Abstract
Peroxisomes are vital eukaryotic organelles that house a variety of metabolic functions. To fully define the proteome of plant peroxisomes, we recently performed a proteomic analysis of peroxisomes from etiolated Arabidopsis seedlings, verified the peroxisomal localization of candidate proteins by in vivo targeting analysis of fluorescent proteins, and subjected the T-DNA mutants of the newly confirmed proteins to a series of phenotypic analysis. Our reverse genetics approach revealed the role of a cysteine protease - Response to Drought 21A-like Cysteine Protease1 (RDL1) - in seed germination, indole-3-butyric acid (IBA) β-oxidation and stress response. Here, we developed a quick assay aimed at identifying peroxisomal proteins involved in the metabolism of 12-oxo-phytodienoic acid (OPDA), which is converted to jasmonic acid in the peroxisome through β-oxidation. We performed a survey of the same mutants analyzed in our previous reverse genetics study with this new assay by measuring the response of mutants to OPDA’s inhibitory effect on root elongation. Mutants of RDL1 and SERINE CARBOXYPEPTIDASE-LIKE20 (SCPL20) exhibited statistically significant hypersensitivity to OPDA, indicating the potential involvement of these proteins in OPDA metabolism. This convenient assay may be used in the future to rapidly screen for mutants defective in OPDA metabolism or signaling.
Keywords: 12-oxo-phytodienoic acid (OPDA), jasmonic acid biosynthesis, peroxisomes, reverse genetics, β-oxidation
Peroxisomes are single-membrane organelles vital for the development of eukaryotic organisms.1-4 Plant peroxisomes are involved in various metabolic functions including photorespiration, fatty acid β-oxidation, the glyoxylate cycle, detoxification of hydrogen peroxide, biosynthesis of jasmonic acid (JA), metabolism of indole-3-butyric acid (IBA) to indole-3-acetic acid (IAA), and metabolism of branched amino acids, urate and polyamines.1-3 To fully define the proteome of this versatile organelle in plants, we performed proteomic analyses of Arabidopsis peroxisomes from green leaves and etiolated seedlings, followed by in vivo targeting verification of candidate proteins.5-7 After proteomic analysis of peroxisomes from etiolated seedlings, we also subjected the T-DNA mutants of the newly identified peroxisomal proteins to a series of phenotypic assays, including sugar-dependent seedling establishment, IBA response, seed germination, and growth under low CO2, which led us to discover the role of Response to Drought 21A-like Cysteine Protease1 (RDL1) in seed germination, indole-3-butyric acid (IBA) β-oxidation, and stress response.6
To develop alternative assays to screen for mutants with peroxisomal defects, we focused our attention on JA biosynthesis, a pathway with the last steps taken place inside the peroxisome. The JA precursor, 12-oxo-phytodienoic acid (OPDA), which is produced in chloroplasts, is transported into the peroxisome, where it is converted to 3-oxo-2-(2'-[Z]-pentenyl)cyclopentane-1-octanoic acid (OPC-8:0) before entering 3 rounds of β-oxidation to produce JA.8 Given that methyl-JA (MeJA) inhibits primary root elongation,9 we reasoned that OPDA, as a biologically active precursor of JA,10 may have similar effect on root. If so, we could use this root assay to quickly screen for mutants deficient in OPDA metabolism or signal transduction.
To test the feasibility of this approach, we first conducted a pilot experiment on wild-type plants to determine the optimal concentrations of OPDA and MeJA to be applied. As previously reported,9 treatment with 10 µM and 20 µM of MeJA resulted in a decrease of ~60% to 70% of the primary root length in comparison with the untreated plants (Fig. 1A). At 250 nM and 500 nM, OPDA inhibited primary root elongation in wild type at similar degrees, i.e., ~25% (Fig. 1A). Therefore, 250 nM OPDA and 10 µM MeJA were subsequently used for the mutant screen.
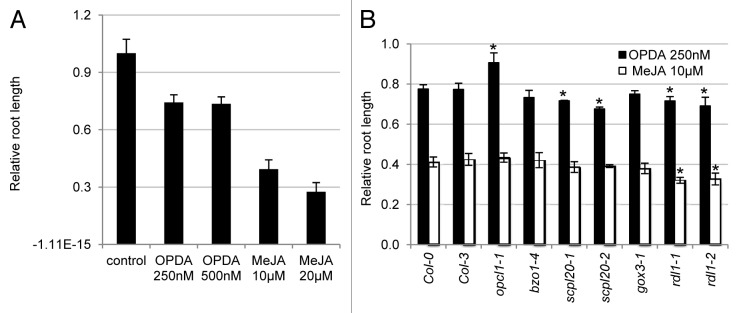
Figure 1. OPDA response assay. (A) Testing the effect of OPDA and MeJA on root elongation in wild-type plants. Y-axis indicates relative root length (ratio between root length under treatment and root length with no treatment) of 7-d Col-0 seedlings grown on ½ LS medium supplemented with 0.5% sucrose and OPDA or MeJA. Data represent means ± se of 3 independent experiments. For each experiment, n ≥ 30. (B) Root response to OPDA and MeJA in peroxisomal mutants. Y-axis indicates relative root length (ratio between root length under treatment and root length with no treatment) of 7-d seedlings grown on ½ LS medium supplemented with 0.5% sucrose and 250 nM OPDA or 10 µM MeJA. Data represent means ± se of 3 independent experiments. For each experiment, n ≥ 30. Asterisks indicate statistically significant changes from the wild type (Col-0 or Col-3). bzo1–4, scpl20–2, gox3–1, and rdl1 mutants are in Col-0 background, and scpl20–1 mutant is in Col-3 background. Student t-test, P < 0.001.
Mutants analyzed in our previous reverse genetics study6 were tested with this new assay. The mutants included one allele each for Benzoyloxyglucosinolate1 (BZO1) and Glycolate Oxidase3 (GOX3), and 2 alleles each for RDL1 and Serine carboxypeptidase-like20 (SCPL20). A previously characterized mutant of OPC-8:0 CoA Ligase1 (OPCL1), which acts in the peroxisomal activation of OPC-8:0,11 served as the positive control. On OPDA, the relative primary root length of the opcl1–1 mutant was ~15% longer than that of the wild-type plants, but on MeJA its relative root length was similar to that of the wild-type (Fig. 1B). These results suggested that opcl1–1 was partially resistant to OPDA, but not to MeJA, which does not need the peroxisome for its biological action. We thus concluded that this OPDA assay was feasible in identifying mutants impaired in OPDA metabolism.
Among the mutants of the newly identified peroxisomal proteins, bzo1–4 and gox3–1 showed similar levels of response to OPDA and MeJA compared with the wild-type (Fig. 1B). In contrast, scpl20 and rdl1 mutants exhibited hypersensitivity to OPDA (Fig. 1B, the Student t-test, P < 0.001). The rdl1 mutants were also hypersensitive to MeJA, which is consistent with our previous finding that RDL1 is involved in multiple aspects of growth and development.6 OPDA interacts with abscisic acid (ABA) to repress seed germination.10,12 Our data, together with our previous finding that rdl1 mutants are hypersensitive to ABA,6 support the view that RDL1 plays a negative role in both ABA and OPDA responses. Given that RDL1 and SCPL20 are both highly expressed during germination and post-germination,6 it is possible that these proteases repress OPDA metabolism in seedlings. However, RDL1 promotes IBA metabolism,6 raising an interesting possibility that this protease plays opposite roles in the metabolism of IBA and OPDA, a hypothesis that will need to be tested more rigorously. Nonetheless, this simple root assay may be used in future screens to identify additional proteins involved in OPDA metabolism or signaling.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants to J.H. from the National Science Foundation (MCB 0618335 and MCB 1330441) to J.H. We also thank Gregg Howe for providing the opcl1–1 mutant.
References
- 1.Beevers H. Microbodies in higher plants. Annu Rev Plant Physiol. 1979;30:159–93. doi: 10.1146/annurev.pp.30.060179.001111. [DOI] [Google Scholar]
- 2.Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK. Plant peroxisomes: biogenesis and function. Plant Cell. 2012;24:2279–303. doi: 10.1105/tpc.112.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaur N, Reumann S, Hu J. Peroxisome biogenesis and function. Arabidopsis Book. 2009;7:e0123. doi: 10.1199/tab.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Bosch H, Schutgens RB, Wanders RJ, Tager JM. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–97. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 5.Quan S, Switzenberg R, Reumann S, Hu J. In vivo subcellular targeting analysis validates a novel peroxisome targeting signal type 2 and the peroxisomal localization of two proteins with putative functions in defense in Arabidopsis. Plant Signal Behav. 2010;5:151–3. doi: 10.4161/psb.5.2.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quan S, Yang P, Cassin-Ross G, Kaur N, Switzenberg R, Aung K, Li J, Hu J. Proteome analysis of peroxisomes from etiolated Arabidopsis seedlings identifies a peroxisomal protease involved in β-oxidation and development. Plant Physiol. 2013;163:1518–38. doi: 10.1104/pp.113.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, Linka N, Switzenberg R, Wilkerson CG, Weber AP, et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 2009;150:125–43. doi: 10.1104/pp.109.137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acosta IF, Farmer EE. Jasmonates. Arabidopsis Book. 2010;8:e0129. doi: 10.1199/tab.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci U S A. 1992;89:6837–40. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dave A, Graham IA. Oxylipin signalling: a distinct role for the jasmonic acid precursor 12-Oxo-Phytodienoic Acid (OPDA) Front Plant Sci. 2012;3:42. doi: 10.3389/fpls.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo AJ, Chung HS, Kobayashi Y, Howe GA. Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem. 2006;281:33511–20. doi: 10.1074/jbc.M607854200. [DOI] [PubMed] [Google Scholar]
- 12.Dave A, Hernández ML, He Z, Andriotis VME, Vaistij FE, Larson TR, Graham IA. 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell. 2011;23:583–99. doi: 10.1105/tpc.110.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]


