Abstract
Subgenic-resolution oligonucleotide microarrays were used to study global RNA degradation in wild-type Escherichia coli MG1655. RNA chemical half-lives were measured for 1036 open reading frames (ORFs) and for 329 known and predicted operons. The half-life of total mRNA was 6.8 min under the conditions tested. We also observed significant relationships between gene functional assignments and transcript stability. Unexpectedly, transcription of a single operon (tdcABCDEFG) was relatively rifampicin-insensitive and showed significant increases 2.5 min after rifampicin addition. This supports a novel mechanism of transcription for the tdc operon, whose promoter lacks any recognizable ς binding sites. Probe by probe analysis of all known and predicted operons showed that the 5′ ends of operons degrade, on average, more quickly than the rest of the transcript, with stability increasing in a 3′ direction, supporting and further generalizing the current model of a net 5′ to 3′ directionality of degradation. Hierarchical clustering analysis of operon degradation patterns revealed that this pattern predominates but is not exclusive. We found a weak but highly significant correlation between the degradation of adjacent operon regions, suggesting that stability is determined by a combination of local and operon-wide stability determinants. The 16 ORF dcw gene cluster, which has a complex promoter structure and a partially characterized degradation pattern, was studied at high resolution, allowing a detailed and integrated description of its abundance and degradation. We discuss the application of subgenic resolution DNA microarray analysis to study global mechanisms of RNA transcription and processing.
Gene regulation is a dynamic process which can be controlled by a number of mechanisms as genetic information flows from nucleic acids to proteins. The study of gene regulation in the steady state, while informative, overlooks the underlying dynamics of the processes. Steady-state transcript levels are a result of both RNA synthesis and degradation, and as such, measurements of degradation rates can be used to determine their rates of synthesis (if their steady-state levels are known) as well as reveal regulation which occurs via changes in RNA stability.
For the genetic regulatory network of Escherichia coli to be understood and eventually modeled, all means of regulation in use by the cell must be given due attention. RNA degradation in eubacteria was once viewed as a nonspecific, unregulated process. Today it is known to involve multiple degradation pathways, a multisubunit protein complex (the degradosome), and to be an important regulatory mechanism for the expression of some genes (for reviews, see Grunberg-Manago 1999; Rauhut and Klug 1999; Regnier and Arraiano 2000). A small number of large-scale RNA degradation analyses have recently been reported in budding yeast (Wang et al. 2002), humans (Lam et al. 2001), and E. coli (Bernstein et al. 2002).
RNA expression analysis with DNA microarrays has allowed transcription to be studied at an unprecedented scale. Nevertheless, the potential of the technology to elucidate the low-level details of the transcription and processing of RNA has been poorly explored. In this study we have taken a first step by identifying global RNA degradation patterns at the operonic, genic, and subgenic levels.
High-density oligonucleotide arrays from Affymetrix were used to study the degradation of RNA over essentially the entire transcriptome of E. coli MG1655 (Selinger et al. 2000). These arrays have subgenic-resolution coverage of the genome (both coding and noncoding regions), allowing us to examine transcription and degradation in a relatively continuous and unbiased manner.
We present RNA half-life measurements for 1036 open reading frames (ORFs) and for 329 known and predicted operons. We present significant over- and underrepresentation of ORF functional categories in the set of most labile RNAs. We identify an unusual rifampicin-insensitive promoter (of the tdc operon) and strengthen the case for its transcription by a novel mechanism. We present evidence for the higher lability of the 5′ ends of operons relative to their 3′ ends, supporting the current model of an overall 5′ to 3′ direction of degradation. Finally, we explore positional patterns of RNA degradation and discuss the current state of the art of high-resolution global transcription analysis.
RESULTS AND DISCUSSION
Half-Life Determination
For the determination of half-lives, all experiments were done in triplicate for each RNA preparation. On average, 23% of the genes were detected at 2.33ς (99% confidence) above negative control probe sets. Half-lives were calculated for 1036 ORFs, of which 479 were calculated exactly and 557 represent upper bounds. Average half-lives were calculated for 329 known and predicted operons (Tables 1,2; see Methods), although these are only a rough approximation, as typically only a subset of the ORFs had measurable half-lives, and there can be considerable differences between the degradation of different operonic regions.
Table 1.
Twenty Most Labile mRNAs
| B# | Name | HL | 0 | 2.5 | 5 | 10 | 20 |
| b4188 | yjfN | 0.8* | 8782 | 744 | 67 | −41 | −270 |
| b3605 | IldD | 0.9* | 7031 | 770 | 1424 | 221 | −109 |
| b3914 | cpxP(2) | 1.0 | 10398 | 1812 | 3530 | 997 | −11 |
| b0990 | cspG | 1.1 | 6302 | 1324 | 935 | 389 | 105 |
| b3913 | cpxP(1) | 1.1 | 10811 | 2352 | 3506 | 790 | 27 |
| b0553 | nmpC | 1.2 | 4704 | 1147 | 742 | 187 | −183 |
| b2398 | yfeC | 1.2 | 5062 | 1218 | 614 | 122 | −107 |
| b3494 | uspB | 1.2* | 4330 | 870 | 366 | −33 | −191 |
| b3556 | cspA | 1.2 | 20403 | 4696 | 3056 | 1556 | 100 |
| b3685 | yidE | 1.2* | 4373 | 651 | 722 | 202 | −150 |
| b0726 | sucA | 1.3 | 4699 | 1236 | 1001 | 701 | −161 |
| b1205 | ychH | 1.3 | 11630 | 2964 | 692 | 67 | −171 |
| b3362 | yhfG | 1.3* | 3959 | 884 | 1014 | 556 | −114 |
| b0162 | cdaR | 1.4* | 3366 | 859 | 473 | 105 | 520 |
| b1060 | yceP | 1.4 | 11780 | 3294 | 5286 | 1886 | 1374 |
| b2080 | yegP | 1.4 | 5355 | 1567 | 2914 | 1769 | 922 |
| b2377 | yfdY | 1.4 | 6525 | 1880 | 1189 | 944 | 53 |
| b3361 | fic | 1.4 | 6270 | 1888 | 2035 | 1267 | 39 |
| b4132 | cadB | 1.4 | 6923 | 2019 | 3046 | 2287 | 238 |
| b4396 | rob | 1.4 | 4685 | 1339 | 734 | −32 | −322 |
The 20 most labile mRNAs with their average difference (AD) intensities at each timepoint. Twelve of 20 have unknown or putative functions. High lability may be an indication of regulation at the level of RNA stability. This is known to be the case for cspA, which is extremely unstable at 37° but transiently stable after a shift to 15° (Goldenberg et al. 1996). The lability of cspG suggests that it may behave similarly. Numbers shaded in gray are below the 99% confidence detection threshold (see Methods). *Half-life represents an upper bound.
Table 2.
Operons with Average Half-Lives ≤2.5 Minutes
| Avg. HL | Operon |
| 1.35 | pabA fic yhfG |
| 1.35 | yfeC yfeD |
| 1.65 | cadA cadBcadC |
| 1.75 | deoC deoA deoB deoD |
| 1.95 | yhcH yhcl nanE nanT |
| 2.05 | ynfB speG |
| 2.1 | thrL thrA thrB thrC |
| 2.2 | sdhC sdhD sdhA sdhB |
| 2.2 | yjbQ yjbR |
| 2.35 | lacA lacY lacZ |
| 2.4 | folX yfcH |
| 2.45 | ybjC mdaA |
| 2.47 | nagD nagC nagA nagB |
A number of these unstable operons enable biosynthesis that is presumably unnecessary in rich media, such as amino acid biosynthesis (thr, cad), alternative carbon sources (lac, sdh), and nucleotide biosynthesis (deo). Underlining indicates half-lives used in the average.
After addition of rifampicin, which prevents initiation of new transcripts by binding to the β subunit of RNA polymerase (Campbell et al. 2001), the total intensity for all mRNAs decreases exponentially with time (R = 0.98) with an estimated overall chemical half-life of 6.8 min. This is in rough agreement with a reported half-life of 7.5 min for total pulse-labeled RNA in comparable conditions (Mohanty and Kushner 1999). Although absolute decay rates are known to vary appreciably across experiments, especially those determined in different laboratories, we observe qualitative agreement with some well studied transcripts, such as ompA, a very stable RNA in fast-growing cells (Nilsson et al. 1984; see Methods), and cspA, an extremely unstable one which is transiently stabilized upon cold shock (Table 1; Goldenberg et al. 1996).
Genes encoding enzymes known to be involved in RNA decay such as pnp, rhlB, and rho show exponential decay patterns starting immediately after rifampicin treatment. The genes rne and rnc also show progressive decay patterns but were expressed at relatively low levels, making half-life measurement difficult. The genes rnb and pcnB were undetected throughout the timecourse.
Average operon half-lives were calculated by taking the mean of the operons' member ORFs for which half-lives had been determined. A number of the most unstable operons (Table 2) enable metabolism that is presumably unnecessary in rich media, such as amino acid biosynthesis (thr, cad), alternative carbon source catabolism (lac, sdh), and nucleotide biosynthesis (deo). It would be interesting to see whether these transcripts are more stable in rich media.
Discovery of a Rifampicin-Insensitive Promoter
Surprisingly, a single operon, tdcABCDEFG, which encodes a pathway for the transport and anaerobic degradation of L-threonine, was relatively rifampicin-insensitive. All seven ORFs of this operon were significantly upregulated at 2.5 min after rifampicin addition. After their initial increase at 2.5 min, the ORFs of the tdc operon show either gradual decay or stability through the 5- and 10-min timepoints, followed by near-complete degradation by the 20-min timepoint (data not shown). Because rifampicin targets the core of the only RNA polymerase (RNAP) in E. coli, we were initially surprised to find an operon which could still be transcribed after rifampicin addition. However, differential sensitivity to rifampicin by RNAP holoenzyme containing different ς subunits (ς70 vs. ς32) was observed previously (Wegrzyn et al. 1998), suggesting that certain holoenzymes may be rifampicin-insensitive. Furthermore, the tdc promoter is unusual in that it doesn't contain any recognizable ς binding sites, but does contain sites for a number of transcription factors, including CRP, IHF, FNR, LysR, TdcA, and TdcR. It has also been suggested that the tdc promoter is controlled by a novel mechanism and can be activated by altering its local topology (Wu and Datta 1995; Sawers 2001).
RNA Decay Related to Function
To determine whether transcripts whose gene products participate in the same cellular processes tended to be degraded at the same rates, we looked at the over- and underrepresentation of 23 gene functional categories (Blattner et al. 1997) within different half-life ranges (Table 3). P-values were calculated using the cumulative hypergeometric distribution, and a 95% confidence level was used as a cutoff (Tavazoie et al. 1999). In the set of short-lived (≤5-min) transcripts, genes annotated as putative enzymes were significantly overrepresented. Rapidly degraded transcripts are good candidates for regulation via RNA stability, and many of these may be transiently stabilized in some environmental condition in which they are needed. The instability of their transcripts, and likely low protein levels, may have been a hindrance to their discovery and/or characterization. Genes involved in translation and posttranslational modification were significantly underrepresented among short-lived (≤5-min) transcripts, reflecting the known stability of the cell's translational machinery. Genes involved in energy metabolism were significantly overrepresented among transcripts with intermediate half-lives of between 10 and 20 min. The genes in this category are, in general, well studied and are regulated by a variety of mechanisms unrelated to RNA stability, although in most cases regulation via transcript stability has not been ruled out.
Table 3.
Functional Category Representation of Half-Life Groups
| Functional category | Experimental group | Rep. | p-value |
| Putative enzymes | HL <= 5 min | over | 6.5 × 10−5 |
| Translation and posttranslational modification | HL <= 5 min | under | 1.8 × 10−5 |
| Energy metabolism | 10 min < HL < 20 min | over | 5.4 × 10−3 |
| Translation, posttranslational modification | ORFs with measured HLs | over | 1.8 × 10−23 |
| Hypothetical, unclassified, unknown | ORFs with measured HLs | under | 1.3 × 10−8 |
| Putative transport proteins | ORFs with measured HLs | under | 2.8 × 10−5 |
Several half-life groupings (HL ≤ 5 min, 5 < HL ≤ 10, 10 < HL ≤ 20, and HL > 20) were tested for over- or underrepresentation of 23 different functional categories (Blattner et al. 1997) relative to all genes whose half-lives were estimated. Categories were also identified which were over- or underrepresented in the set of all ORFs with measured half-lives. P values were calculated using the cumulative hypergeometric distribution (Tavazoie et al. 1999). A 95% confidence level was achieved using a cutoff of 2.2 × 10−3 to account for multiple hypotheses. Transcripts displaying no preference were those encoding proteins involved in transport and binding, structure and membrane proteins, carbon compound metabolism, amino acid and nucleotide biosynthesis and metabolism, and central intermediary metabolism, transcription, and posttranscriptional regulation. Despite the rapid degradation of some well studied genes (such as pnp, rhlB, and rho), as a whole, genes involved in RNA degradation were not significantly enriched in any half-life group.
To assess whether our experiment preferentially measured the half-lives of some groups of genes relative to others, we looked for differential representations of genes with measured half-lives relative to all genes on the array. Genes whose half-lives could be determined in our experiment were significantly overrepresented for those involved in translation and posttranslational modification, which are generally very highly expressed and easy to detect. Those classified as “hypothetical, unclassified, unknown” or as putative transport proteins were significantly underrepresented, suggesting that both of these classes in general are expressed at a very low level and/or may contain a number of spuriously predicted ORFs. These two uncharacterized groups stand in contrast to putative enzymes and putative regulatory proteins, which were detected at a rate indistinguishable from those of other groups.
5′ to 3′ Directionality of Degradation
RNA is degraded within the cell by the combined action of RNA exo- and endonucleases. The precise way in which this process occurs has been a subject of intense study (Grunberg-Manago 1999; Regnier and Arraiano 2000). Stable 5′ secondary structures have been shown to confer stability on downstream sequences (Emory et al. 1992), whereas 3′ polyadenylation targets transcripts for degradation (Sarkar 1997). To investigate whether degradation is targeted preferentially towards the 5′ or 3′ end of the mRNA, we measured the variability of degradation rates at different positions of predicted and known operons containing at least two ORFs. Each operon coding region was divided into three equal regions (5′, middle, and 3′), whereas 30 bases upstream and downstream of the operons were denoted 5′ and 3′ UTRs, respectively. The UTR was chosen to be relatively short to increase the probability that it was in fact cotranscribed with the operon. The average log2 ratio of probes in each region was calculated for each operon (see Methods).
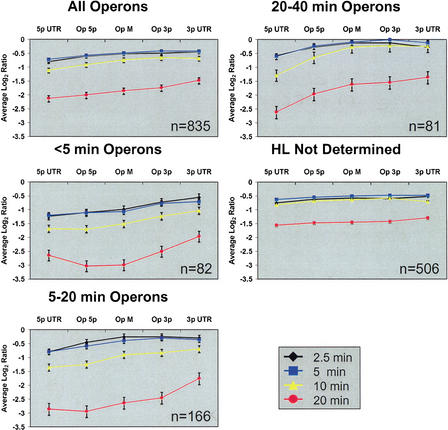
Log2 ratios of each region were averaged for all operons, as well as for subsets with specified half-lives, to compare the degradation rates of different transcript regions (Fig. 1). In the set of all operons, the log2 ratios were most negative for the 5′ UTR and became less negative in a 5′ to 3′ direction, consistent with a predominantly 5′ to 3′ directional mechanism of degradation.
Figure 1.
Positional differences in operon degradation. Operon regions are plotted on the x-axis, and average log2 ratios (compared to the 0 min timepoint) are plotted on the y-axis. Vertical bars indicate standard error. Operons were divided into five regions: 30 bases upstream (5p UTR) and downstream (3p UTR), and three equal-length regions of the coding region: 5 prime (Op 5p), middle (Op M), and 3 prime (Op 3p). Patterns of operons with different average half-lives were compared. A 5′ to 3′ directionality is observable in the coding regions of all operon subsets. This directionality generally extends at least 30 bases into the UTRs, although the 5′ UTR of quickly degrading operons (<5 min) seems to be more stable than the coding region. All curves in this figure have significant variation between means by one-way ANOVA at α = 0.001, with the following exceptions: 2.5 min of the ‘20–40 min’ graph, and the 5- and 20-min curves of the ‘half-life not determined’ graph, which were significant at α = 0.05, 0.05, and 0.10, respectively. P-values for timepoints on the ‘all operons’ graph were all below 1×10−12.
To determine whether positional patterns varied depending on overall stability, operons were grouped based on their average half-lives (Fig. 1). The same trend of 3′-increasing stability was seen for all groups, regardless of overall half-life. This trend was most consistent for the 20–40-min operons, whereas for the <5 min and the 5–20-min operons, there were some discrepancies at their 5′ ends, especially at the later timepoints.
To assess the significance of the differential degradation rates, we used a one-way ANOVA to test whether the differences between average degradation rates of different operon regions could be accounted for by chance. Significant differences between regional mean degradation rates were found for almost all timepoints in all half-life sets using α = 0.001. Three cases were significant only at α = 0.05 or 0.10, as detailed in the Figure 1 legend. The results for the analysis of all 835 operons were especially significant, with all P-values below 1×10−12. We conclude that the observed variation in the rate of degradation of different operonic regions is significant.
Clustering of Degradation Patterns
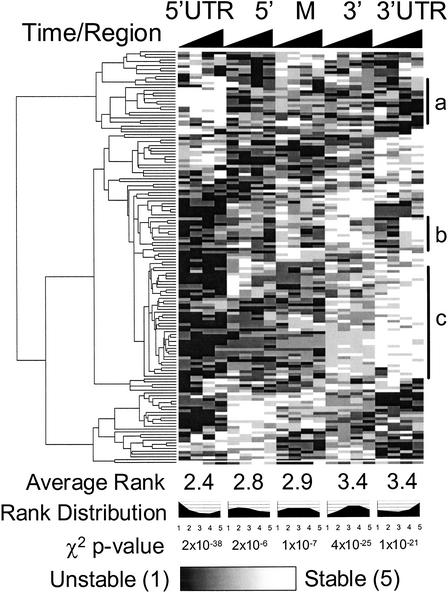
It is important to note that although the 5′ to 3′ directionality illustrated by Figure 1 indicates that in general, the 5′ ends of operons are degraded more quickly than their 3′ ends, it does not indicate whether this is the only pattern of operon degradation, or simply the most common one. To distinguish between these two possibilities, the degradation patterns of all operons were clustered using a hierarchical clustering algorithm and displayed as a tree (Fig. 2; Eisen et al. 1998). Here, 149 known and predicted operons for which complete data was available were divided into five operon regions: 5′ and 3′ UTR (representing 30 bases up- and downstream of the translation start and stop, respectively), and equal-length 5′, middle, and 3′ coding regions. Within each operon, each region was ranked from most stable (5) to least stable (1) based on the average log2 ratio of oligos in that region at each timepoint. This within-operon normalization allows operons with similar patterns to be grouped together regardless of their overall rate of degradation. The results of the clustering analysis indicate that although there is a clear predominance of a 5′ to 3′ degradation pattern, other patterns are also present. Nevertheless, the degradation ranks for each region, when averaged over all operons, show a clear trend consistent with an overall 5′ to 3′ directionality of degradation.
Figure 2.
Whole-genome cluster analysis of operon degradation. The degradation patterns of 149 operons (containing two or more ORFs, and oligo probes in all targeted regions) were hierarchically clustered after ranking the relative degradation rate of each region. The algorithm was implemented using the GeneCluster/TreeView package (Eisen et al. 1998). Transcript regions are on the x-axis, with each region split into 2.5-, 5-, 10-, and 20-min timepoints. The average rank increases from 5′ to 3′, supporting a predominant 5′ to 3′ directionality of degradation (cluster c). The clustering also reveals that a variety of degradation patterns are present, such as operons with relatively stable 5′ UTRs (cluster a). One group of operons (cluster b) is initially degraded most quickly at its 3′ UTR at 2.5 and 5 min, but then by the 10 min timepoint is more quickly degraded at its middle and 3′ coding regions. χ2 goodness of fit tests show that the distributions of degradation ranks are highly nonrandom, with 5′ regions more likely to be degraded quickly and 3′ regions more likely to be degraded slowly. The complete clustering file, including gene names, is available at http://arep.med.harvard.edu/rna_decay/.
To assess the statistical significance of the observed directionality, we performed a χ2 goodness of fit test on each transcript region. We are easily able to reject the null hypothesis that each region has an equiprobable distribution of ranks, with P-values ranging from 2×10−6 to 2×10−38 (Fig. 2). From inspection of the rank distributions we conclude that 5′ regions of operons are significantly more likely to be degraded quickly and 3′ regions more likely to be degraded slowly.
Because certain transcript features, such as the ompA stabilizer (Emory et al. 1992), are known to exert their effects along an entire transcript, we analyzed the extent to which the degradation of one region is correlated to other regions. The average Pearson's linear correlation coefficient (R) between the degradation of adjacent regions was 0.38, and the average correlation between any two operon regions was 0.26. These weak but statistically significant (P < 0.005) correlations suggest that although there are important operon-wide determinants of stability, local determinants may play a larger role in the stability of RNAs. This emphasizes the need to scrutinize transcription and degradation at a higher level of resolution.
It should be noted that despite the difficulties of defining transcript boundaries, as well as the existence of operons with multiple promoters and terminators, we were still able to identify significant patterns. As our knowledge of these confounding factors increases, we may expect to see even clearer patterns emerge.
High-Resolution Analysis of the dcw Gene Cluster
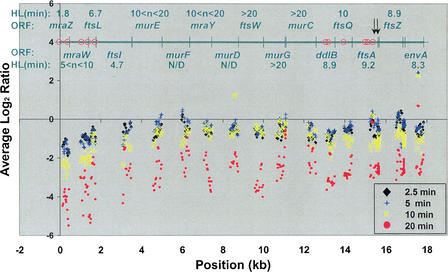
The dcw gene cluster, important for cell envelope biosynthesis and cell division, contains 16 ORFs and has a complex promoter structure (Fig. 3; Vicente et al. 1998; Dewar and Dorazi 2000). It is transcribed mainly from two clusters of promoters located at the 5′ end (∼ORFs 1–3), and near the 3′ end (ORFs 12–14). We observe a complex degradation pattern for this operon, with three primary domains of stability (Figs. 3,4). The 5′ end is degraded most rapidly, consistent with the most commonly observed pattern. The central region is relatively stable from murE to murC. The 3′ end, from ddlB to envA, has an intermediate stability, with ftsA and ftsZ having nearly identical half-lives, as has been reported previously (Cam et al. 1996).
Figure 3.
High-resolution analysis of the dcw gene cluster transcripts. Average log2 ratios (y-axis) were plotted against operon position (x-axis) for three-probe sliding windows of the 156 probes in the dcw gene cluster, including 30 bases up- and downstream of the first and last ORFs. The positions of known promoters (red circles) and the ORFs with their estimated half-lives are given in the upper part of the graph. Arrows indicate known RNase E processing sites. An additional weak promoter is thought to be present in either murD or ftsW (Mengin-Lecreulx et al. 1998). rho-independent terminators are present in the 5′ region of mraZ and immediately downstream of envA. Degradation is fastest at the 5′ end of the operon, with three apparent regions of distinct degradation rates: The 5′ region (mraZ–ftsI) is degraded the fastest, the middle (murE–murC) is relatively stable, and the 3′ region (ddlB–envA) is degraded at an intermediate rate.
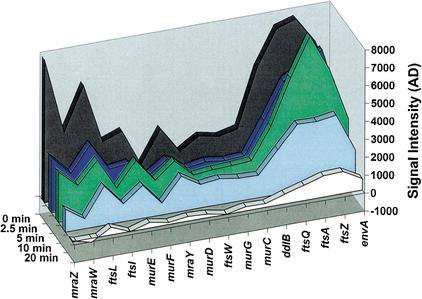
Figure 4.
Transcript abundance and degradation of the dcw gene cluster. The dcw gene cluster contains 16 ORFs involved in cell envelope biosynthesis and cell division. Several promoters have been described (see Fig. 3), and it is likely that they are all used, to varying extents. It has also been speculated that the cluster may sometimes be transcribed in its entirety. The ORFs were plotted here in the order they are transcribed, showing their array signal intensities (average differences) throughout the timecourse. Although average difference is only an approximate indicator of transcript abundance, relatively high levels of steady-state RNA are observed downstream of the mraZ and ddlB promoters, at the 5′ end and about two-thirds of the way into the transcript, respectively. The middle portion of the operon has lower steady-state RNA levels and is degraded more slowly (see Fig. 3).
These domains of stability roughly coincide with the clusters of promoters, suggesting that they represent somewhat independent units which the cell chooses to regulate simultaneously by both transcriptional initiation and degradation. Interestingly, the relatively high signal intensity at mraZ and ddlB corresponds to the positions of the two major promoters Pmra and ftsQ2p1p, respectively (Fig. 4; Flardh et al. 1997; Mengin-Lecreulx et al. 1998). This suggests that the regions downstream of these promoters are maintained at higher steady-state RNA levels in the cell, although we are cautious about making a firm conclusion in this regard due to the only semiquantitative nature of the relationship between microarray signal intensity and absolute RNA abundance. Nevertheless, this observation is consistent with previous measurements which show that about one-third of the transcription of ftsZ originates at promoters located within and between ddlB and ftsA, with the other two-thirds originating upstream of ddlB (Flardh et al. 1998; de la Fuente et al. 2001).
The Future of High-Resolution Transcriptome Analysis
The type of transcriptome data presented here enables genome-wide analyses, which until now have only been done on a small scale. For example, the relationship between RNA degradation and RNA sequence features such as RNase sites and known and predicted secondary structures can be assessed, as well as the effects of mutations, especially to the RNA degradation machinery. These data are also useful in the empirical definition of transcription boundaries (Selinger et al. 2000; Tjaden et al. 2002) and promoter usage.
We expect such high-resolution analyses to increase in precision. Probe-to-probe variation, which can mask local changes in RNA abundance, can be improved by smoothing or, perhaps, by more sophisticated model-based (Li and Hung Wong 2001) or correlation-based methods (Cohen et al. 2000). High-resolution mapping of human exon boundaries using oligonucleotide arrays has also been reported (Shoemaker et al. 2001; Kapranov et al. 2002). Microarrays could be designed with probes more evenly spaced throughout the ORFs and the intergenic regions to allow more comprehensive coverage of the transcriptome. The continually increasing density of oligonucleotide arrays suggests that transcriptome data, and our resulting understanding of transcriptional regulation, will increase not only in scope, but also in detail.
METHODS
Growth of Bacterial Strains and Transcript Inhibition
E. coli wild-type strain MG1655 was grown in LB broth medium in shaken flasks at 37°C to mid-logarithmic phase (A600 = 0.8) and then split into five flasks of 20 mL each. To initiate transcription inhibition, four of these samples were treated with rifampicin (Sigma) at a concentration of 50 μg mL−1 and incubated for an additional 2.5, 5, 10, and 20 min respectively, followed by immediate harvesting of the cells. The fifth sample was used as a control, and cells were harvested immediately (at timepoint zero). All RNA isolation procedures were accomplished with the MasterPure Complete DNA and RNA Purification kit from Epicentre Technologies, as described (Rosenow et al. 2001).
RNA Labeling and Hybridization
The cDNA synthesis method was described (Rosenow et al. 2001). Briefly, 10 μg of total RNA was reverse-transcribed using the Superscript II system for first-strand cDNA synthesis from Life Technologies. The remaining RNA was removed using 2 U RNase H (Life Technologies) and 1 μg RNase A (Epicentre) for 10 min at 37°C in 100 μL total volume. The cDNA was purified using the Qiaquick PCR purification kit from QIAGEN. Isolated cDNA was quantitated based on the absorption at 260 nm and fragmented using a partial DNase I digest. The fragmented cDNA was 3′ end-labeled using terminal transferase (Roche Molecular Biochemicals) and biotin-N6-ddATP (DuPont/NEN). The fragmented and end-labeled cDNA was added to the hybridization solution without further purification. Three microarray hybridizations were carried out for each timepoint.
Chip Scaling, Transcript Detection
To account for experimental and chip variations, all intensities were normalized according to the variations of the cRNA controls, which were added before the RNA labeling reaction and contained four probe sets targeting RNAs not present in the E. coli genome. The controls show a variation of less than 10% before scaling for all 15 labeling reactions (data not shown). Transcript abundances for each RNA were calculated in GAPS by taking a mean of the perfect match (PM) minus mismatch (MM) probes, after removing the highest and two lowest (2–13 max.; Selinger et al. 2000) and are referred to here simply as “average difference” (AD; Lockhart et al. 1996). Each RNA is typically targeted by 15 unique oligonucleotide probe pairs. A transcript was considered “detected” if it was 2.33ς (99% confidence) above the negative controls (90 probe sets for genes not present in the MG1655 genome). For the five timepoints (0, 2.5, 5, 10, and 20 min), mRNA detection rates were 24%, 27%, 27%, 18%, and 6%, respectively, with detection cutoffs of 1766, 1014, 975, 1202, and 1327 AD units. The mean of the negative controls has been subtracted from all reported values, so that values greater than 0 signify an average difference greater than the negative controls. For high-resolution analysis (including the directionality analysis), we calculated log2 ratios as log2([PM−MM of time t]/[PM−MM of time 0]). We only used probe pairs in which PM−MM at time 0 was greater than 100 normalized fluorescent units.
RNA Chemical Half-Life Determination
Probe pairs (perfect match − mismatch) were averaged over the triplicates of each timepoint (0, 2.5, 5, 10, and 20 min after rifampicin addition), resulting in an average probe set intensity for each ORF. RNA abundances were determined using the average difference metric implemented by GAPS©. Chemical half-life was determined for each RNA by the following “twofold” algorithm: (1) The earliest timepoint at which the transcript was detected was used as the baseline abundance. (2) The earliest successive timepoint for which a twofold decrease was detected was used as the experimental abundance, and the half-life was calculated assuming exponential decay. When the baseline but not the experimental timepoint was detected, the half-life was estimated (yielding an upper-bound estimate) using the noise value in place of the experimental value. Other categories were defined, such as “stable” (transcript is detected but no change as great as twofold observed), “possible increase” (a minimum twofold change between any two timepoints), “erratic” (both a twofold increase and decrease observed), and “possibly stable” (at least a twofold decrease observed, but later returns to baseline level). Slot blots for four genes were carried out as a validation of the array-measured RNA half-lives, giving the following results (slot blot/array): ompA 20.2 min/stable; cspC 17.2 min/possibly stable; fldA 10 min/6.7 min; sodA 9.5 min/6.9 min. Half-lives were alternatively calculated by fitting an exponential decay curve to all timepoints, regardless of fold change or signal-to-noise thresholds. This approach was deemed inferior to the twofold algorithm, because it gave considerably poorer agreement with slot blot data, showed less sensitivity to rapidly degrading transcripts, and gave spurious results for RNAs whose signal dropped below the detection threshold at later timepoints (data not shown). Average half-lives were calculated for predicted and observed operons from RegulonDB (Salgado et al. 2001) by taking a mean for all operon members whose half-life had been determined. Half-lives with estimated upper bounds of greater than 40 min were set equal to 40 min to avoid skewing the results. The complete list of transcripts, calculated half-lives (of both ORFs and operons), and pattern categories are available at http://arep.med.harvard.edu/rna_decay/. The dataset was also deposited in ExpressDB (Aach et al. 2000) at http://arep.med.harvard.edu/ExpressDB/.
WEB SITE REFERENCES
http://arep.med.harvard.edu/rna_decay/; RNA decay data and half-life.
http://arep.med.harvard.edu/ExpressDB/; Relational database containing yeast and E. coli RNA expression data.
Acknowledgments
We thank Sidney Kushner for advice and the provision of mutants (not used in this study), and Kenn Rudd and Joel Belasco for critical reviews of the manuscript. D.S. was graciously hosted in the lab of Minoru Kanehisa for part of this work. This work was supported by grants from the NSF-MEXT Monbusho program, Lipper Foundation, NSF, and DOE.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL carsten_rosenow@affymetrix.com; FAX (408) 481-0422.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.912603. Article published online before print in January 2003.
REFERENCES
- 1.Aach J., Rindone, W., and Church, G.M. 2000. Systematic management and analysis of yeast gene expression data. Genome Res. 10: 431-445. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein J.A., Khodursky, A.B., Lin, P.H., Lin-Chao, S., and Cohen, S.N. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. 99: 9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner F.R., Plunkett, G., III, Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Cam K., Rome, G., Krisch, H.M., and Bouche, J.P. 1996. RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 24: 3065-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell E.A., Korzheva, N., Mustaev, A., Murakami, K., Nair, S., Goldfarb, A., and Darst, S.A. 2001. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 104: 901-912. [DOI] [PubMed] [Google Scholar]
- 6.Cohen B.A., Mitra, R.D., Hughes, J.D., and Church, G.M. 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 26: 183-186. [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente A., Palacios, P., and Vicente, M. 2001. Transcription of the Escherichia coli dcw cluster: Evidence for distal upstream transcripts being involved in the expression of the downstream ftsZ gene. Biochimie 83: 109-115. [DOI] [PubMed] [Google Scholar]
- 8.Dewar S.J. and Dorazi, R. 2000. Control of division gene expression in Escherichia coli. FEMS Microbiol. Lett. 187: 1-7. [DOI] [PubMed] [Google Scholar]
- 9.Eisen M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emory S.A., Bouvet, P., and Belasco, J.G. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes & Dev. 6: 135-148. [DOI] [PubMed] [Google Scholar]
- 11.Flardh K., Garrido, T., and Vicente, M. 1997. Contribution of individual promoters in the ddlB-ftsZ region to the transcription of the essential cell-division gene ftsZ in Escherichia coli. Mol. Microbiol. 24: 927-936. [DOI] [PubMed] [Google Scholar]
- 12.Flardh K., Palacios, P., and Vicente, M. 1998. Cell division genes ftsQAZ in Escherichia coli require distant cis-acting signals upstream of ddlB for full expression. Mol. Microbiol. 30: 305-315. [DOI] [PubMed] [Google Scholar]
- 13.Goldenberg D., Azar, I., and Oppenheim, A.B. 1996. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol. Microbiol. 19: 241-248. [DOI] [PubMed] [Google Scholar]
- 14.Grunberg-Manago M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33: 193-227. [DOI] [PubMed] [Google Scholar]
- 15.Kapranov P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916-919. [DOI] [PubMed] [Google Scholar]
- 16.Lam L.T., Pickeral, O.K., Peng, A.C., Rosenwald, A., Hurt, E.M., Giltnane, J.M., Averett, L.M., Zhao, H., Davis, R.E., Sathyamoorthy, M., et al. 2001. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2: Research 0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C. and Hung Wong, W. 2001. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2: 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockhart D.J., Dong, H., Byrne, M.C., Follettie, M.T., Gallo, M.V., Chee, M.S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., et al. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14: 1675-1680. [DOI] [PubMed] [Google Scholar]
- 19.Mengin-Lecreulx D., Ayala, J., Bouhss, A., van Heijenoort, J., Parquet, C., and Hara, H. 1998. Contribution of the Pmra promoter to expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes. J. Bacteriol. 180: 4406-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohanty B.K. and Kushner, S.R. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34: 1094-1108. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson G., Belasco, J.G., Cohen, S.N., and von Gabain, A. 1984. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature 312: 75-77. [DOI] [PubMed] [Google Scholar]
- 22.Rauhut R. and Klug, G. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23: 353-370. [DOI] [PubMed] [Google Scholar]
- 23.Regnier P. and Arraiano, C.M. 2000. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays 22: 235-244. [DOI] [PubMed] [Google Scholar]
- 24.Rosenow C., Saxena, R.M., Durst, M., and Gingeras, T.R. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: Comparison of two approaches. Nucleic Acids Res. 29: E112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salgado H., Santos-Zavaleta, A., Gama-Castro, S., Millan-Zarate, D., Diaz-Peredo, E., Sanchez-Solano, F., Perez-Rueda, E., Bonavides-Martinez, C., and Collado-Vides, J. 2001. RegulonDB (version 3.2): Transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29: 72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66: 173-197. [DOI] [PubMed] [Google Scholar]
- 27.Sawers G. 2001. A novel mechanism controls anaerobic and catabolite regulation of the Escherichia coli tdc operon. Mol. Microbiol. 39: 1285-1298. [DOI] [PubMed] [Google Scholar]
- 28.Selinger D.W., Cheung, K.J., Mei, R., Johansson, E.M., Richmond, C.S., Blattner, F.R., Lockhart, D.J., and Church, G.M. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18: 1262-1268. [DOI] [PubMed] [Google Scholar]
- 29.Shoemaker D.D., Schadt, E.E., Armour, C.D., He, Y.D., Garrett-Engele, P., McDonagh, P.D., Loerch, P.M., Leonardson, A., Lum, P.Y., Cavet, G., et al. 2001. Experimental annotation of the human genome using microarray technology. Nature 409: 922-927. [DOI] [PubMed] [Google Scholar]
- 30.Tavazoie S., Hughes, J.D., Campbell, M.J., Cho, R.J., and Church, G.M. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22: 281-285. [DOI] [PubMed] [Google Scholar]
- 31.Tjaden B., Haynor, D.R., Stolyar, S., Rosenow, C., and Kolker, E. 2002. Identifying operons and untranslated regions of transcripts using Escherichia coli RNA expression analysis. Bioinformatics (Suppl.) 18: 337-344. [DOI] [PubMed] [Google Scholar]
- 32.Vicente M., Gomez, M.J., and Ayala, J.A. 1998. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell. Mol. Life Sci. 54: 317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Liu, C.L., Storey, J.D., Tibshirani, R.J., Herschlag, D., and Brown, P.O. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 99: 5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegrzyn A., Szalewska-Palasz, A., Blaszczak, A., Liberek, K., and Wegrzyn, G. 1998. Differential inhibition of transcription from ς70- and ς32-dependent promoters by rifampicin. FEBS Lett. 440: 172-174. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y. and Datta, P. 1995. Influence of DNA topology on expression of the tdc operon in Escherichia coli K-12. Mol. Gen Genet. 247: 764-767. [DOI] [PubMed] [Google Scholar]