Abstract
The activation of transcription factors in response to environmental conditions is fundamental to cellular regulation. Recent work has revealed that some transcription factors are activated in stochastic pulses of nuclear localization, rather than at a constant level, even in a constant environment. In such cases, signals control the mean activity of the transcription factor by modulating the frequency, duration, or amplitude of these pulses. Although specific pulsatile transcription factors have been identified in diverse cell types, it has remained unclear how prevalent pulsing is within the cell, how variable pulsing behaviors are between genes, and whether pulsing is specific to transcriptional regulators or employed more broadly. To address these issues, we performed a proteome-wide movie-based screen to systematically identify localization-based pulsing behaviors in Saccharomyces cerevisiae. The screen examined all genes in a previously developed fluorescent protein fusion library of 4159 strains in multiple media conditions. This approach revealed stochastic pulsing in 10 proteins, all transcription factors. In each case, pulse dynamics were heterogeneous and unsynchronized among cells in clonal populations. Pulsing is the only dynamic localization behavior we observed, and it tends to occur in pairs of paralagous and redundant proteins. Taken together, these results suggest that pulsatile dynamics play a pervasive role in yeast and may be similarly prevalent in other eukaryotic species.
Results
A 4-phase screen enables the identification of proteins that exhibit dynamic localization pulsing
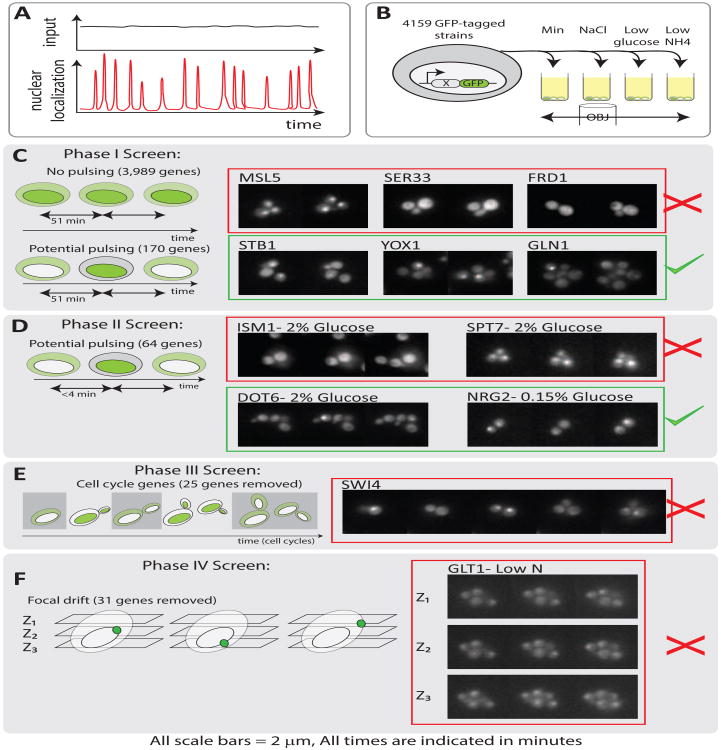
We designed a sequential screening strategy to identify genes from the GFP protein fusion library [1] that showed pulses of localization under constant media conditions (Fig. 1). The screen was conducted in four phases: First, we performed an initial low time resolution movie-based screen to identify candidate genes that showed heterogeneous localization patterns across a population. Second, we performed a higher time resolution movie-based screen to confirm or reject candidate proteins from the first phase. Third, we performed an additional screening step to discriminate pulsing from cell-cycle correlated localization. Fourth, we used a final set of more detailed 3-D (z-stack) movies of the remaining candidates to exclude proteins in which apparent pulsing was only an artifact of fluctuations in the z-position of the localized protein.
Figure 1. A 4-phase screen identifies 9 pulsing proteins, all of which are transcriptional regulators.
(A) Pulsing is defined as the coherent translocation of many molecules of a protein in and out of an organelle in response to a constant input. (B) Time-lapse movies of the GFP library were acquired at low time resolution (∼51 minutes) under four environmental conditions. (C) Examination of these low time-resolution movies revealed 170 potential pulsing proteins. (D) Repeats of movies of these 170 proteins at higher time resolution showed 64 pulsing proteins. (E) 25 of these 64 pulsing proteins were cell-cycle related while (F) 31 were due to focal drift, leaving 9 pulsing proteins remaining. 7 pulsing proteins are sequence-specific transcriptional regulators while 2 are histone deacetylase complex members. Scale bars are 2 μm. Times are indicated in minutes.
Phase I of the screen produced filmstrips for 4159 strains, under four different media conditions, with a time resolution of about ∼51 minutes between frames (Fig. 1B, Figure S1). Visual inspection revealed that most proteins exhibit relatively homogeneous localization patterns, with cells showing similar types of localization across time (7-12 hours) and condition (3989 strains, Fig. 1C). In contrast, 170 strains showed apparently heterogeneous localization patterns (See STB1, YOX1, and GLN1 in Fig. 1B, and Table S1).
In phase II, we re-screened candidate proteins that were positively identified as heterogeneous in phase I. We imaged them at a higher time resolution of ≤4 minutes between frames, choosing a single media condition for each protein. Because we did not observe condition-dependence of heterogeneity in Phase I, we screened for pulsatility in a single condition in subsequent phases. Of the 170 candidates, 64 appeared to behave in a pulsatile fashion in Phase II, as judged by manual inspection of all movies (e.g. DOT6 and NRG2 in Fig. 1D; Table S1). For non-pulsatile proteins, localization patterns were observed to be stable over the ∼4 hour duration of the movie (Fig. 1D, ISM1 and SPT7).
In phase III, our goal was to exclude proteins whose pulsatile dynamics were driven by the cell cycle, which is known to regulate the nuclear localization of many proteins such as Msa1 and Whi5 [2]. We used two strategies to exclude cell cycle driven pulsing: First, we synchronized cell cycles using a transient hydroxyurea DNA replication block prior to the start of movie acquisition [3] and imaged them for ∼6 hours at a time resolution of ≤4 minutes between frames. Second, we acquired 12-14 hour movies, also at a time resolution of ≤4 minutes, that included multiple cell divisions, and visually inspected the correlation of nuclear localization pulses with cell cycle phase, as measured by the time between successive cell division events. Together, these results were used to eliminate 25 proteins from our visual analysis (Fig 1E, Table S1).
39 proteins remained after phase III, all of which showed pulsing apparently uncorrelated with cell cycle (Table S1). Many of these proteins, such as Glc3 and Gln1, were localized to smaller organelles rather than to the nucleus [1]. We reasoned that proteins localized to small organelles could appear to pulse due to small drifts in z-position relative to the focal plane. To eliminate such artifacts, we performed a fourth phase of screening to specifically test for this issue with non-nuclear localized proteins. We acquired 3-D movies of these proteins across a set of 3 focal planes, spaced 0.5 μm apart, with a time resolution of 3 min for 4-6 hours (Fig 1F). This visual analysis revealed that for all non-nuclear proteins, apparent localization pulses could be attributed to z-position fluctuations. By contrast, the 3D analysis did not exclude nuclear localized proteins such as Crz1, which showed clear pulsatile behavior.
The entire four-part screen identified 9 proteins that showed pulsatile localization dynamics not explained by cell cycle or positional fluctuations. All previously known pulsatile proteins (Msn2, Crz1, and Mig1) were recovered in the screen [4-10], validating its ability to detect pulsing.
Screen of transcription factor nuclear localization dynamics
A striking feature of the proteins identified in the screen is that they were all directly involved in transcriptional regulation, either as sequence-specific gene regulatory proteins (7), or as general transcription factors, i.e. histone deacetylase complex members (2), strongly suggesting that pulsatile spatial regulation of localization is predominantly, or even exclusively, used in the control of transcription (p<10-13, Fisher's exact test, See Supplemental Information).
Because all proteins identified in the original screen were involved in transcriptional regulation, we reasoned that there might be additional pulsatile transcription factors in the library that were not activated or not pulsatile under the media conditions examined. Therefore, we set up a transcription factor screen to explore the dynamic single-cell behavior of each transcription factor in the library under conditions expected to activate it (Fig. 2) [11]. We examined 121 transcription factors out of 143 in the GFP library [1] (Table S2), omitting the 22 transcription factors for which we could not find readily available inducers [11]. An additional 90 transcription factors were not analyzed because they are not included in the yeast GFP library [1, 11, 12].
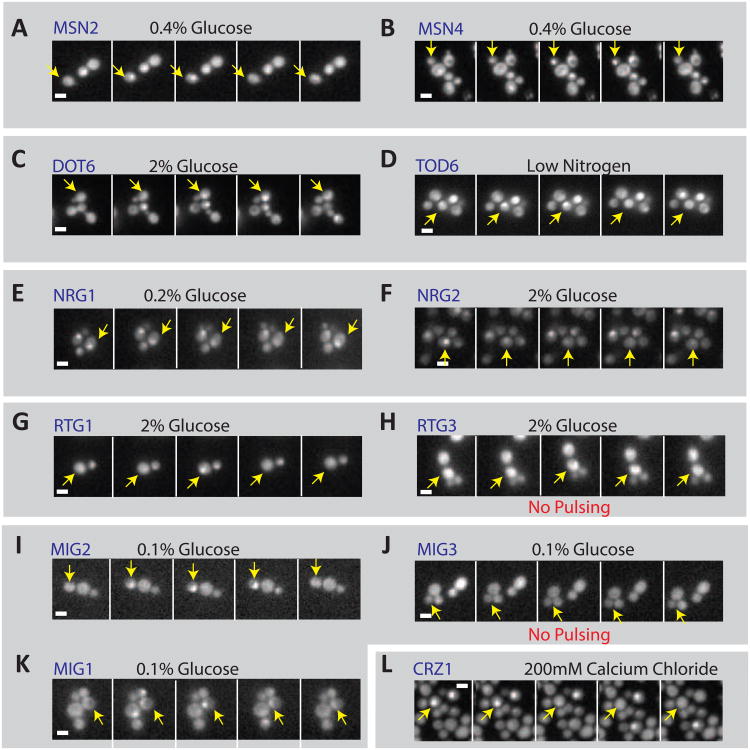
Figure 2. A transcription factor screen confirms 10 pulsing transcription factors.
(A-L) Filmstrips of 12 transcription factors reveal pulses in nuclear localization across varying timescales and conditions. Many, but not all of the proteins that pulse have a duplicate or redundant protein that also pulses. These proteins are grouped accordingly in grey boxes. Filmstrips are labeled with the protein name and condition. Scale bars are 2 μm. Times are indicated in minutes.
For each transcription factor, we selected a corresponding inducer known to modulate its activity [11], and titrated the inducer concentration over a broad and physiologically relevant range (at least 10-fold). At each inducer concentration, we acquired a 4-6 hour time-lapse movie, with intervals between frames ranging from 30 seconds to 4 minutes. We then visually analyzed nuclear localization dynamics in each of these movies (Table S2; Figs. 2-3; Movie S1).
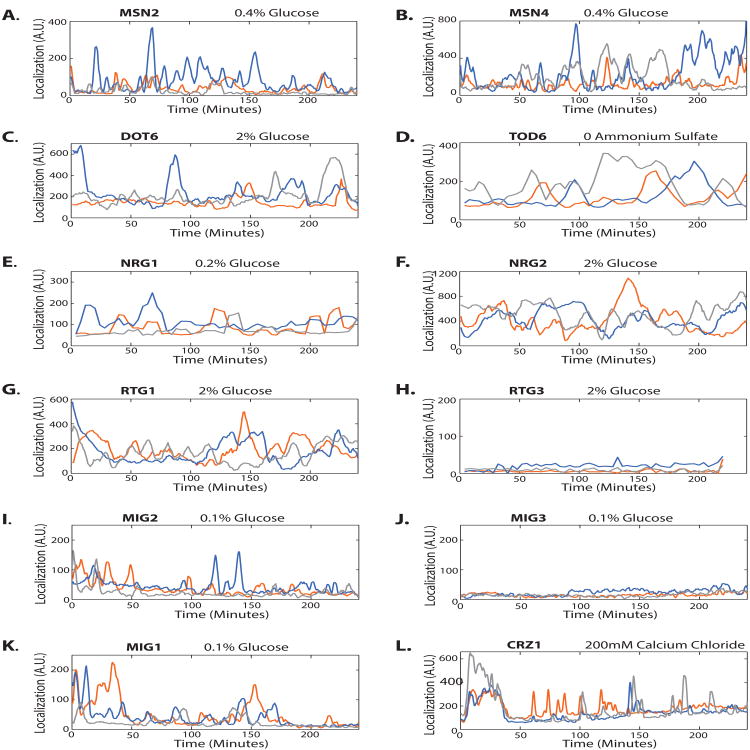
Figure 3. Pulsing is variable.
(A-L) Single-cell traces show that pulses vary from cell to cell (across colors on the same trace), from paralog to paralog (across columns) and from protein to protein. All traces are from the same movie that generated corresponding filmstrips in Figure 2. Scale bars are 2 μm. Times are indicated in minutes. All traces have been smoothed.
Based on these movies, we confirmed the pulsatile behavior of the 9 transcription factors identified in the original 4-phase screen, and further identified the glucose-dependent regulator Mig2 as an additional pulsatile gene regulatory protein (Figures 2I, 3I), bringing the total number of pulsatile proteins to 10 (Table 1). This validated the original proteome-wide screen and suggested its false-negative rate was quite low; only Mig2, or 1/112 additional pulsing transcription factors, were identified. Traces from these movies (Figure 3) revealed a wide range of pulse frequencies and durations suggesting that pulsatility operates on multiple timescales.
Table 1.
| Pulsing Regulator | Conditions In Which Pulsing Observed |
|---|---|
| Crz1 | Calcium |
| Msn2 | All Stresses |
| Msn4 | All Stresses |
| Mig1 | Low Glucose |
| Mig2 | Low Glucose |
| Nrg1 | Low Glucose |
| Nrg2 | Low Glucose |
| Rtg1 | Wild-Type Media Only |
| Dot6 | All Conditions Tested |
| Tod6 | All Conditions Tested |
These data also permit detection of other dynamic behaviors such as exact adaptation to a step change in input. For example, Hog1 nuclear localization increases in response to a step increase in osmotic stress and then returns to its pre-stimulus level [13, 14]. While the movies acquired here confirmed this previously characterized behavior in Hog1, they did not identify additional examples of it.
Finally, we observed that most transcription factors (92/121) are constitutively nuclear and that with the exception of cell-cycle transcription factors, pulsing was the only type of sustained dynamic behavior we observed. It should be noted that in principle, other transcription factors could be regulated in a dynamic fashion through mechanisms (e.g. phosphorylation) that do not influence their nuclear localization. This type of dynamic regulation, lacking observable changes in localization, would not be detectable using this screen.
Regulatory dynamics of paralogs
Yeast has maintained many paralogous protein pairs produced by a whole genome duplication event [15], enabling us to ask whether the pulsatility of one transcription factor is informative about the pulsatility of its paralog.
With the exception of Crz1, all pulsatile transcription factors are members of a pair of either functionally redundant or paralogous transcription factors [11]. Paralogs Msn2 and Msn4 both pulsed in response to glucose deprivation and other stress conditions, as did Dot6 and Tod6 (Fig 3A-D). The Nrg1 and Nrg2 glucose repressor paralogs both pulsed as well, (Fig 3E-F). In contrast, Mig3 did not pulse in the conditions investigated [16], although its paralog Mig2 did (Fig. 3I,J). Thus, the property of pulsatility is conserved in at least 3 out of 4 paralog pairs.
Two pulsatile transcription factors had functionally redundant, but non-paralogous, partners. Among these, Mig1 and Mig2 both pulsed in low glucose (Fig. 3 J,K), while Rtg1 pulsed; its redundant counterpart Rtg3 did not (Fig. 3 G,H).
Although the sample sizes are small, these results suggest that pulsatility is generally correlated between paralogs and redundant partners. To examine whether pulse dynamics across paralogs are also correlated, we constructed a two-color strain in which the localization of both paralogs Msn2 and Msn4 can be examined in individual cells. We took movies of this strain in low glucose and found that Msn2 and Msn4 pulses were generally correlated (Figure S2); both proteins pulse together in most cases though Msn2 sometimes pulses while Msn4 does not (for example, Figure S2H at ∼200 minutes, Figure S2I at ∼350 minutes).
Discussion
Pulsing appears in diverse contexts but has not been examined systematically at a genomic scale. Hence, it had remained unclear how prevalent pulsatile dynamics are in the eukaryotic proteome. Our systematic approach identified 10 proteins that show nuclear localization pulsing, all transcription factors, suggesting that this regulatory mode is used predominantly to regulate transcription. From our results, the fraction of transcription factors that utilize pulsatile nuclear localization is ∼8% (10/121 examined). The yeast GFP library is incomplete [1] but extrapolation from the frequency of pulsatile proteins among those examined to the remaining ∼110 transcription factors not examined here suggests that we might expect an additional ∼9 pulsatile proteins yet to be discovered. Moreover, since most transcription factors appear constitutively nuclear, it remains possible that these proteins are also activated in pulses that do not involve changes in spatial localization, and therefore could not be detected by this screen. Thus, this study provides only a lower limit on the full extent of pulsatile dynamics in the cell.
Since most pulsing proteins are members of a pair of paralogous or functionally redundant transcription factors, one explanation for the evolution of pulsing is one in which pulsing is ancient and existed prior to the whole genome duplication (estimated to be ∼80 million years ago, [15]). Since then, pulsing appears to have been lost only in some proteins (Mig3, Rtg3), while the paralogs that have retained the ability to pulse have changed in their dynamics (Figure 3). Alternatively, paralogs that both pulse could have acquired pulsatile regulation through shared regulatory inputs that later became pulsatile. Further work analyzing whether proteins orthologous to the pulsing transcription factors described here also pulse, specifically in species that diverged prior to the whole genome duplication such as S. castelli or K. lactis, will discriminate between these hypotheses.
Recent work shows that pulsatile regulation occurs in diverse mammalian systems including NF-AT, [17] p53 [18], Erk signaling ([19], TGF-β signaling [20] and NF-κB [21-23]. Moreover, many bacterial systems such as persistence in Mycobacterium smegmatis [24] and bacterial competence [25], sporulation [26] and stress response in Bacillus subtilis [27], employ pulsing. The presence of pulsing in so many systems across a wide range of species is suggestive that pulsing may be a common solution to many biological problems. For example, pulsing has already been shown to proportionally regulate entire regulons of target genes [4, 9] implement transient differentiation [25, 28] enable a multi-cell-cycle timer [26]; and promote bet-hedging [24]. Pulsing may provide a time-based mode of regulation that facilitates these and other functions [29].
Taken together, these observations reveal that pulsatility is surprisingly pervasive in cells. It will now be critical to determine its mechanisms and functions and understand how these dynamics are integrated into the core functions of living cells. Although recent work has provided new insights into Msn2 pulsing [5, 6, 9, 10, 30, 31], and other work has provided a mechanism for pulsatile activation of a sigma factor in bacteria [27], we still lack a full understanding of the mechanisms of pulse generation and modulation for any yeast transcription factor. Do different pulsing systems use a common type of mechanism for pulsing, or are there many distinct mechanisms that can generate similar pulse dynamics? Pulsatility appears to be a core regulatory mechanism in yeast and likely in other cell types as well [17]. The pulsatile proteins identified here should provide a starting point for understanding the roles that this dynamic regulatory mechanism plays in diverse cell types.
Experimental Procedures
Strains and Media
All GFP strains were obtained from the GFP C-terminal protein fusion library, available from Invitrogen [1].
S. cerevisiae were grown in synthetic complete or the appropriate drop-out media made using low-fluorescence yeast nitrogen base, adapted from previous work [4, 32]. This media is yeast nitrogen base without riboflavin or folic acid: 5 g/l (NH4)2 SO4, 1 g/l KH2PO4, 0.5 g/l MgCl2, 0.05 g/l NaCl, 0.5 mg/l H3BO4, 0.04 mg/l CuSO4, 0.1 mg/l KI, 0.2 mg/l FeCl3, 0.4 mg/l MnCl2, 0.2 mg/l Na2MoO4, 0.4 mg/l ZnSO4, 2 μg/l biotin, 0.4 mg/l calcium pantothenate, 2 mg/l inositol, 0.4 mg/l niacin, 0.2 mg/l PABA, 0.4 mg/l pyridoxine HCl, 0.4 mg/l thiamine, 0.1 g/L CaCl2, and 20 g/l dextrose.
Just prior to imaging (∼10-20 minutes before movie acquisition), various permutations were made to the media, ranging from changing the concentration or identity of the sugarsource, changing the concentration or identity of the nitrogen source and/or adding various chemicals or stressors.
Time-Lapse Microscopy
Cells were attached to 96-well or 384-well plates with glass-bottom dishes (Matrical) that were functionalized with 0.1 mg/ml Concanavalin-A (Sigma #C275). Fluorescence images were taken at room temperature on an Olympus IX81 with the ZDC autofocus option and an Andor Ikon (DU-934) camera. Automation was controlled by Andor IQ software. Time resolution of movies varied from 15 seconds to 51 minutes.
Analysis
To generate traces in Figure 3 and Figure S2, fluorescence cell images were segmented using a Hough transformation algorithm in Matlab, provided by Sharad Ramanathan [33]. Localization score was determined by the difference between the mean intensity of the 5 brightest pixels in the cell and mean intensity of the rest of the pixels in the cell [4]. These localization scores were smoothed and plotted using MATLAB.
Target genes of pulsing transcription factors were downloaded from Yeastract [12, 34, 35]. Both direct and indirect target genes were included in tabulating total target genes in Table S3. Interaction partners of pulsing transcription factors were downloaded from the BioGrid [36] and tabulated in Table S3. Gene Ontology analysis of transcription factors was conducted using the GO Slim Mapper Process tool at the Saccharomyces Genome Database; www.yeastgenome.org [11]. These results are tabulated in Table S2.
Supplementary Material
Highlights.
Pulsing is prevalent in the yeast proteome.
Pulsing is specific to transcription factors.
Pulsing regulates a large fraction of the genome.
Acknowledgments
We thank T.F. Chou for initial assistance in obtaining and culturing the yeast GFP library collection. We thank A. Moses, J.G. Ojálvo and P. Swain for comments on the manuscript and acknowledge NIH grants P50 GM068763 and R01 GM079771B for funding.
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 2.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forsburg SL, Nurse P. Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annual review of cell biology. 1991;7:227–256. doi: 10.1146/annurev.cb.07.110191.001303. [DOI] [PubMed] [Google Scholar]
- 4.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao N, Budnik BA, Gunawardena J, O'Shea EK. Tunable signal processing through modular control of transcription factor translocation. Science. 2013;339:460–464. doi: 10.1126/science.1227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao N, O'Shea EK. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nature structural & molecular biology. 2012;19:31–39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquet M, Renault G, Lallet S, De Mey J, Goldbeter A. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. The Journal of cell biology. 2003;161:497–505. doi: 10.1083/jcb.200303030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS biology. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart-Ornstein J, Nelson C, DeRisi J, Weissman JS, El-Samad H. Msn2 coordinates a stoichiometric gene expression program. Current biology: CB. 2013;23:2336–2345. doi: 10.1016/j.cub.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Molecular cell. 2012;45:483–493. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic acids research. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdulrehman D, Monteiro PT, Teixeira MC, Mira NP, Lourenco AB, dos Santos SC, Cabrito TR, Francisco AP, Madeira SC, Aires RS, et al. YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic acids research. 2011;39:D136–140. doi: 10.1093/nar/gkq964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319:482–484. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muzzey D, Gomez-Uribe CA, Mettetal JT, van Oudenaarden A. A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell. 2009;138:160–171. doi: 10.1016/j.cell.2009.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 16.Lewis JA, Gasch AP. Natural variation in the yeast glucose-signaling network reveals a new role for the Mig3p transcription factor. G3 (Bethesda) 2012;2:1607–1612. doi: 10.1534/g3.112.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yissachar N, Sharar Fischler T, Cohen AA, Reich-Zeliger S, Russ D, Shifrut E, Porat Z, Friedman N. Dynamic response diversity of NFAT isoforms in individual living cells. Molecular cell. 2013;49:322–330. doi: 10.1016/j.molcel.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Molecular cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warmflash A, Zhang Q, Sorre B, Vonica A, Siggia ED, Brivanlou AH. Dynamics of TGF-beta signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1947–1956. doi: 10.1073/pnas.1207607109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 23.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 24.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 25.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 26.Levine JH, Fontes ME, Dworkin J, Elowitz MB. Pulsed feedback defers cellular differentiation. PLoS biology. 2012;10:e1001252. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locke JC, Young JW, Fontes M, Hernandez Jimenez MJ, Elowitz MB. Stochastic pulse regulation in bacterial stress response. Science. 2011;334:366–369. doi: 10.1126/science.1208144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 29.Levine JH, Lin Y, Elowitz MB. Functional roles of pulsing in genetic circuits. Science. 2013;342:1193–1200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrenko N, Chereji RV, McClean MN, Morozov AV, Broach JR. Noise and interlocking signaling pathways promote distinct transcription factor dynamics in response to different stresses. Molecular biology of the cell. 2013;24:2045–2057. doi: 10.1091/mbc.E12-12-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roetzer A, Gregori C, Jennings AM, Quintin J, Ferrandon D, Butler G, Kuchler K, Ammerer G, Schuller C. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Molecular microbiology. 2008;69:603–620. doi: 10.1111/j.1365-2958.2008.06301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 33.Nachman I, Regev A, Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–556. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 34.Monteiro PT, Mendes ND, Teixeira MC, d'Orey S, Tenreiro S, Mira NP, Pais H, Francisco AP, Carvalho AM, Lourenco AB, et al. YEASTRACT-DISCOVERER: new tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic acids research. 2008;36:D132–136. doi: 10.1093/nar/gkm976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, Sa-Correia I. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic acids research. 2006;34:D446–451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic acids research. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.