer-membrane β-barrel porin ompF(Graeme-Cook et al. 1989).
Here we demonstrate that increased negative supercoiling is necessary for proper elicitation of the E. coli osmotic shock response. Using a whole-genome approach (Lockhart et al. 1996; DeRisi et al. 1997; Richmond et al. 1999; Selinger et al. 2000), we have characterized the genetic subprograms activated in response to osmotic shock, clustered by expression profiles under salt stress and perturbed with three antibiotics: novobiocin, pefloxacin, and chloramphenicol. Statistical assignment of cellular function using the GenProtEC E. coli database (Riley and Serres 2000) identifies enrichment for several major functional categories and one subset of genes with transcriptional kinetics consistent with supercoiling-dependent regulation. This subset is composed of genes implicated in the adaptation of E. coli to osmotic stress, including genes in the major osmoprotectant synthesis/transport families, betaine (betABIT), trehalose (otsAB), and proline (proVWX). Statistical analyses of known transcription factor binding sites support the involvement of rpoS, and computational predictions implicate other global regulators including crp and fis that are sensitive to supercoiling. We propose a functional role for supercoiling in the osmotic stress response, and suggest that supercoiling may have broader significance in stress-induced transcription.
RESULTS
Microarray Studies of Genome-Wide E. coli Transcription in Response to Salt and a Panel of Microbial Antibiotics
We examined whole-genome mRNA expression in E. coli MG1655 (Blattner et al. 1997) following exposure to five different conditions: novobiocin, high salt, novobiocin plus salt, pefloxacin, and chloramphenicol (Table 1). Samples obtained at 0 and 10 min after treatments with novobiocin or salt, at 0, 7, and 10 min with pefloxacin, and at 0, 10, and 40 min with chloramphenicol were fluorescently labeled and hybridized to microarrays.
Table 1.
List of Experimental Conditions
| Condition | Time point (min) | Growth media |
| Baseline | 0 | Minimal |
| Novobiocin | 10 | Minimal |
| Salt | 10 | Minimal |
| Novobiocin + salt | 10 | Minimal |
| Baseline | 0 | Rich |
| Pefloxacin | 1 | Rich |
| Pefloxacin | 7 | Rich |
| Baseline | 0 | Rich |
| Chloramphenicol | 10 | Rich |
| Chloramphenicol | 40 | Rich |
All conditions were sampled in duplicate with the exception of pefloxacin at 1 min and 7 min, and chloramphenicol at 10 min. Pearson correlation coefficients between replicates over the set of all mRNAs and noncoding RNAs ranged from 0.90 to 0.99.
The microbial antibiotics novobiocin and pefloxacin were chosen for their effects on supercoiling in the cell (Maxwell 1997). E. coli topoisomerase II, also known as gyrase, is unusual among topoisomerases in that it can introduce negative supercoils into DNA by ATP hydrolysis (Luttinger 1995; Wang 1996; Champoux 2001). Novobiocin binding decreases the affinity of gyrB (the β subunit of topoisomerase II) for the ATP nucleotide, which is required for DNA breakage and strand passage (Gellert et al. 1976). Novobiocin therefore causes increased positive supercoiling. The quinolone antibiotic pefloxacin also targets gyrase, but possesses a different mechanism of inhibition. Pefloxacin, in contrast to novobiocin, stabilizes the transition state of gyrase after DNA breakage, leading to the formation of a cleavable ternary complex (Drlica and Zhao 1997). This complex can form a barrier, which on collision with replication forks, leads to double-stranded breaks. Joint treatment with novobiocin and salt produces an intermediate response (Conter et al. 1997). Chloramphenicol blocks the 23S subunit of the ribosome and thus protein synthesis. This antibiotic is not reported to have direct effects on supercoiling, but we include it to facilitate in the discovery of coregulated genes.
Total RNA samples were enriched for mRNA, biotinylated, hybridized to Affymetrix E. coli arrays, and scanned according to the Affymetrix protocol. Genes below detection threshold as reported by Affymetrix software were eliminated from consideration. For further stringency, a z-test was used to eliminate probes near threshold as defined by negative controls found on the microarrays. Using these criteria, 2146 of 4249 mRNAs and untranslated RNAs were deemed above detection with P values ≤0.01.
gyrB and topA Transcription Match Predicted Effects of Supercoiling
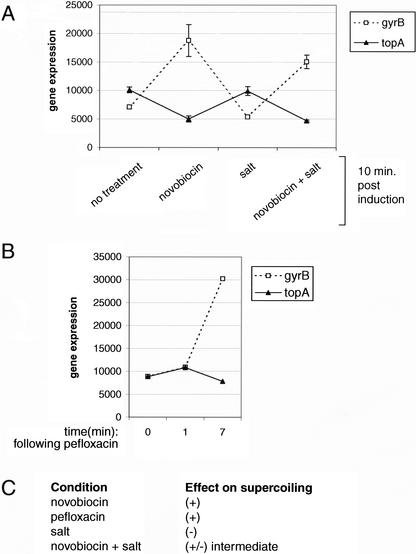
E. coli topoisomerases are regulated by supercoiling in a negative feedback loop (Menzel and Gellert 1987). Positive supercoiling stimulates gyrase expression, whereas negative supercoiling stimulates topoisomerase I expression. Results shown in Figure 1A and B support this model. gyrB mRNA expression increases with novobiocin and pefloxacin treatments, but decreases with salt. In contrast, expression of the topA gene (encoding topoisomerase I) increases with salt, and is unchanged with novobiocin. These results are consistent with the known effects of these treatment conditions on DNA supercoiling (Fig. 1C).
Figure 1.
topA and gyrB gene expression mirror predicted effects of supercoiling. topA encodes topoisomerase I, and gyrB encodes the β subunit of gyrase. (A) Escherichia coli MG1655 was cultured to optical density (OD) 0.4 and sampled (no treatment), or induced with novobiocin, salt, or novobiocin + salt. Treated cultures were sampled 10 min postinduction. (B) E. coli was cultured in a separate experiment to OD 0.6 and treated with pefloxacin. (C) Predicted changes in DNA supercoiling from salt or gyrase inhibitors based on the literature.
Functional Characterization of the Osmotic Stress Regulon
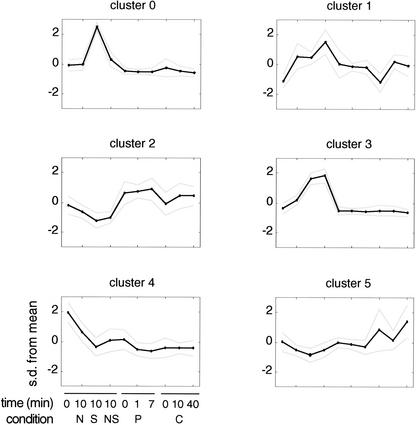
Genes significantly induced or repressed by salt were identified by changes in expression greater than twofold, and in the same direction consistently between replicates. One hundred seventy-five genes satisfied these criteria. We list the top 5 up-regulated and top 5 down-regulated genes in Table 2. In orderto functionally characterize the osmotic stress response, we used a two-stage approach. Genes were first clustered using self-organizing maps. Six-cluster partitioning was chosen for optimal balance between separation and number of clusters (Fig. 2). Second, the resulting clusters were assigned cellular functions using the GenProtEC: E. coli genome and protein database (Riley and Labedan 1997; Riley and Serres 2000) (Table 3). Following the method of Tavazoie et al. (1999), we estimated the probability that an observed set of genes with a common functional role could have segregated into an individual cluster at random (Sokal and Rohlf 1995). P values below the Bonferroni-corrected significance threshold (P value <0.01) are suggestive of functional enrichment (see Methods).
Table 2.
Top Five Up- and Down-Regulated Genes in the Osmostic Stress Response
| Gene | Product | log2 fold change |
| proX | High-affinity transport system for glycine betaine and proline | 5.9 |
| proW | High-affinity transport system for glycine betaine and proline | 5.7 |
| proV | ATP-binding component of transport system for glycine, betaine, and proline | 4.5 |
| otsA | Trehalose-6-phosphate synthase | 4.5 |
| b1481 | Orf, hypothetical protein | 4.0 |
| ykgM | Putative ribosomal protein | −2.9 |
| upp | Uracil phosphoribosyltransferase | −3.2 |
| treC | Trehalase 6-P hydrolase | −3.5 |
| yifE | Orf, hypothetical protein | −4.0 |
| lamB | Phage lambda receptor protein; maltose high-affinity receptor | −4.5 |
The log2 fold change was calculated as log2 (gene expression with salt treatment)/(gene expression with no treatment).
Figure 2.
The osmotic stress response partitions into distinct clusters of drug sensitivity. Mean-variance normalized expression profiles for the 175 genes changed greater than twofold with salt treatment were partitioned into six clusters using self-organizing maps. The black line denotes the mean expression profile of the cluster, and the gray lines indicate one standard deviation above and below this mean (N) Novobiocin; (S) salt; (NS) novobiocin + salt; (P) pefloxacin; (C) chloramphenicol.
Table 3.
The Osmotic Stress Response Partitions into Distinct Classes of Cellular Function
| Cluster | p value | GenProt designation | Description | Observed (F∩C) | Expected (F∩C) |
| 0 | 7.06E-05 | 5.5 | Adaptation to stress | 12 | 4.9 |
| 0 | 5.38E-04 | 5.5.1 | Osmotic pressure | 10 | 4.2 |
| 1 | 8.99E-05 | 1.7.34 | Peptidoglycan (murein) turnover, recycling | 3 | 0.1 |
| 1 | 1.93E-04 | 6 | Cell structure | 6 | 1.5 |
| 2 | 4.32E-03 | 1.6 | Macromolecules (cellular constituent) biosynthesis | 6 | 2.0 |
| 2 | 5.03E-03 | 1.1.1 | Carbon compounds | 11 | 5.3 |
| 2 | 9.11E-03 | 6.4 | Flagellum | 4 | 1.1 |
| 2 | 9.11E-03 | 1.6.12 | Flagella | 4 | 1.1 |
| 2 | 9.11E-03 | 5.3 | Motility (incl. chemotaxis, energytaxis, aerotaxis, redoxtaxis) | 4 | 1.1 |
| 2 | 9.23E-03 | 1.1 | Carbon utilization | 12 | 6.5 |
| 3 | 9.29E-04 | 1.7.1 | Unassigned reversible reactions | 3 | 0.3 |
| 4 | 8.13E-06 | 1.5 | Building block biosynthesis | 13 | 4.2 |
| 4 | 4.41E-04 | 1.5.1 | Amino acids | 9 | 2.9 |
| 5 | 7.06E-04 | 4.S | Substrate | 11 | 4.4 |
| 5 | 8.77E-04 | 4 | Transport | 12 | 5.2 |
| 5 | 2.31E-03 | 4.4.A.6 | The PTS Mannose-Fructose-Sorbose (Man) family | 3 | 0.4 |
| 5 | 6.45E-03 | 1.1 | Carbon utilization | 9 | 4.0 |
| 5 | 6.70E-03 | 1.1.1 | Carbon compounds | 8 | 3.3 |
| 5 | 7.34E-03 | 4.4 | Group translocators | 4 | 1.0 |
| 5 | 7.34E-03 | 4.4.A | Phosphotransferase systems (PEP-dependent PTS) | 4 | 1.0 |
The six clusters were screened using a hypergeometric test measuring statistical enrichment for functional categories from the GenProtEC Escherichia coli database. Observed (F∩C) denotes the number of genes in the specified functional category that are found in the cluster. Expected (F∩C) denotes the number of genes expected by chance to lie within the cluster. P values below 0.01 were deemed significant using Bonferroni correction for multiple hypothesis testing phosphoenolpyruvate (PEP).
As shown in Table 3, several major functional classes of genes govern the osmotic stress response. These include adaptation to stress and osmoregulation (cluster 0), peptidoglycan biosynthesis (cluster 1), macromolecular biosynthesis (cluster 2), amino acid biosynthesis (cluster 4), and the PTS Mannose-Fructose-Sorbose family (cluster 5). A majority of the known osmoregulated genes as listed by GenProtEC are found in cluster 0 (P value ≤5.4 × 10−4). These results provide evidence that the transcriptional response to osmotic shock greatly extends beyond what is currently known about osmoregulated genes, and provides a promising set of candidates for future experimental validation.
Potassium and Osmoprotectant Genes Segregate into Different Clusters
The immediate effect of hyperosmotic stress is plasmolysis, leading to reduced respiration and growth arrest (Ingraham and Marr 1996). Restoration of intracellular water by potassium influx and glutamate synthesis occurs within minutes, but is only an interim solution because high intracellular ion concentrations are also unfavorable for cell growth (Record Jr. et al. 1998). Osmoprotectants, in contrast, allow resumption of growth, but must be synthesized or brought into the cell by active transport. Thus, two emergency systems, one fast and one slow, are triggered by osmotic stress (Wood 1999). In support of this model, our data demonstrate that Trk family and Kdp family potassium transport genes (clusters 1 and 3) cocluster separately from osmoprotectant genes (cluster 0). Major components for synthesis/transport of osmoprotectants betaine (betABIT), trehalose (otsAB and treF), and proline transport (proVWX) are all located in cluster 0. In contrast, trkH is located in cluster 1, whereas kdpABCD, although outside the twofold threshold, exhibits expression profiles similar to cluster 3 (r = 0.89–0.99). Likewise, the low affinity proline transporter (proP, cluster 3) and components of the murein oligopeptide transporter (oppA, cluster 3; oppDEF, cluster 1) segregate outside of cluster 0. proP is activated in medium hyperosmolarity and its synthesis coincides with potassium influx. Glutamate is a necessary building block in the synthesis of the peptidoglycan wall (Van Heijenoort 1996). Up-regulation of murein oligopeptide transporters (Goodell and Higgins 1987) may therefore reflect coupling of peptidoglycan degradation with potassium counter-ion synthesis.
Salt Stress Causes a Reduction in Metabolic Synthesis
Clusters 2, 4, and 5 also demonstrate the presence of a significant down-regulated component in the osmotic stress response. These include biosynthetic genes for amino acids (cluster 4), flagellar biosynthetic genes (cluster 2), and genes encoding components for galactitol and maltose transport (clusters 2 and 5, respectively). Maltose transport has been previously linked to trehalose synthesis, and is repressed by increased osmolarity in the absence of inducer (Boos and Shuman 1998). Galactitol, however, has not been described in the context of osmotic stress (Nobelmann and Lengeler 1996). Peptidoglycan-associated lipoprotein, pal, and peptidoglycan synthetase, mrdA, are also repressed by salt (cluster 2), as is trehalase 6-phosphate hydrolase (treC), which degrades intracellular trehalose. These findings are consistent with the mirror-image up-regulation seen for murein oligopeptide uptake and trehalose synthesis genes, respectively.
Cluster 0 is Enriched for Supercoiling-Dependent Transcription
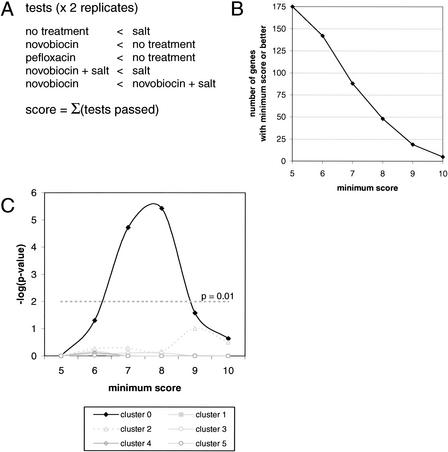
If supercoiling regulates the expression of osmotic stress genes, then we would predict that nonphysiological supercoiling would cripple the osmotic stress response. Genes regulated by supercoiling should be down-regulated in novobiocin, and pefloxacin, and up-regulated in salt, or vice versa. Conversely, genes that fit this pattern are candidates for regulation by a supercoiling-dependent mechanism. Topoisomerase genes gyrB and topA (Fig. 1), for example, have expression profiles satisfying this criterion. Using this working model, we scored the 175 genes for their consistency with this expression pattern (Fig. 3A). Scores were based on 10 criteria or equivalently two replicates of five constraints. Higher scores correspond to greater consistency with our predicted model. For a given score, we calculated the number of genes with at least this score or better for each cluster, and tested the null hypothesis that this distribution could have occurred by chance. These calculations demonstrate that at all score cutoffs, only cluster 0 is statistically enriched for supercoiling-dependent transcription (Fig. 3B and Table 4). Given the functional significance of cluster 0 in osmoprotectant synthesis and stress adaptation (Table 3), we propose that negative supercoiling is a physiologically necessary activation signal for osmotically induced transcription.
Figure 3.
Cluster 0 is enriched for supercoiling-dependent gene regulation. (A) Scoring function for supercoiling-dependent transcription. A total of five tests with two replicates were incorporated into our scoring function. Because genes could be up-regulated or down-regulated in response to supercoiling, we took the better score assuming either of these two cases. Thus, scores could range from 5–10. (B) The 175 genes in the putative osmotic stress response were assayed for their consistency with a supercoiling-dependent profile using the scoring function described in A. The minimum score denotes the lowest score or better that is included in the set of supercoiling-regulated genes. (C) For each minimum score, a hypergeometric test was used to gauge statistical enrichment over all clusters. Only cluster 0 shows statistically significant enrichment.
Table 4.
Cluster 0 is Enriched for Supercoiling-Dependent Gene Regulation
| Gene | Product |
| acnA | Aconitate hydrase 1 |
| b1664 | Possible enzyme |
| b1724 | Orf, hypothetical protein |
| b2809 | Orf, hypothetical protein |
| bax | Putative ATP-binding protein |
| btuE | Vitamin B12 transport |
| dps | Global regulator, starvation conditions |
| gcd | Glucose dehydrogenase |
| grxB | Glutaredoxin 2 |
| nlpD | Lipoprotein |
| osmE | Activator of ntrL gene |
| otsA | Trehalose-6-phosphate synthase |
| otsB | Trehalose-6-phosphate phosphatase, biosynthetic |
| poxB | Pyruvate oxidase |
| proV | ATP-binding component of transport system for glycine, betaine, and proline |
| proW | High-affinity transport system for glycine, betaine, and proline |
| proX | High-affinity transport system for glycine, betaine, and proline |
| rpoS | RNA polymerase, sigma S (sigma38) factor; synthesis of many growth phase-related proteins |
| sugE | Suppresses groEL, may be chaperone |
| tktB | Transketolase 2 isozyme |
| wrbA | Trp repressor binding protein; affects association of trp repressor and operator |
| yacK | Orf, hypothetical protein |
| ybaL | Putative transport protein |
| ybaY | Glycoprotein/polysaccharide metabolism |
| ybeL | Putative alpha helical protein |
| ygaM | Orf, hypothetical protein |
| yggB | Putative transport protein |
| ykfE | Orf, hypothetical protein |
| ynhG | Orf, hypothetical protein |
| yrbL | Orf, hypothetical protein |
Thirty of 61 genes in cluster 0 have scores of 8 or better using the metric described in Fig. 3A.
The Global Transcription Factor rpoS Is Implicated in the Regulation of Cluster 0
Cluster 0 expression (Fig. 2) is significantly lower in the absence of salt stress, in support of a model in which supercoiling does not act alone, but rather is a coactivator of gene expression. Several lines of evidence indicate that rpoS, encoding the stationary phase/stress-activated ς factor ςS, may be a partner in this interaction (Hengge-Aronis 1996). First, rpoS is a member of cluster 0, and by our assay, shows supercoiling-dependent regulation (see Table 4). In addition, the rpoS transcript has a short half-life that is dramatically stabilized by high osmolarity (Muffler et al. 1996). Second, cluster 0 is enriched for stationary phase genes. rpoS was initially characterized as a critical regulator of the stationary phase response; rpoS mutants demonstrate both reduced transcription of a number of stationary phase-activated genes as well as decreased viability (Lange and Hengge-Aronis 1991). Cross-referencing stationary and log phase data by Selinger et al. (2000), we have found that, of 50 genes significantly increased in stationary phase relative to log phase, 29 are found in cluster 0, corresponding to a P value of enrichment of 1.2 × 10−4. This result indicates overlap between stationary phase and osmotic stress responses, very likely through rpoS. Third, a number of genes in cluster 0 have been shown to be directly controlled by rpoS. Of 13 rpoS-regulated genes described by Hengge-Aronis (1996) that are changed greater than twofold in the salt condition, 11 are also located in this cluster (P value = 1 × 10−4). Interestingly, the lipoprotein nlpD, found in cluster 0, has been shown to be part of an operon with rpoS (Lange and Hengge-Aronis 1994). More recently, glutaredoxin 2 (grxB), also in this cluster, has been shown to be rpoS-dependent (Potamitou et al. 2002). We therefore consider it likely that the other genes in this cluster are also regulated by a similar mechanism. Fourth, transcription by ςS (rpoS) is enhanced by both high salt conditions (Ohnuma et al. 2000) and by more positively supercoiled templates in in vitro studies (Kusano et al. 1996). This indicates that rpoS may be sensitive to supercoiling topology in vivo. These findings are consistent with the hypothesis that the effects of supercoiling are mediated through rpoS.
We have also calculated enrichment for predicted transcription factor binding sites within upstream noncoding regions of genes in the osmotic stress response (Table 5). Predicted binding sites were generated computationally from a library of DNA-binding site matrices built from known transcription factor binding sites (Robinson et al. 1998; Roth et al. 1998). Eleven DNA-binding motifs were enriched at P values below 0.05, with three motifs corresponding to global regulators known to interact with supercoiling. These include ςS (rpoS), Fis (fis), and cyclic AMP receptor protein (crp). We were unable to resolve motifs further to individual clusters by this method. Many of the global transcription factors linked to supercoiling recognize structural features in addition to and sometimes preferentially over explicit sequence motifs. Structural recognition presents a challenge to computational binding site prediction by sequence data alone. In these cases, genome-wide in vivo protein–DNA cross-linking experiments (genome-wide ChIP) will be expected to be particularly informative (Lieb et al. 2001; Simon et al. 2001).
Table 5.
Upstream Regions of Genes in the Osmotic Stress Response Are Enriched for Regulatory Binding Sites
| DNA-binding protein | p value | # of putative motifs found in OSR | # expected in OSR |
| crp | 9.13E-05 | 86 | 62.4 |
| soxS | 1.09E-04 | 94 | 70.3 |
| fls | 2.03E-04 | 120 | 97.5 |
| glpR | 2.85E-04 | 125 | 103.4 |
| ompR | 4.00E-04 | 93 | 71.4 |
| rpoS | 1.11E-03 | 124 | 104.8 |
| lrp | 8.88E-03 | 139 | 125.3 |
| cytR | 1.28E-02 | 34 | 23.5 |
| tyrR | 1.97E-02 | 85 | 71.6 |
| malT | 4.15E-02 | 8 | 4.0 |
| cpxR | 5.00E-02 | 22 | 15.4 |
Regulatory binding sites predicted by 55 DNA-binding site matrices found on our website (www.arep.med.harvard.edu) were tallied among genes in the putative osmotic stress response. Statistical enrichment was gauged as described previously (osmotic stress response [OSR]).
Expression Profiles of Temperature Stress Genes Indicate Supercoiling-Dependent Regulation
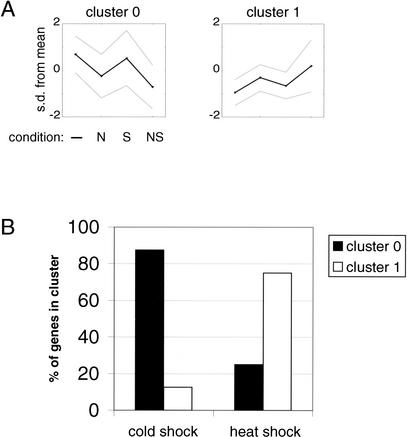
We observed that the heat shock gene, clpB, was among the highest fold up-regulated with novobiocin treatment (2.7-fold), whereas the cold shock gene, cspA, was among the highest fold down-regulated (0.57-fold). It has been reported that transient increases in positive supercoiling follow heat shock; conversely, increased negative supercoiling is observed following cold shock (Ogata et al. 1996; Tse-Dinh et al. 1997; Phadtare et al. 1999; Yura and Nakahigashi 1999). To examine this further, we clustered known temperature stress genes into two categories (see Fig. 4). Strikingly, cold shock genes predominate in cluster 0, whereas heat shock genes predominate in cluster 1. Among heat shock genes found in cluster 0, rpoE, rseA, and htrA are all members of the ςE regulon (Pallen and Wren 1997) involved in envelope protein stress, and mseB is a suppressor of htrB, found in cluster 1. Thus, at least four of six heat shock genes found in our putative cold shock cluster (cluster 0) would not be expected a priori to cluster with up-regulated heat shock genes. Similarly, the single cold shock gene found in cluster 1 is gyrA. gyrA synthesis is stimulated following cold stress. A short lag phase, however, preludes this increased transcription, and, in fact, gyrA synthesis actually decreases during this lag phase (Jones et al. 1992). These results offer the possibility that supercoiling-driven regulation is a general property of bacterial stress responses.
Figure 4.
Heat shock and cold shock genes segregate into supercoiling-responsive profiles. A total of 32 temperature stress genes were selected from the 2146 genes above noise as defined by Blattner notation (Blattner et al. 1997), GenProtEC (Karp et al. 2002), and EcoGene (Rudd 2000). Genes were partitioned into two clusters using the conditions denoted in Fig. 2. (A) Expression profiles for novobiocin (N) and salt (S) conditions are plotted. (B) The distribution of cold shock and heat shock genes are shown for each cluster.
DISCUSSION
Here we demonstrate that increased negative supercoiling is necessary for induction of a functionally significant set of genes activated in the osmotic stress response. Through a series of genome-wide expression profiling experiments under conditions of high salt and perturbation with drug antibiotics, we have identified several major categories of cellular function involved in the osmotic stress response (Fig. 2, Table 3) including adaptation to stress and known osmoregulation (cluster 0), peptidoglycan biosynthesis (cluster 1), macromolecular biosynthesis (cluster 2), amino acid biosynthesis (cluster 4), and the PTS Mannose-Fructose-Sorbose family (cluster 5). Our choice of experimental conditions permitted identification of supercoiling-dependent gene transcription. These supercoiling-dependent transcription profiles are enriched in cluster 0 (Fig. 3B). Significantly, cluster 0 constitutes 35% of the OSR (61/175), and is enriched for genes involved in stress protection such as osmoprotectant synthesis. We propose that increased negative supercoiling is critical for recovery because it mediates the activation of an important subcomponent of the osmotic stress response program.
Computational analysis of predicted DNA regulatory binding domains found upstream of genes in the osmotic stress response also indicates involvement of a number of global regulators known to interact with supercoiling, including fis, crp, and rpoS (Table 5). Comparison of known downstream targets with their distribution among our cluster supports regulation of cluster 0 by rpoS transcriptional activation. We consider it likely that supercoiling exerts its effects through these transcription factors. More complex modeling of the genetic network will be expedited by algorithms that incorporate the combinatorial interactions of these transcription factors, as well as binding site data derived from genome-wide ChIP analysis, and in vitro binding specificity assays (Bulyk et al. 2001; Pilpel et al. 2001; Simon et al. 2001).
It is an interesting question as to what causes negative supercoiling to increase following osmotic shock. ATP/ADP ratios are observed on treatment with salt (Hsieh et al. 1991). Because gyrase activity is coupled to the phosphorylation potential of the cell (van Workum et al. 1996), increased ATP concentration following salt stress is hypothesized to stimulate gyrase activity. In support of this mechanism, an increased ATP/ADP ratio is also observed following nutritional upshift, with a similar increase in negative supercoiling (Balke and Gralla 1987; Travers et al. 2001). Because long-term survival is contingent on adaptation to a new environment, transcriptional activity of the genes found in cluster 0 may be hardwired for optimal expression at high levels of negative supercoiling. We acknowledge that our osmotic shock experiments are performed in a rough E. coli K12 strain. Future experiments should include a comparison of our microarray data with that derived from an E. coli strain with full-length lipopolysaccharide (LPS) or alternatively, Salmonella typhimurium. Interestingly, the his operon in S. typhimurium has been shown to be induced by novobiocin and repressed by high osmolarity. An analogous role for supercoiling may therefore exist in this bacterium as well.
Our data provide a number of bioinformatics insights. These include a genome-wide characterization of the osmotic shock response and tentative assignments for 60 uncharacterized genes in this transcriptional program. The 30 genes in cluster 0 with supercoiling-dependent transcription (Table 4) should expand the number of gene targets available for future study of this regulatory mechanism. Gmuender et al. (2001) have previously examined the genome-wide effects of novobiocin and ciprofloxacin (an antibiotic in the same class as pefloxacin) in Haemophilus influenzae. The microarray data described here allow comparative study of drug inhibition in both H. influenzae and E. coli (Table 6), and should facilitate the discovery of potential drug targets. For example, gmhA mutants show increased outer membrane permeability from aberrant lipopolysaccharide synthesis, leading to increased sensitivity to hydrophobic compounds such as novobiocin (Brooke and Valvano 1996). As shown in Table 6, however, both novobiocin and quinolone classes of antibiotics repress gmhA synthesis. An interesting possibility is that the efficacy of these drugs can be attributed in part to their ability to increase membrane permeability. Finally, we present here a set of computational tools to gauge statistical enrichment for cellular function drawn from GenProtEC as well for transcription factor binding sites. These technologies should help accelerate the functional annotation of the genome. Both programs and datasets are available on our Web site, http://arep.med.harvard.edu/supercoiling/supplement.htm.
Table 6.
Common Effects of Novobiocin and Quinolone Antibiotic Treatment for Both Escherichia coli and Haemophilus influenzae
| In Escherichia coli MG1655 | In Haemophilus influenzaeKW20 | %ident | Drug effect | ||
|---|---|---|---|---|---|
| gene | description | gene | description | ||
| b2255 | Putative transformylase | hi0623 | Methionyl-tRNA formyltransferase | 28 | ↑ |
| fmt | 10-Formyltetrahydrofolate:L-methionyl-tRNA(fMet) N-formyltransferase | hi0623 | Methionyl-tRNA formyltransferase | 65 | ↑ |
| b2392 | Putative transport system permease | hi1728 | Conserved hypothetical protein | 25 | ↓ |
| gyrA | DNA gyrase, subunit A, type II topoisomerase | hi1264 | DNA gyrase, subunit A | 71 | ↑ |
| gyrB | DNA gyrase subunit B, type II topoisomerase, ATPase activity | hi0567 | DNA gyrase, subunit B | 75 | ↑ |
| nusB | Transcription termination; L factor | hi1304 | N utilization substance protein B | 55 | ↑ |
| topA | DNA topoisomerase type I, omega protein | hi1365 | DNA topoisomerase I | 72 | ↓ |
| ygjU | Putative transport protein | hi1545 | Sodium dicarboxylate symporter protein | 62 | ↓ |
| dctA | Uptake of C4-dicarboxylic acids | hi1154 | Proton glutamate symporter protein, putative | 25 | ↓ |
| gmhA | Phosphoheptose isomerase | hi1181 | Phosphoheptose isomerase | 74 | ↓ |
| yraO | Orf, hypothetical protein | hi1181 | Phosphoheptose isomerase | 45 | ↓ |
H. influenzae genes that were changed greater than two-fold in both novobiocin and ciprofloxacin treatments (Gmuender et al. 2001) were compared against E. coli genes with similar changes in both novobiocin and pefloxacin. Sequence homology scores (%Ident) were obtained from the Comprehensive Microbial Resource at TIGR (Peterson et al. 2001). P values for all gene pairs were better than 10−8.
We propose that transiently induced changes in supercoiling may be relevant in environmental challenges beyond osmotic stress. Clustering of temperature stress genes yields expression profiles consistent with reported changes in supercoiling under heat shock and cold shock (Fig. 4). We have also found that oxyR transcription increases with positive supercoiling. Hydrogen peroxide stress induces a transient increase in positive supercoiling; oxyR mutants show delayed resupercoiling and peroxide response (Weinstein-Fischer et al. 2000). An intriguing question is whether bacteria will fare worse if challenged with multiple stresses that require conflicting supercoiling responses. Identification of pharmacological effects on supercoiling may therefore aid in the design of drug combinations with synergistic potency.
METHODS
Array Design
Using an array of oligos capable of specifically hybridizing to their target sequence (RNA or DNA), an entire mRNA population can be probed in parallel (Lockhart et al. 1996). The oligonucleotide array used here is a 544 × 544 grid divided into 24 × 24-μm regions (Affymetrix). Each region contains ∼107 copies of a 25-mer oligonucleotide probe. Photolithography and combinatorial chemical methods are used to synthesize the oligonucleotides directly on a derivatized glass plate. Probe oligonucleotides are grouped into pairs consisting of a perfect match (PM) probe and a mismatch (MM) probe. The PM probe is complementary to the target sequence, whereas the MM probe contains a single base pair mismatch. The MM oligo serves as a control used in identifying cross-hybridization. Probe pairs are, in turn, grouped into probe sets that correspond to mRNA transcripts.
Experimental Design
Ten conditions were assayed in our experiments (see Table 1) using E. coli MG1655 (provided by Fred Blattner, University of Wisconsin, Madison, WI). For novobiocin and salt treatments, E. coli was grown to optical density (OD) 0.4 in M9 minimal media supplemented with 0.4% glucose at 37°C, and aliquoted into two separate flasks. In the first flask, prewarmed novobiocin/dH2O was added for a final concentration of 300 μg/mL of novobiocin. In the second flask, prewarmed NaCl/dH2O was added for a final osmolarity of 0.8. All samples were spun down, flash-frozen in dry ice-ethanol, and stored at −70°C in accordance to the procedures outlined by DeRisi et al. (1997). For pefloxacin and chloramphenicol treatments, E. coli MG1655 was grown in rich media to OD 0.6 at 37°C. Pefloxacin was added to a final concentration of 1 μg/mL. The chloramphenicol concentration was 0.32 μg/uL. Samples were taken directly to phenol. All conditions are averages of two biological replicates (i.e., replicates of separate independent experiments) except for pefloxacin 1-min, pefloxacin 7-min, and chloramphenicol 10-min time points, which are generated from single experiments only.
Sample Preparation/Labeling/Measurement
RNA was isolated by phenol-chloroform extraction and prepared following the Affymetrix protocol for E. coli arrays. This protocol includes an mRNA enrichment step using RNase H enzyme to reduce cross-hybridization from ribosomal RNA. Chips were read using an HP-Affymetrix scanner, and quantified using Affymetrix GeneChip 3.2 software. The expression of each probe set was quantified based on the intensities of its PM and MM probes by a composite statistic, the “Average Difference” intensity. The Average Difference (AvgDiff) is calculated as the mean of all PM–MM pairs after removal of outliers. The AvgDiff is a more representative measure of target sequence concentration than PM intensities alone, because it accounts for cross-hybridization. The resultant data were then background subtracted and total intensity was normalized to 5000 using Affymetrix GeneChip version 3.2 software.
Normalization Specifications
We eliminated noisy gene expression data by a significance test developed previously (Selinger et al. 2000). We assumed that (1) intensities observed among 80 negative control probe sets on the chip are representative of cross-hybridization and other forms of noise only, and (2) noise is approximately normally distributed. For each gene, we calculated its mean expression across all conditions (MEC). We then estimated a mean MEC and standard deviation based on 80 negative control probes on the array. For each gene on the chip, we performed the following procedure. If the MEC was 2.33 standard deviations above the mean MEC of our estimated noise distribution, we rejected the null hypothesis that the observed gene expression was noise with a P value ≤0.01. Genes that were below the 2.33 standard deviation cutoff were called “absent” by Affymetrix GeneChip software in greater than eight conditions, or that contained negative values in any condition were eliminated from further analysis.
Functional Enrichment Analysis
Functional categories from the genProtEC database (Riley et al. 1997; Riley and Serres 2000) were downloaded from http://genprotec.mbl.edu. The actual scoring algorithm is described as follows. Given a file of clustered genes, the perl script catscore.pl (CATegory Score) counts over all functional categories and clusters the number of genes in a given cluster that are members of a particular functional category. Tabulated data are subsequently analyzed using the statistical test developed by Tavazoie et al (1999):
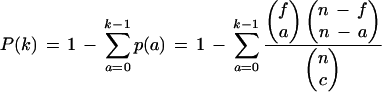 |
where P(k) = the cumulative probability of observing at least k genes in a functional category within a given cluster, f = the number of genes within the functional category, n = the total number of genes, a = the number of genes that match the functional category within the cluster, and c = the number of genes inside the cluster.
We note that this test is mathematically equivalent to a one-tailed Fisher's exact test for 2 × 2 contingency tables (Sokal and Rohlf 1995). With multiple clusters, we use the Bonferroni correction for reducing experimental error rate. The procedure is simply:
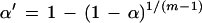 |
where α is the desired error rate, α‘ is the necessary cutoff to achieve this error rate, and m is the number of clusters. For example, if α = 0.05, and there are six clusters, α‘ = 1 − (1 − 0.05)1/5 = 0.01.
Computational Analysis of Motif Enrichment
Motifs were generated with the AlignACE program (Robinson et al. 1998; Roth et al. 1998), using known footprinting sites from the DPInteract database. The highest scoring motif for each protein was then scanned against the E. coli genome using the program ScanACE. High-scoring sequences were defined to be two standard deviations below the mean of the input binding sites or greater, and constituting greater than 50% noncoding sequence. The enrichment for high-scoring sequences was then assayed using catscore.pl.
WEB SITE REFERENCES
http://arep.med.harvard.edu/supercoiling/supplement.htm; complete datasets and software as described in manuscript.
http://genprotec.mbl.edu; functional category assignments for theE. coli MG1655 genome.
www.arep.med.harvard.edu; resources and supplementary material for publications from the Church laboratory.
Acknowledgments
We thank F. Ausubel and S. Lory for helpful comments in a preliminary draft of this work, as well as members of the Church lab for their generous comments and support. This research was supported by grants from the DOE, ONR, and NSF to G.M.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL church@arep.med.harvard.edu; FAX (617) 432-7266.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.401003. Article published online before print in January 2003.
REFERENCES
- 1.Ali N., Herron, P.R., Evans, M.C., and Dyson, P.J. 2002. Osmotic regulation of the Streptomyces lividans thiostrepton-inducible promoter, ptipA. Microbiology 148: 381-390. [DOI] [PubMed] [Google Scholar]
- 2.Alice A.F. and Sanchez-Rivas, C. 1997. DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr. Microbiol. 35: 309-315. [DOI] [PubMed] [Google Scholar]
- 3.Balke V.L. and Gralla, J.D. 1987. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J. Bacteriol. 169: 4499-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F., Plunkett, G., III, Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Boos W. and Shuman, H. 1998. Maltose/maltodextrin system of Escherichia coli: Transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62: 204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooke J.S. and Valvano, M.A. 1996. Molecular cloning of the Haemophilus influenzae gmhA (lpcA) gene encoding a phosphoheptose isomerase required for lipooligosaccharide biosynthesis. J. Bacteriol. 178: 3339-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulyk M., Choo, Y., Huang, X., and Church, G.M. 2001. Exploring DNA binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. 98: 7158-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champoux J.J. 2001. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70: 369-413. [DOI] [PubMed] [Google Scholar]
- 9.Conter A., Menchon, C., and Gutierrez, C. 1997. Role of DNA supercoiling and rpoS ς factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J. Mol. Biol. 273: 75-83. [DOI] [PubMed] [Google Scholar]
- 10.DeRisi J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-686. [DOI] [PubMed] [Google Scholar]
- 11.Dorman C.J. 1996. Flexible response: DNA supercoiling, transcription and bacterial adaptation to environmental stress. Trends Microbiol. 4: 214-216. [DOI] [PubMed] [Google Scholar]
- 12.Drlica K. and Zhao, X. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61: 377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkel S.E. and Johnson, RC. 1992. The Fis protein: It’s not just for DNA inversion anymore. Mol. Microbiol. 6: 3257-3265. [DOI] [PubMed] [Google Scholar]
- 14.Gellert M., O'Dea, M.H., Itoh, T., and Tomizawa, J. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. 73: 4474-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gmuender H., Kuratli, K., Di Padova, K., Gray, C.P., Keck, W., and Evers, S. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: Combined transcription and translation analysis. Genome Res. 11: 28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodell E.W. and Higgins, C.F. 1987. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J. Bacteriol. 169: 3861-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graeme-Cook K.A., May, G., Bremers, E., and Higgens, C.F. 1989. Osmotic regulation of porin expression: A role for DNA supercoiling. Mol. Microbiol. 3: 1287-1294. [DOI] [PubMed] [Google Scholar]
- 18.Hengge-Aronis R., et al. 1996. Regulation of gene expression during entry into stationary phase. In Escherichia coli and Salmonella: cellular and molecular biology (ed. F.C. Neidhardt), pp. 1497–1512. American Society for Microbiology, Washington D.C.
- 19.___, 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2: 148-152. [DOI] [PubMed] [Google Scholar]
- 20.Higgins C.F., Dorman, C.J., Stirling, L., Waddell, L., Booth, I.R., May, G., and Bremer, E. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52: 569-584. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh L., Rouviere-Yaniv, J., and Drlica, K. 1991. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: Changes associated with salt shock. J. Bacteriol. 173: 3914-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingraham J.L., Marr, A.G., et al. 1996. Effect of Temperature, Pressure, pH, and Osmotic Stress on Growth. In Escherichia coli and Salmonella: cellular and molecular biology. (ed. F.C. Neidhardt), pp. 1570–1577. American Society for Microbiology, Washington D.C.
- 23.Jones P., Krah, R., Tafuri, S.R., and Wolffe, A.P. 1992. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 174: 5798-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordi B. and Higgins, C.F. 2000. The downstream regulatory element of the proU operon of Salmonella typhimurium inhibits open complex formation by RNA polymerase at a distance. J. Biol. Chem. 275: 12123-12128. [DOI] [PubMed] [Google Scholar]
- 25.Jordi B.J., Owen-Hughes, T.A., Hulton, C.S., and Higgins, C.F. 1995. DNA twist, flexibility and transcription of the osmoregulated proU promoter of Salmonella typhimurium. EMBO J. 14: 5690-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karp P.D., Riley, M., Saier, M., Paulsen, I.T., Collado-Vides, J., Paley, S.M., Pellegrini-Toole, A., Bonavides, C., and Gama-Castro, S. 2002. The EcoCyc database. Nucleic Acids Res. 30: 56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kempf B. and Bremer, E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolarity environments. Arch. Microbiol. 170: 319-330. [DOI] [PubMed] [Google Scholar]
- 28.Kusano S., Ding, Q., Fujita, N., and Ishihama, A. 1996. Promoter selectivity of Escherichia coli RNA polymerase E ς 70 and E ς 38 holoenzymes. Effect of DNA supercoiling. J. Biol. Chem. 271: 1998-2004. [DOI] [PubMed] [Google Scholar]
- 29.Lange R. and Hengge-Aronis, R. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5: 49-59. [DOI] [PubMed] [Google Scholar]
- 30.___, 1994. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13: 733-743. [DOI] [PubMed] [Google Scholar]
- 31.Lieb J.D., Liu, X., Botstein, D., and Brown, P.O. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28: 327-334. [DOI] [PubMed] [Google Scholar]
- 32.Lockhart D.J., Dong, H., Byrne, M.C., Follettie, M.T., Gallo, M.V., Chee, M.S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., et al. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14: 1675-1680. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Garcia P. and Forterre, P. 2000. DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. Bioessays 22: 736-746. [DOI] [PubMed] [Google Scholar]
- 34.Luttinger A. 1995. The twisted ‘life’ of DNA in the cell: Bacterial topoisomerases. Mol. Microbiol. 15: 601-606. [DOI] [PubMed] [Google Scholar]
- 35.Maxwell A. 1997. DNA gyrase as a drug target. Trends Microbiol. 5: 102-109. [DOI] [PubMed] [Google Scholar]
- 36.Menzel R. and Gellert, M. 1987. Modulation of transcription by DNA supercoiling: A deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc. Natl. Acad. Sci. 84: 4185-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muffler A., Fischer, D., and Hengge-Aronis, R. 1996. Posttranscriptional osmotic regulation of the s subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 178: 1607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobelmann B. and Lengeler, J.W. 1996. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J. Bacteriol. 178: 6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogata Y., Mizushima, T., Kataoka, K., Kita, K., Miki, T., and Sekimizu, K. 1996. DnaK heat shock protein of Escherichia coli maintains the negative supercoiling of DNA against thermal stress. J. Biol. Chem. 271: 29407-29414. [DOI] [PubMed] [Google Scholar]
- 40.Ohnuma M., Fujita, N., Ishihama, A., Tanaka, K., and Takahashi, H. 2000. A carboxy-terminal 16-amino-acid region of ς(38) of Escherichia coli is important for transcription under high-salt conditions and ς activities in vivo. J. Bacteriol. 182: 4628-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pallen M. and Wren, B.W. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26: 209-221. [DOI] [PubMed] [Google Scholar]
- 42.Peterson J.D., Umayam, L.A., Dickinson, T.M., Hickey, E.K., and White, O. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29: 123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phadtare S., Alsina, J., and Inouye, M. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2: 175-180. [DOI] [PubMed] [Google Scholar]
- 44.Pilpel Y., Sudarsanam, P., and Church, G.M. 2001. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 29: 153-159. [DOI] [PubMed] [Google Scholar]
- 45.Potamitou A., Neubauer, P., Holmgren, A., and Vlamis-Gardikas, A. 2002. Expression of Escherichia coli glutaredoxin 2 is mainly regulated by ppGpp and ςS. J. Biol. Chem. 277: 17775-17780. [DOI] [PubMed] [Google Scholar]
- 46.Record T.M., Courtenay, E.S., Cayley, D.S., and Guttman, H.J. 1998. Responses of E. coli to osmotic stress: Large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 23: 143-148. [DOI] [PubMed] [Google Scholar]
- 47.Richmond C., Glasner, J.D., Mau, R., Jin, H., and Blattner, F.R. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27: 3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riley M. and Labedan, B. 1997. Protein evolution viewed through Escherichia coli protein sequences: Introducing the notion of a structural segment of homology, the module. J. Mol. Biol. 268: 857-868. [DOI] [PubMed] [Google Scholar]
- 49.Riley M. and Serres, M.H. 2000. Interim report on genomics of Escherichia coli. Annu. Rev. Microbiol. 54: 341-411. [DOI] [PubMed] [Google Scholar]
- 50.Robinson K., McGuire, A.M., and Church, G.M. 1998. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 284: 241-254. [DOI] [PubMed] [Google Scholar]
- 51.Rohde J.R., Fox, J.M., and Minnich, S.A. 1994. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol. Microbiol. 12: 187-199. [DOI] [PubMed] [Google Scholar]
- 52.Roth F.P., Hughes, J.D., Estep, P.W., and Church, G.M. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantification. Nat. Biotechnol. 16: 939-945. [DOI] [PubMed] [Google Scholar]
- 53.Rudd K. 2000. EcoGene: A genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28: 60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider R., Travers, A., Kutateladze, T., and Muskhelishvili, G. 1999. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 34: 953-964. [DOI] [PubMed] [Google Scholar]
- 55.Selinger D.W., Cheung, K.J., Mei, R., Johansson, E.M., Richmond, C.S., Blattner, F.R., Lockhart, D.J., and Church, G.M. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18: 1262-1268. [DOI] [PubMed] [Google Scholar]
- 56.Simon I., Barnett, J., Hannett, N., Harbison, C.T., Rinaldi, N.J., Volkert, T.L., Wyrick, J.J., Zeitlinger, J., Gifford, D.K., Jaakkola, T.S., et al. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697-708. [DOI] [PubMed] [Google Scholar]
- 57.Sokal R.R. and Rohlf, F.J., 1995. Biometry: The principles and practice of statistics in biological research. W.H. Freeman, New York.
- 58.Tavazoie S., Hughes, J.D., Campbell, M.J., Cho, R.J., and Church, G.M. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22: 281-285. [DOI] [PubMed] [Google Scholar]
- 59.Travers A., Schneider, R., and Muskhelishvili, G. 2001. DNA supercoiling and transcription in Escherichia coli: The FIS connection. Biochimie 83: 213-217. [DOI] [PubMed] [Google Scholar]
- 60.Tse-Dinh Y., Qi, H., and Menzel, R. 1997. DNA supercoiling and bacterial adaptation: Thermotolerance and thermoresistance. Trends Microbiol. 5: 323-326. [DOI] [PubMed] [Google Scholar]
- 61.Van Heijenoort J., et al. 1996. Murein biosynthesis. In Escherichia coli and Salmonella: cellular and molecular biology (ed. F.C. Neidhardt), pp. 1025–1034. American Society for Microbiology, Washington, D.C.
- 62.van Workum M., van Dooren, S.J., Oldenburg, N., Molenaar, D., Jensen, P.R., Snoep, J.L., and Westerhoff, H.V. 1996. DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol. Microbiol. 20: 351-360. [DOI] [PubMed] [Google Scholar]
- 63.Wang J.C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65: 635-692. [DOI] [PubMed] [Google Scholar]
- 64.Weinstein-Fischer D., Elgrably-Weiss, M., and Altuvia, S. 2000. Escherichia coli response to hydrogen peroxide: A role for DNA supercoiling, topoisomerase I and Fis. Mol. Microbiol. 35: 1413-1420. [DOI] [PubMed] [Google Scholar]
- 65.Wood J.M. 1999. Osmosensing by bacteria: Signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63: 230-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yura T. and Nakahigashi, K. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2: 153-158. [DOI] [PubMed] [Google Scholar]