Abstract
Structural genetic alterations in cancer often involve gene loss or gene amplification. With the advent of microarray approaches for the analysis of the genome, as exemplified by array–CGH (Comparative Genomic Hybridization), scanning for gene-dosage alterations is limited only by issues of DNA microarray density. However, samples of interest to the pathologist often comprise small clusters of just a few hundred cells, which do not provide sufficient DNA for array–CGH analysis. We sought to develop a simple method that would permit amplification of the whole genome without the use of thermocycling or ligation of DNA adaptors, because such a method would lend itself to the automated processing of a large number of tissue samples. We describe a method that permits the isothermal amplification of genomic DNA with high fidelity and limited sequence representation bias. The method is based on strand displacement reactions that propagate by a hyperbranching mechanism, and generate hundreds, or even thousands, of copies of the genome in a few hours. Using whole genome isothermal amplification, in combination with comparative genomic hybridization on cDNA microarrays, we demonstrate the ability to detect gene losses in yeast and gene dosage imbalances in human breast tumor cell lines. Although sequence representation bias in the amplified DNA presents potential problems for CGH analysis, these problems have been overcome by using amplified DNA in both control and tester samples. Gene-dosage alterations of threefold or more can be observed with high reproducibility with as few as 1000 cells of starting material.
There are many instances in which the amount of genomic DNA available from a biological sample becomes a limiting factor for genomic analysis. An example is the study of gene dosage in tumor DNA by comparative genomic hybridization (CGH), a procedure that requires the use of several hundred nanograms of genomic DNA for fluorescent labeling. A variety of methods have been devised to amplify total genomic DNA, many of them based on thermocycling protocols (Telenius et al. 1992; Zhang et al. 1992; Cheung and Nelson 1996). The yield obtained with such methods comprises a few hundred copies of the genome, and the size of the DNA product ranges from 200 to 3000 bases. More elaborate methods are capable of generating higher amplification yield; however, such methods require the ligation of adaptors for PCR (Lüdecke et al. 1989; Saunders et al. 1989; Klein et al. 1999).
We sought to develop a very simple method that would permit direct amplification of the whole genome without the use of thermocycling or DNA adaptors, because such a method would lend itself to the automated processing of a large number of tissue samples. Strand-displacement reactions are known to permit DNA amplification in very high yields (Walker et al. 1992; Lizardi et al. 1998; Dean et al. 2001). We have adapted the use of the strand-displacing polymerases of phage φ29 and Bacillus stearothermophilus (Bst DNA polymerase large fragment, 5′ → 3′ exo−) for random-primed amplification of human genomic DNA. Here we report the properties of these whole genome amplification reactions, and evaluate their usefulness for genome-wide analysis of allele dosage alterations in small samples using DNA microarrays.
RESULTS
Properties of the Amplification Reaction and the Need for Special Primers
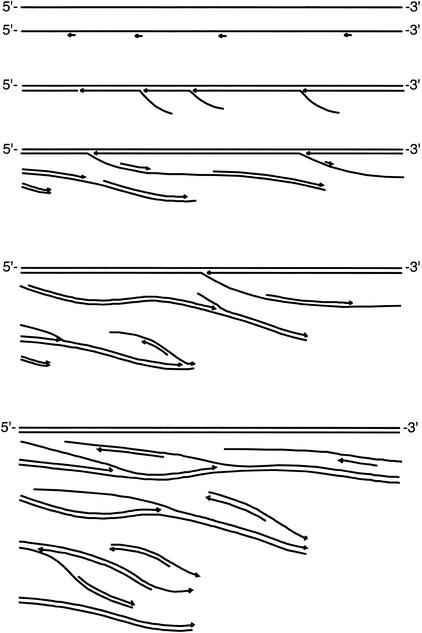
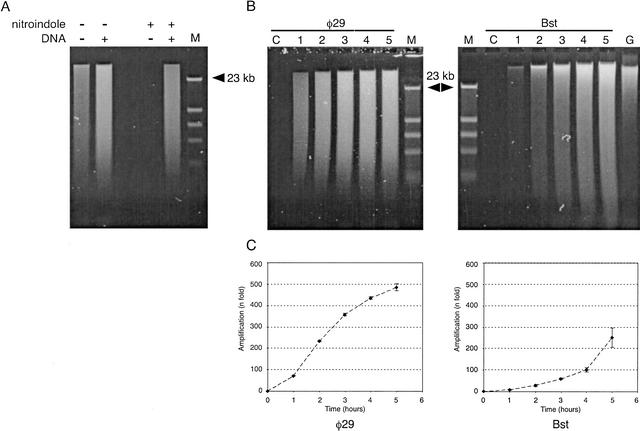
The amplification method is based on random priming of denatured DNA, followed by strand-displacement synthesis at constant temperature. Multiple primers are extended over tens of kilobases, and the resulting DNA products are of high molecular weight. As more DNA is generated by strand displacement, an increasing number of random priming events occur, forming a network of hyperbranched DNA structures (Fig. 1). The reaction is catalyzed by φ29 DNA polymerase, or by the large fragment of Bst DNA polymerase. Both reactions generate large DNA products, and require high primer concentrations for maximum amplification yield. Relatively short random primers (6–8 bases) are more effective than longer primers. The reaction catalyzed by Bst DNA polymerase requires T4 gene 32 protein for efficient strand-displacement synthesis. Bst DNA polymerase large fragment does not have the 3′ → 5′ exonuclease domain (Aliotta et al. 1996), and therefore is expected to have error rates in the range of other modified Pol I polymerases without proofreading activity. The error rate for Klenow 3′ → 5′ exo− DNA polymerase has been reported to be 1 × 10−4 (Bebenek et al. 1990). Because φ29 is an enzyme with 3′ → 5′ proofreading activity (Blanco and Salas 1985), the reaction is more efficient if one uses primers with a single exonuclease-resistant phosphorothioate residue just before the 3′-terminal base (Skerra 1992; Dean et al. 2001). The DNA amplification yield of reactions catalyzed by φ29 DNA polymerase can reach more than a millionfold in an overnight incubation. However, a problem with both enzymes is their capability for primer-directed DNA synthesis in the absence of DNA template (Fig. 2A). Thus, although it is possible to obtain very high amplification yields with standard random primers, reactions initiated with very small inputs of genomic DNA tend to be contaminated with spurious DNA sequences. We explored the use of modified primers with one or two 5′-terminal nitroindole (universal base) residues, which in theory will stabilize primer binding without increasing the sequence complexity of the primer (Loakes and Brown 1994; Loakes et al. 1997). Primers containing nitroindole residues have been used to obtain improved signal intensity in cycle sequencing reactions (Ball et al. 1998). Surprisingly, we observed that background synthesis was completely suppressed when we used primers containing two nitroindole bases at the 5′ end (Fig. 2A). The mechanism responsible for this interference with background synthesis is not understood. Using nitroindole-modified primers, reactions without DNA input may be incubated 5 h or more without detectable background synthesis (control lanes in Fig. 2B).
Figure 1.
Schematic representation of the hyperbranched strand-displacement amplification reaction. Single-stranded genomic DNA serves as the target for multiple random priming events. Growing strands are propagated by a DNA polymerase with strand-displacement activity. The 5′ end of each strand is displaced by another upstream strand, growing in the same direction. Displaced strands, which are single-stranded, are now targeted by new random priming events, and these new strands are elongated in the opposite direction. As the reaction proceeds, the hyperbranched network expands dramatically, generating thousands or even millions of copies of the original DNA.
Figure 2.
Gel electrophoresis analysis of amplified DNA. (A) Control reactions were incubated for 5 h in a 30-μL volume containing no DNA, or 7.5 ng of human DNA. Samples representing 5% of the reaction were denatured in alkaline buffer and analyzed on a 0.5% alkaline agarose gel, and stained with SYBR-green II (Molecular Probes). Lanes labeled M contained phage λ DNA digested with restriction endonuclease HindIII. Reactions catalyzed by φ29 polymerase were incubated with (+) or without (−) input of denatured human DNA. Random heptamers contained standard (−) DNA, or were modified by the addition of two nitroindole groups (+) at the 5′ end. (B) Time-course reactions for φ29 and Bst DNA polymerases. Reactions were performed using nitroindole-modified primers. Every hour (from 1–5 h), 1.5 μL was removed, denatured in alkaline buffer, and analyzed in 0.5% alkaline agarose gel. Lanes labeled C correspond to control samples incubated for 5 h without input DNA. The lane labeled G corresponds to a gel load of genomic DNA equivalent to 100× the original DNA input of the amplification reactions. (C) Plots under gel images display the time course (fold amplification vs. time) of both polymerase reactions, generated by quantification of DNA yield with the PicoGreen Quantitation Kit. Background fluorescence at time 0 was subtracted for all time points. Each point represents the mean (±1 SD) of four independent analyses.
Time-course experiments over 5 h demonstrate a relatively slow increase in the amount of DNA over the first 1–2 h of incubation, and a more rapid, nonlinear increase in DNA from 2–5 h (Fig. 2B,C). DNA fluorescence assays using PicoGreen indicated that the DNA amplification obtained after 5 h of incubation with Bst DNA polymerase was 251 ± 46-fold, and with φ29, 485 ± 17-fold (averages of four independent experiments, Fig. 2C). The highest level of amplification obtained in a 5-h incubation with φ29 DNA polymerase was 1200-fold, but higher yields are obtainable using buffers with pyrophosphatase (Dean et al. 2001). The mitochondrial genome of human cells is circular, and would be expected to be amplified by a highly efficient hyperbranched Rolling Circle Amplification (RCA) mechanism, as described by Lizardi et al. (1998) and Dean et al. (2001). We compared the amplification yield of mitochondrial DNA with chromosomal DNA by quantitative TaqMan PCR, and found no significant differences in sequence representation.
Incubations longer than 5 h generated many thousands of copies of the input genomic DNA. However we have focused our attention on reactions that amplify DNA 1000-fold or less because of concerns about sequence representation bias, which is expected to increase in a time-dependent manner. Preliminary mathematical modeling of a random-primed hyperbranched reaction, assuming a DNA polymerization rate of 15 nucleotides per second and an average interprimer distance of 2000 bases (shorter distances generate a higher yield), predicts that over a 5-h period the polymerase will engage in approximately six or seven hyperbranched copying cycles. Because interprimer distance is likely to vary over the genome in a sequence-dependent manner, one would expect that longer incubation times, and more hyperbranched copying cycles, should lead to larger and larger differences in amplification yield for different gene loci. Another prediction of mathematical modeling is that longer DNA molecules will be amplified more than short DNA molecules. This is the case because reduced DNA template length translates into fewer hyperbranched copying cycles.
Evaluation of Amplification Bias Using Yeast cDNA Microarrays
DNA microarrays provide a powerful tool for assessing DNA sequence representation in genomic DNA amplified by hyperbranched strand displacement. Comparative genomic hybridization (Kallioniemi et al. 1992) can be performed on microarrays, at a level of resolution determined by the number and spacing of DNA sequences spotted in the microarrays (Pinkel et al. 1998; Snijders et al. 2001). Although microarrays of Bacterial Artificial Chromosome (BACs) generate stronger signals, it is possible to perform array–CGH analysis using the more readily available cDNA microarrays constructed by spotting of PCR products (Pollack et al. 1999). An advantage of standard cDNA microarrays is the high resolution among closely linked gene loci, and the ability to measure representational bias on a gene-by-gene basis.
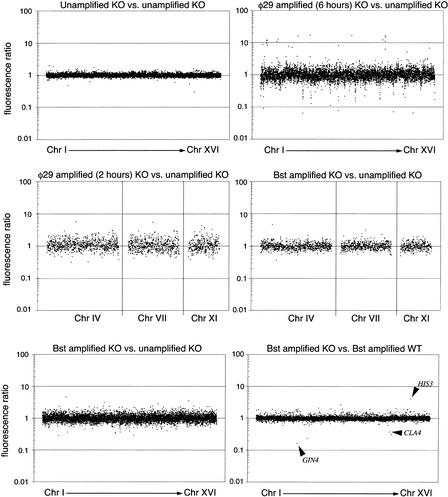
For an initial evaluation of relative sequence representation in the amplified DNA, we performed array–CGH using yeast strains with well-defined deletions, taking advantage of the availability of high-quality commercial microarrays containing all yeast open reading frames. The reference experiment, performed without DNA amplification, involved a self-self comparison of Cy3-labeled DNA from Saccharomyces cerevisiae strain his3-delta200 & leu2-3 (whose genes GIN4 and CLA4 were replaced by HIS3 and LEU2, respectively) and Cy5-labeled DNA of the same strain. After mixing of the labeled DNA preparations, hybridization was performed on microarrays containing 6372 spots corresponding to 6135 known yeast ORFs (Corning, Inc.). As expected, a plot of Cy3/Cy5 ratios shows data points aligned along the line corresponding to ratios close to 1:1 (Fig. 3, panel in upper left). We then compared DNA amplified 1000-fold by φ29 DNA polymerase to unamplified DNA from the same source (Fig. 3, panel in upper right). In this experiment the ratios show considerable scatter, indicating significant over-representation or under-representation of many ORFs. Notably, small groups of adjacent ORFs are under-represented, as indicated by dots aligned vertically at the same chromosome map coordinates. A large number of these under-represented loci map near the ends of yeast chromosomes. These loci are, indeed, expected to be bounded by fewer potential DNA priming sites toward the telomeric side of each locus. Six under-represented genes in Chromosome 5 are within a 6-kb region at the end of the chromosome, whereas two under-represented genes in Chromosome 8 are within 10 kb from the chromosome end. A small number of ORFs are over-represented, presumably because of a higher-than-average frequency of priming events. In contrast, when the same type of experiment was performed using DNA amplified 250-fold by Bst DNA polymerase, we also found evidence of moderate distortion of the ratios, but fewer instances of markedly over-represented or under-represented clusters of loci (Fig. 3, panel in lower left). We then performed an experiment in which the amplification reaction with φ29 DNA polymerase was limited to 2 h, to generate ∼250 copies of the input DNA. Comparison of the CGH profiles obtained with 250-fold amplification by φ29 DNA polymerase and Bst DNA polymerase (Fig. 3, middle panels) shows that at the same level of amplification there is less distortion of the ratios from the expected 1:1 when using the latter enzyme.
Figure 3.
Evaluation of amplification bias using array–CGH on yeast cDNA microarrays. Microarrays contained 6135 unique yeast ORFs. Fluorescence ratios were measured and plotted against the order of the genes in the genome, starting from Chromosome I to Chromosome XVI. (Upper left panel) Analysis of a microarray hybridized with the same DNA, labeled with Cy3 and Cy5. (Upper right panel) DNA from the yeast KO strain was amplified using φ29 DNA polymerase, labeled with Cy3, and hybridized against unamplified (Cy5) DNA from the same strain. (Lower left panel) DNA from the yeast KO strain was amplified using Bst DNA polymerase, labeled with Cy3, and hybridized against unamplified (Cy5) DNA from the same strain. (Center left panel) DNA from the yeast KO strain was amplified using φ29 DNA polymerase for only 2 h, labeled with Cy3, and hybridized against unamplified (Cy5) DNA from the same strain. (Center right panel) Equivalent experiment using Bst DNA polymerase. (Lower right panel) DNAs from the two different yeast strains were amplified to the same extent using Bst and hybridized together. The three genes known to be deleted appear as outlier data points indicated by arrows. The other two outlier data points, near genes GIN4 and CLA4, have abnormally low area values of 48 and 21 according to the Spot analysis software, compared with the average of 255 for all the spots in the array. This abnormality could be produced by a fluorescent speckle over the spot, resulting in unreliable ratios.
Remarkably, in an experiment in which two different yeast genomes were amplified by Bst DNA polymerase, and compared by array–CGH (Fig. 3, panel in lower right), the Cy3/Cy5 ratios were found to be very close to 1:1, indicating that the over-representation and under-representation of ORFs is reproducible, and thus balances out when both genomes are amplified, and then compared by array–CGH. In this experiment we were able to observe precisely the known allelic imbalances corresponding to the GIN4 and CLA4 deletions in the KO strain and the HIS3 deletion in the so called wild-type (WT) strain (Fig. 3, panel in lower right). The ratios at these deleted loci are not infinitely large (or small), because the signals observed at the deleted ORFs are not zero, most likely because of a small amount of cross-hybridization of unrelated sequences. We interpret these observations as indicative of a deterministic mechanism for amplification bias, which may be based on priming frequencies that are different, albeit reproducibly so, across the genome. Analogous observations have been reported in conventional CGH experiments, in which DOP-PCR-amplified DNA was used for both test and reference samples, resulting in reduced distortion of ratios (Voullaire et al. 1999; Huang et al. 2000). Thus, if amplification bias is not excessive, distortions in representation can be compensated for by using a reference genomic DNA amplified under identical conditions.
Quantitative Assessment of Dynamic Range Compression in Human cDNA Microarrays
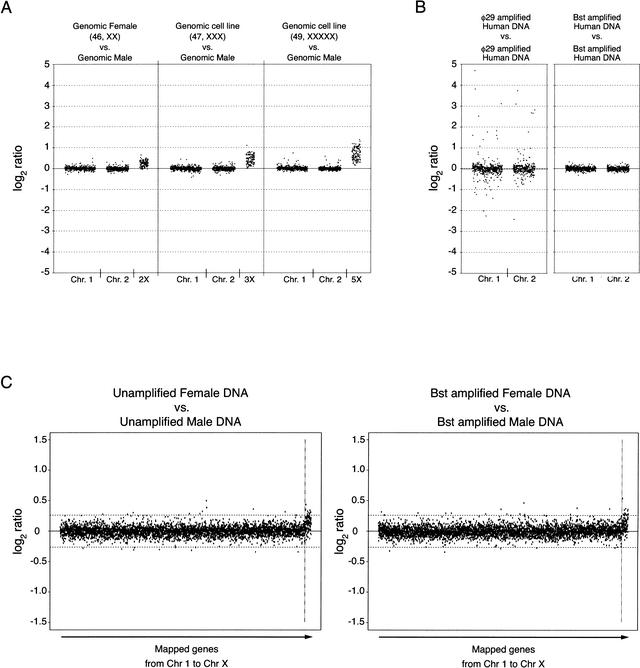
A feature of microarray experiments performed with very complex genomes, such as the human genome, is the presence of dynamic range compression. To determine the extent of dynamic range compression in our cDNA-based microarray–CGH system, we performed control experiments with DNA inputs from cell lines with an abnormal number of X chromosomes (3X and 5X), hybridizing against a reference unamplified genomic DNA from a normal male. We observed ratios for X-chromosome probes that correlate with gene dosage increments for the known karyotypes, with significant ratio distortion due to dynamic range compression (shown in Fig. 4A). A true gene dosage ratio of 3:1 is represented in the microarray data as an observed average ratio of 1.341 (0.423 in log2 scale, average of 113 probes). Likewise, a true dosage ratio of 5:1 is represented in the microarray data as an observed average ratio of 1.603 (0.681 in log2 scale, average of 115 probes). Ratios for autosomal genes, shown for Chromosomes 1 and 2, are themselves subject to dynamic range compression, and their overall distribution is quite narrow. The left panel in Figure 4A shows that twofold changes in gene dosage are not measurable with good reliability. On the other hand, changes of threefold are detectable for the majority of loci (Fig. 4A, middle panel), and changes of fivefold are detectable for practically all loci in the X-chromosome (Fig. 4A, right panel). These observations establish a gene amplification level of threefold as the limit of detection for human gene dosage alterations using our microarray–CGH method.
Figure 4.
Evaluation of DNA amplification bias using array–CGH on human cDNA microarrays. (A) To assess the extent of dynamic range compression, three microarray–CGH experiments were performed in duplicate using genomic female DNA (46, XX) versus genomic male DNA, or DNA from cell lines containing 3 X-chromosomes (47, XXX) and 5 X-chromosomes (49, XXXXX) with a normal number of autosomes against genomic male DNA. Autosomal genes located in Chromosomes 1 and 2 are compared with genes located in Chromosome X for the same set of experiments. Ratio values correspond to the average of two independent experiments, and are displayed in a log2 scale. Averaging the log2 ratios for the X-linked probes in the three different experiments gives values of 0.234 ± 0.143 (1.176 in linear scale, average of 112 probes), 0.423 ± 0.220 (1.341 in linear scale, average of 113 probes), and 0.681 ± 0.290 (1.603 in linear scale, average of 115 probes) for the 2X, 3X, and 5X experiments, respectively. Dynamically compressed ratios can be converted to actual ratios by fitting log2 values to a power-law mathematical formula as described by Yuen et al. (2002). (B) Comparison of DNA polymerase-induced representational distortion using human DNA samples. Normal human DNA was amplified with either φ29 or Bst DNA polymerase, labeled with Cy3, and hybridized against similarly amplified human DNA labeled with Cy5. Plots for Chromosomes 1 and 2 are shown in the same scale as the plot in A. (C) Confidence limits for array–CGH analysis of human DNA. Plots correspond to unamplified human female versus male DNAs and whole genome Bst-amplified human female versus Bst-amplified male DNAs. Average log2 fluorescence ratios for replicate spots are ordered according to the chromosome number and the position in the chromosome. Ratio values for X-linked genes show a similar distribution to that observed for the 2X dosage in A. Confidence limits (horizontal dashed lines) for 99.9% of data for autosomal genes are between −0.262 and 0.262 (0.833 and 1.199 when expressed as linear ratios) for the unamplified experiment. The same confidence bounds calculated for the unamplified experiment are replicated in the plot of ratios generated by microarray analysis of amplified DNA.
Evaluation of DNA Amplification Bias Using Human cDNA Microarrays
We then performed array–CGH experiments using human DNA generated by whole genome amplification, to determine if the observations made in yeast would be valid for a much larger and repeat-rich genome. We used microarrays containing 4592 human cDNA clones, with two replicates for each clone. Figure 4B shows a comparison of array–CGH experiments performed using human genomic DNA amplified with φ29 DNA polymerase or Bst DNA polymerase. An important feature of these two experiments is that a single DNA preparation was split into two samples, and each sample was amplified independently, using the same enzyme and the same reaction conditions. Thus, the experiments are designed to measure the reproducibility of sequence representation during separate amplification reactions. If representation bias were random for many human loci, we would expect considerable noise in the self–self experiment. Notably, as was the case with yeast, the ratios obtained with Bst DNA polymerase are close to 1:1, whereas the ratios obtained with φ29 DNA polymerase deviate from the expected 1:1 at numerous loci in all chromosomes (data shown for Chromosomes 1 and 2 in Fig. 4B). We thus chose to use whole genome amplification with Bst DNA polymerase for all subsequent array–CGH experiments, because this enzyme generates the least representation noise.
To further evaluate the extent of sequence representation bias using amplified human DNA, we performed a control experiment hybridizing unamplified female DNA labeled with Cy3 against unamplified male DNA labeled with Cy5. We compared these results to an experiment performed under identical conditions but using two amplified DNAs. Log2 ratios for the two experiments were plotted in a scale of −1.5 to 1.5 (two panels in Fig. 4C), and 99.9% confidence bounds (−0.262 and 0.262) of the ratios were calculated for the unamplified DNA experiment, after removing genes associated with Chromosome X. We then artificially applied the same confidence bounds to the graph generated in the experiment with the amplified DNA (Fig. 4C, right panel). The number of autosomal data points outside the 0.262 confidence bounds in the experiment with unamplified DNA is 20, whereas the number of points outside the same bounds in the experiment with amplified DNA is also 20. We showed previously that changes of threefold or more were detected for the majority of loci analyzed in a dosage-calibration control experiment (Fig. 4A, middle panel). Given that the experiment in the right panel of Figure 4C does not show any increase whatsoever in the number of points outside the confidence bounds, we conclude that representational distortion introduced by whole genome amplification and measured by array–CGH is at most threefold, or even less, for those human genes surveyed in the microarray.
Application of Hyperbranched Whole Genome Amplification to the Study of Cancer Genetics
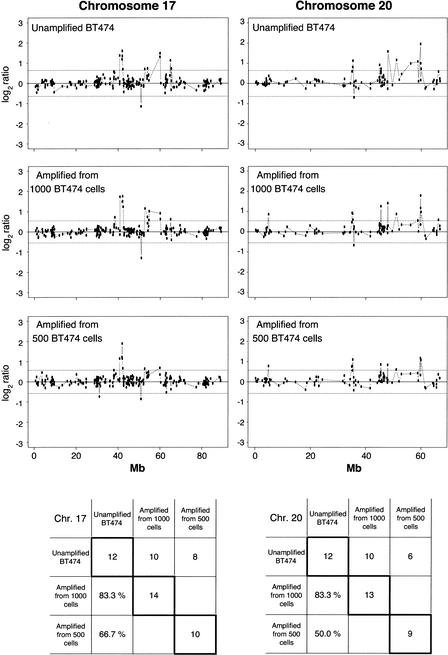
To demonstrate the utility of whole genome amplification for studies on cancer genetics, we analyzed DNA from the breast cancer cell line BT474, which has previously been demonstrated to harbor gene-dosage alterations by array–CGH (Pinkel et al. 1998; Pollack et al. 1999, 2002; Monni et al. 2001; Snijders et al. 2001). In a reference experiment, we performed array–CGH using unamplified DNA from the BT474 cell line, compared with reference unamplified DNA from a normal human female. Two reference experiments with unamplified DNA, one of which is shown in Figure 5A, reproduced most of the known genetic alterations previously described for this cell line by conventional CGH (Kallioniemi et al. 1994) and further corroborated by array–CGH (Pollack et al. 1999, 2002; Snijders et al. 2001). We performed array–CGH experiments with DNA amplified from 1000 and 500 BT474 cell equivalents, and used as a reference amplified DNA from the same normal female source. Figure 5B shows the array–CGH profiles obtained with amplified DNA samples from 1000 cells, indicating several of the gene-dosage alterations that were also observed in unamplified DNA. Among the 500- and 1000-cell experiments, only the 1000-cell experiment showed high concordance of alterations across the entire genome, relative to those alterations observed in the unamplified controls. The concordance between the 1000-cell experiment and the first unamplified control is 53.6% (30 out of 56 loci flagged as displaying significantly altered ratios). The concordance between the 1000-cell experiment and the second unamplified control is 59.1% (26 out of 44 loci). In contrast, the concordance values for the 500-cell experiment are 33.9% (19 out of 56) and 38.6% (17 out of 44). Some of the most dramatic alterations are observed in Chromosomes 17 and 20, for unamplified as well as amplified DNA from 1000 and 500 cells (Fig. 6). For the altered genes in Chromosomes 17 and 20, the concordance is very high (83.3%) for the 1000-cell experiment for both chromosomes. The corresponding values for the 500-cell experiment are 66.7% and 50% for Chromosomes 17 and 20, respectively.
Figure 5.
Array–CGH of cancer cell line BT474. (A) DNA equivalent to 300,000 cells from breast cancer cell line BT474 was cohybridized with a similar amount of normal human female DNA from peripheral blood lymphocytes to a cDNA microarray containing replicate probes for 4600 genes. BT474 to female fluorescence log2 ratio profiles were ordered by chromosomes, and into each chromosome, according to the position as assessed by SOURCE (http://genome-www4.stanford.edu/cgi-bin/SMD/source/sourceSearch). Ratio values were connected by lines. A log2 ratio near 0 indicates an equal copy-number representation of that gene in both genomes. Dashed lines at −0.642 and 0.642 represent nonparametric bounds for log2 ratios as described in Methods. (B) DNA from 1000 cells, obtained from the BT474 cell line, or DNA from peripheral lymphocytes of a normal human female was amplified using large fragment Bst DNA polymerase and cohybridized to the microarray in the same conditions as before. Nonparametric bounds for this experiment are denoted with dashed lines at −0.546 and 0.546. Arrows indicate gene-dosage alterations observed in both the unamplified and the whole genome-amplified experiments except for shaded areas in Chromosomes 17 and 20 in both figures. These chromosomes are very rich in altered genes, and can be observed in more detail in Figure 6.
Figure 6.
Enlarged representation of array–CGH data for Chromosomes 17 and 20. Chromosomes 17 and 20 are known to contain notorious amplifications in the breast cancer cell line BT474. Chromosome 17 is represented in the microarray by 221 cDNAs, and Chromosome 20 by 99 cDNAs. Note that the profiles for Chromosomes 17 and 20 are mostly conserved for the whole genome-amplified samples from different inputs of DNA (equivalent to 500 and 1000 cells) compared with one of the unamplified experiments. The number of altered genes and percentage of concordance of the amplified experiments to the unamplified control are presented in the tables below the plots. The values in diagonal show the total number of altered genes in every experiment for Chromosomes 17 and 20 separately. The first row of each table contains the number of altered genes in common for every amplified experiment compared with the unamplified control. Likewise, the first column of each table gives the percentage of concordance for altered genes found in common between each amplified experiment and the total number of altered genes in the unamplified control. Nonparametric bounds are −0.642 and 0.642 for the unamplified experiment, −0.546 and 0.546 for the experiment using amplified DNA from 1000 cells, and −0.574 and 0.573 for the experiment using amplified DNA from 500 cells.
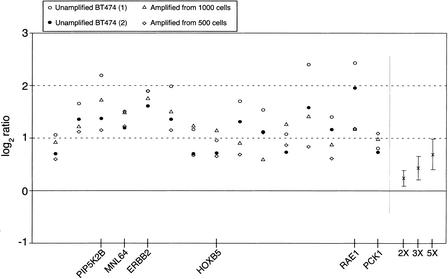
Notably, altered loci with relatively high gene-dosage alterations are detected with high reproducibility among different experiments, although the observed ratios are somewhat variable. Figure 7 shows all the data points for 15 loci that were recorded as showing the most distinct copy-number gains across four experiments, two of which include amplified DNA. Dosage gains, shown in a log2 scale, are detectable in all four experiments for all 15 gene loci, and can be compared with the 2X, 3X, and 5X dosage calibration points generated by averaging the ratios of all X-linked probes. For this set of loci, all ratios lie above the 3X calibrator reference point. Many of the genes in this set of 15 altered loci have a well-established association with cancer, such as ERBB2. This gene, at 17q12, is highly amplified in the breast cancer cell line BT474, as well as in breast tumors, where it is used as a prognostic marker (Riou et al. 2001). ERBB2 amplification is also associated with high gene expression levels, as demonstrated using array–CGH and array mRNA expression analysis (Pollack et al. 1999). MNL64 (Tomasetto et al. 1995) is a gene included in the ERBB2 amplicon, also with elevated gene-copy number in the BT474 cell line. Other genes found to be notably amplified are RAE1 (Bharathi et al. 1997), implicated in the export of poly(A)+ RNA from nucleus to cytoplasm, and CSE1L (Brinkmann et al. 1996), both on Chromosome 20. Another gene found to be amplified is nuclear receptor NR4A3/NOR1 (Maruyama et al. 1995) at 9q31.
Figure 7.
Repeatability of observed log2 ratios for the set of 15 genes that were identified as significantly amplified in all BT474 experiments. For each experiment, genes were selected using nonparametric methods that flagged log2 ratios beyond the third quartile plus 2.5 times the Interquartile Range of the observed distribution. (Unamplified BT474) Corresponds to the comparison of unamplified BT474 DNA versus unamplified female DNA. (Amplified from 1000 cells) and (Amplified from 500 cells) correspond to the comparison of amplified DNA from inputs equivalent to 1000 and 500 BT474 cells, respectively, against amplified normal female DNA from similar inputs. On the right side of the plot are shown the actual average ratios and standard deviations for X-chromosome copy-number variations corresponding to ratios of 2:1, 3:1, and 5:1. Using the average value for all four ERBB2 ratios (1.791 in log2 scale) and the calibration curve derived from the average ratios for 2X, 3X, and 5X (0.234, 0.423, and 0.681, respectively, in log2 scale), we calculated a hypothetical ratio of 12.5 for this gene. This ratio value is in agreement with amplification values determined by other methods (Lucito et al. 1998), that reported copy number increases in the range of 10–15 for ERBB2 in the breast cancer cell line BT474. Seven of the amplified genes are located in the chromosomal arm 17q (among them PIP2KB, MNL64, ERBB2, and HOXB5), and six probes are located in the chromosomal arm 20q (among them RAE1 and PCK1).
Evaluation of the Linearity of DNA Amplification for Different Gene Loci Using Human BAC Arrays
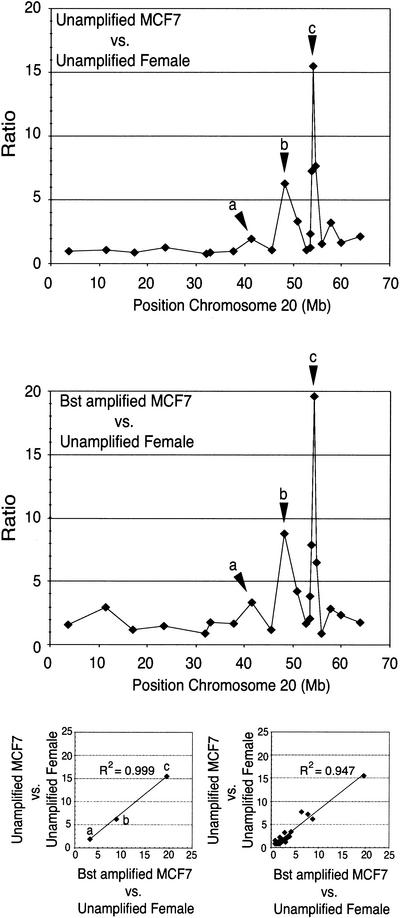
We carried out an additional experiment to assess with more quantitative precision the performance of the hyperbranched whole genome amplification method, taking advantage of BAC arrays. Using a small BAC array, containing clones in Chromosome 20, we performed CGH using amplified DNA from the cell line MCF7 (Pinkel et al. 1998). The results show that the profile of ratios obtained with amplified DNA is remarkably similar to the profile obtained with unamplified DNA as observed in Figure 8. Furthermore, a scatter plot of the ratios at the three loci showing the largest amplification shows a very high correlation coefficient (R2 = 0.999 ). The correlation coefficient for the scatter plot that included all the loci was R2 = 0.947.
Figure 8.
Analysis of breast cancer cell line MCF7 using BAC clones located in Chromosome 20. The graph on the top shows the copy-number variation for the array–CGH experiment corresponding to the hybridization of genomic DNA from the cell line MCF7 against female DNA. The graph in the middle shows the equivalent experiment performed with isothermal whole genome-amplified DNA from MCF7 against female DNA. Regions of copy number increase are remarkably conserved for the isothermal whole genome-amplified DNA from MCF7. The lower left scatter plot shows the high correlation obtained for the ratios of the three loci showing the largest amplification. The lower right scatter plot shows the correlation coefficient for all the ratios in both experiments.
DISCUSSION
A key feature distinguishing hyperbranched whole genome amplification from PCR-based methods is the length of the DNA replication products, which is in the range of 10–20 kb or more. This reaction mechanism favors equal representation of sequences because each priming event is propagated over very long distances in the genome, and, as mentioned earlier, the number of cycles of strand replication in a 5-h incubation is lower than for PCR. It should be possible to improve the overall yield of hyperbranched whole genome amplification by optimizing components of the reaction mixture to reduce pyrophosphate accumulation. Dean et al. (2002) have recently reported the use of optimized buffers for hyperbranched whole genome amplification (also known as MDA; Lizardi 2000) to achieve amplification yields as high as 100,000 copies of the original DNA. In agreement with our observations, they report that representational distortion, assessed by quantitative PCR at eight different loci, is less than threefold, provided that amplification is limited to no more than 10,000-fold. They also demonstrate the utility of the Multiple Displacement Amplification (MDA) reaction for accurate Single Nucleotide Polymorphism (SNP) genotyping. Among potential future applications of this method that deserve to be explored is the amplification of DNA from individual chromosomes. We are exploring the use of the method for amplification of BAC DNA, to facilitate the construction and reduce the cost of BAC microarrays.
CGH on metaphase chromosomes, pioneered by Kallioniemi et al. (1992), and array–CGH enable genome-wide assessment of genetic alterations in cancer cells. The power of array–CGH will undoubtedly become greater as the density and count of genomic probes on DNA microarrays increases, eventually attaining full coverage of the genome at high resolution. Important insights about the extent of heterogeneity of genetic alterations in tissue have been dramatically demonstrated by in situ studies using FISH probes (Thompson and Gray 1993; Fiegl et al. 1995) to interrogate a limited number of loci in interphase nuclei. These studies revealed a surprising degree of heterogeneity in gene dosage over relatively small domains of tumors. The significance of the capability of array–CGH analysis of tissue samples containing a few hundred cells, enabled by whole genome amplification, lies in the greatly expanded potential for discovery of novel genetic alterations that may be limited to small clonal patches in tumors, or even present in small preneoplastic lesions. Such alterations would be undetectable when larger, heterogeneous samples are analyzed. Samples obtained by laser-capture microdissection will be an ideal source of DNA for studies at high tissue resolution, enabled by whole genome amplification and array–CGH. These capabilities may lead to the discovery of novel oncogenes or tumor suppressors that map to regions of gene amplification or gene loss (Albertson et al. 2000; Bruder et al. 2001).
To exploit fully the benefits of isothermal whole genome amplification, it will be necessary to combine the method with more precise analytical tools. Microarray tools for whole genome analysis are still at a relatively early stage of development. Array–CGH using only two replicates for each cDNA clone is sufficiently accurate to detect genetic alterations at the whole genome level, provided that the dosage change is over twofold. We observed that threefold and fivefold gene-dosage changes are detected reliably for cell lines with increased X-chromosome dosage, indicating that similar-fold changes occurring in cancer cells should be detectable with high reliability. In our hands, the signal-to-noise ratio of CGH experiments on cDNA microarrays is not sufficient to permit reliable detection of heterozygous deletions, as the resulting 1:2 gene dosage ratio lies within experimental error. Cross-hybridization of unrelated sequences in the highly complex genomic DNA introduces noise in the signals at each array element, and causes most data points of heterozygous deletions to fall within the variance range of observations for genes with a 1:1 ratio. The use of arrays of BACs can provide more precise measurements (Pinkel et al. 1998; Snijders et al. 2001). However, because BAC probes are ∼100 kb long, the resolution of very closely spaced gene loci can be inferior to that of cDNA arrays, and could obscure subtle dosage alterations. The use of genomic representations (Lucito et al. 2000) and special arrays designed for detection of such representations might improve measurement precision for specific subsets of the genome. Using the new whole genome amplification method in combination with cDNA arrays, our best data for detection of genetic alterations in cancer cells were obtained with DNA samples amplified from 1000 cells. Rough calculations indicate that, after amplification, the 1000-cell experiment generated ∼1.43 μg of DNA. This amount of DNA is similar to the 2 μg of DNA used previously by Pollack et al. (1999). We believe that in the future, the use of BAC arrays, which have superior signal-to-noise characteristics, would enable the generation of high-quality array–CGH data starting with DNA amplified from as few as 200 cells. A challenge for translation of array–CGH technology to clinical applications is to find a viable combination of sequence coverage, reproducible data with sufficient statistical robustness, and relatively low cost.
Cancer risk assessment is a potential future clinical application of whole genome amplification and array–CGH. Loss of heterozygosity at multiple loci has been reported to occur early in tumorigenesis in a number of cancers (Watson et al. 1998; Lakhani et al. 1999; Kittiniyom et al. 2001). Assessment of metastatic potential will be another exciting application, as demonstrated by the recent findings that gene-dosage increases for PRL3 at 8q24.3 correlate with metastasis in colon cancer (Saha et al. 2001). The simplicity of isothermal whole genome amplification will make it possible to perform gene dosage analysis in large numbers of small samples, and should reveal whether gene-dosage changes can be used as a biomarker for assessment of cancer risk or risk of metastasis. The capability for generating sufficient DNA from samples derived from a few hundred cells will facilitate the implementation of genomic analysis by array–CGH in surrogate samples. Among such samples are cell-containing fluids obtained by noninvasive or minimally invasive procedures, as exemplified by epithelial cells released by breast duct lavage (Dooley et al. 2001), cellular samples from pancreatic duct fluids (Kondoh et al. 1998), or peripheral blood cell fractions isolated by cell sorting based on surface markers. Any of these samples can be a reliable source of DNA, and, in contrast to the complex issue of RNA quality, the integrity of DNA in a biopsy sample should be less sensitive to tissue physiological state or storage time.
A significant limitation of the whole genome amplification methods described here is that the DNA yield is reduced as the molecular weight of the starting material decreases, owing to the occurrence of fewer priming and hyperbranching events in each molecule of denatured DNA. Thus, the reaction is not ideal for analysis of formalin-fixed archival DNA or low-molecular-weight DNA from deteriorated forensic samples. Nonetheless, for studies in cancer biology, fresh tissue, tissue preserved by freezing, or by ethanol fixation, will provide excellent material for amplification. We have observed that the nucleotide sequence of DNA loci of interest is very accurately preserved in DNA amplified using φ29 DNA polymerase. Recent studies on related reactions catalyzed by φ29 DNA polymerase (Dean et al. 2001) have demonstrated that whole genome amplification of circular genomes by randomly primed hyperbranched rolling-circle amplification (HRCA) generates high-quality DNA suitable for analysis by DNA sequencing. The error rate of φ29 DNA polymerase has been reported to be in the range of 2.2 × 10−5 to 4 × 10−6 (Esteban et al. 1993). The high fidelity of replication by φ29 DNA polymerase ensures that DNA generated by whole genome amplification is suitable for further analysis by cloning and sequencing procedures. Such samples may be analyzed for alterations in microsatellite markers, alterations detectable by PCR-SSCP, as well as by DNA sequencing or cloning, in order to obtain information about DNA slippage, point mutations, translocation events, and so on.
On the other hand, Bst DNA polymerase is the enzyme of choice for applications in which consistent and relatively unbiased sequence representation of the amplified genome is required, as is the case for array–CGH. Hybridization on the cDNA microarrays is insensitive to single-base errors in DNA replication generated by the lack of proofreading activity. A striking observation is the very high molecular weight of DNA generated by hyperbranched strand-displacement amplification with Bst DNA polymerase. With regard to this issue, it is relevant to note that in the array–CGH experiments comparing amplified yeast DNA with unamplified yeast DNA, the plots generated with material amplified by φ29 DNA polymerase showed marked reductions in ratios for genes near the telomeres, as expected for a reaction that is inefficient near the terminus of a DNA fragment, with the consequent representational drop-off. This is in sharp contrast to the results of array–CGH for the identical experiment performed using Bst DNA polymerase, instead of φ29. In this case, there is little indication of representation drop-off near the telomeres, indicating that DNA replication is less affected by the presence of a DNA terminus. We speculate that Bst DNA polymerase may be capable of template-switching, as has been described for Thermus aquaticus DNA polymerase (Odelberg et al. 1995). Template-switching may explain the generation of larger DNA strands during amplification with Bst DNA polymerase, because the DNA product will be much larger if the polymerase can continue polymerization as it reaches a DNA terminus, by switching to the product strand, and using it as a new template. It is important to note, however, that template-switching need not cause sequence representation bias; on the contrary, it may reduce the drop-off of sequence representation for loci near telomeres.
METHODS
Yeast, Human, and Cell-Line Genomic DNAs
The Saccharomyces cerevisiae strain designated as KO harbors GIN4 and CLA4 deletions, and the strain designated WT harbors an HIS3 deletion. Yeast cultures were harvested and lysed using glass beads. Yeast genomic DNA was isolated by phenol-chloroform extraction and ethanol precipitation. Human DNA was obtained from peripheral lymphocytes using a standard guanidine-HCl protocol (Ciulla et al. 1988). DNA from BT474 cell line (American Type Culture Collection) was extracted using the kit Blood and Cell Culture DNA Maxi Kit (QIAGEN). DNA from cell lines with three and five X-chromosomes (NA04626, NA06061) was obtained from NIGMS Human Genetic Cell Repository, Coriell Institute for Medical Research. The average size of the DNA was >20 kb for all the samples as assessed by agarose gel electrophoresis.
DNA Quantitation
Quantitations were done using the PicoGreen DNA quantitation kit (Molecular Probes) according to the manufacturer's specifications. In those cases in which quantification was performed in amplified DNA at different times, a sample at time 0 was analyzed, and the background fluorescence was subtracted from the other time points.
Isothermal Amplification
Appropriate amounts of DNA (see figure legends) were mixed with primers (random 7-mers with an additional two nitroindole residues at the 5′ end and a phosphorothioate linkage at the 3′ end) at a concentration of 100 μM in 9 μL of 1× buffer. For φ29 polymerase, 1× Buffer Y+/Tango (MBI Fermentas) was used supplemented with Tween-20 at a final concentration of 0.12%. The buffer for Bst reactions was 1× ThermoPol buffer (New England Biolabs) with DMSO at 4% final concentration. DNA mix was denatured at 96°C for 4 min, let cool at room temperature for 10 min, and then placed on ice. The reaction mixture was then brought up to 30 μL containing 400 μM dNTPs in 1× buffer and the polymerase. φ29 was added at a final concentration of 0.1 units/μL, and large fragment Bst DNA polymerase (New England Biolabs) at 0.35 units/μL. T4 gene 32 protein (or G32P, Amersham Pharmacia) was added in the reactions performed with Bst at a final concentration of 30 ng/μL. Reactions were carried out at 32°C for φ29 or 50°C for Bst. Reaction products were analyzed in 0.5% agarose alkaline gels.
DNA Labeling and Hybridization to Microarrays
Genomic or whole genome amplified DNAs were digested with MnlI (New England Biolabs) and purified using microcon 50 filters (Millipore). MnlI cleaves single- and double-stranded DNA. Human and cell-line DNAs were labeled following a protocol described elsewhere (Pollack et al. 1999) with minor modifications. For yeast experiments, DNA was labeled with allylamine-dUTP (Sigma), and reactive Cy3 and Cy5 succinamide ester monofunctional dyes (Amersham Pharmacia) were coupled to the DNA following a protocol described at www.microarrays.org. CMT-Yeast microarrays (version 1.32, Corning) containing 6135 unique ORFs were prehybridized in 35% formamide, 0.5% SDS, 4× SSPE, 2.5× Denhardt's, and 0.2 mg/mL herring sperm DNA for at least 2 h. In the meantime, ∼200 ng of Cy3- and Cy5-labeled yeast DNA was mixed with 15 μg of yeast tRNA (GIBCO BRL) in a final hybridization volume of 60 μL (35% formamide, 0.5% SDS, 4× SSPE, 2.5× Denhardt's; protocol adapted from Cheung et al. 1999). After denaturation at 95°C for 2 min, hybridization was performed under a coverslip (LifterSlip) at 50°C for 18–19 h. Following hybridization, the coverslip was removed in 1× SSC, 0.1% SDS, then the microarray was washed in 0.2× SSC, 0.1% SDS for 10 min and in 0.2× SSC for 20 min (2×). In the case of human experiments, cDNA microarrays containing a total of 4600 duplicated genes (Keck facility, Yale University) were prehybridized with 48% formamide, 0.4% SDS, 3.2× SSPE, 2× Denhardt's, and 0.2 mg/mL at 50°C for at least 30 min. Then differentially labeled DNAs were combined with 1 μg of poly(dA)–poly(dT) (Amersham Pharmacia), 2 μg of yeast tRNA, and 5 μg of Cot-1 DNA (GIBCO BRL) in a final volume of 13 μL (37% formamide, 0.5% SDS, 4.6× SSPE, 2.5× Denhardt's). Once denatured (95°C, 2 min), the solution was hybridized under a coverslip at 42°C for 18–20 h. After hybridization, the coverslip was removed by soaking in 2× SSC, 0.1% SDS, and then the microarray was washed in 2× SSC, 0.1% SDS for 10 min; 0.2× SSC, 0.1% SDS for 10 min (2×); and 0.2× SSC for 10 min (2×). Microarrays were dried out by spinning for 5 min at 200g. Labeling and hybridization to BAC arrays were carried out as described by Pinkel et al. (1998)
Microarray Imaging and Analysis
Slides were scanned with an Axon GenePix 4000A scanner (Axon Instruments), and the resulting 16-bit TIFF images were analyzed with the program Spot (Buckley 2001). Foreground and background intensities for both Cy3 and Cy5 were calculated for each spot and exported as a tab-delimited text file. Background fluorescence intensities were obtained using Spot's morphological opening function (Buckley 2000), which has been shown to provide a more accurate estimate of background intensity than other methods such as valley, histogram, fixed circle, or adaptive shape segmentation (Yang et al. 2002). The intensity data were then combined with gene information for each spot, such as the name, GenBank accession number, chromosome, and mapped chromosome start position. Data analysis was conducted with a suite of functions written in R (Ihaka and Gentleman 1996) and S-Plus. Ratios of background-subtracted Cy3 = G and Cy5 = R intensities were converted to log2 ratios M = log2(R/G), and plotted against A, where A = 1/2 log2(R * G). The use of M versus A plots has been advocated by Dudoit et al. (2000). Microarray spots with undefined ratios (such as Cy-dye controls) and unmapped chromosome locations were removed from the data sets.
Statistical Methods
Because both the unamplified and amplified baseline data sets compared a normal human female genome to a normal human male genome, genes located on the X- and Y-chromosomes were also removed from both data sets when calculating confidence intervals. These data sets provided a perfect data model with an expected ratio of 1:1. To locate potential outliers and sources of systematic variation caused by array artifacts or experimental error in the baseline data sets for human DNA, the two replicate measurements for each spot were compared. For the unamplified data set, analysis revealed a systematic difference in ratios between the two replicate measurements for a large number of contiguous spots, and it was determined that the leftmost columns of the array had been affected by experimental error (most likely a shifting coverslip during hybridization). The spots in the affected area were removed from consideration. Following this modification, the final data set consisted of pairs of observations for 3878 genes. Likewise, the amplified data set consisted of pairs of observations for 3886 genes.
The log ratios for both baseline data sets were normalized using a pinwise lowest curve fit (Yang et al. 2001) based on the summed log signal intensities, and each pair of normalized log ratios was averaged to produce a single mean value for each gene. Then 99.9% confidence intervals for the distribution of the adjusted mean log ratios for the unamplified dataset were computed under the assumption of near normality, with a critical value estimated from the t distribution with 3877 degrees of freedom.
Experimental data included two arrays corresponding to the experiment for the unamplified BT474 cell line versus unamplified human female DNA, one array with amplification from 1000 BT474 cells, and one array with amplification from 500 BT474 cells. Each array was checked for spots with significantly low intensity, resulting in the removal of between 10 and 24 genes per experiment, and additional spots were removed because of the presence of scratches or smudges in the immediate area of the spot. The number of pairs of observations remaining after low-intensity and error filtering were as follows: unamplified, Array 1, 3799 clone pairs; unamplified, Array 2, 3756 clone pairs; amplified from 1000 cells, 3879 clone pairs; amplified from 500 cells, 3805 clone pairs.
The reduced data sets were corrected for spatial effects and normalized to account for intensity-related variability, and for each experiment the two replicate spots per gene were averaged to produce a single mean value. Because it is not appropriate to assume normality for the distribution of mean log2 ratios for the BT474 cell line experiments because of the expected presence of amplified and deleted genes, a nonparametric approach for estimating conservative bounds for the set of unaltered genes was used. For each experiment, we computed the Interquartile Range (IQR) of the distribution of log ratios, in which the IQR is defined to be the distance between the first and third quartiles (denoted Q1 and Q3, respectively) of the observed values, or the values between which the middle half of the distribution is observed to fall. Values less than Q1 − 2.5(IQR) or greater than Q3 + 2.5(IQR) were flagged as significantly altered, where the coefficient 2.5 was chosen to account for the effect of long tails in the distributions of log2 ratios and reduce the number of false-positive identifications.
WEB SITE REFERENCES
http://genome-www4.stanford.edu/cgi-bin/SMD/source/sourceSearch; The Stanford Online Universal Resource for Clones and ESTs.
http://www.microarrays.org; Is a public source for microarray protocols and software, built and maintained by the DeRisi Lab, Department of Biochemistry & Biophysics, Univ. of California at San Francisco.
Acknowledgments
We are indebted to Junhyong Kim for providing the bioinformatics tools for chromosome map ordering of the yeast and human clones. We thank Antonio Casamayor and Michael Snyder for a gift of two well-characterized yeast strains, and David Stern for critical reading of this manuscript. The enzyme φ29 DNA polymerase was a gift of APBiotech, Inc. We are also indebted to the Keck Foundation Biotechnology Resource at Yale University for supplying human DNA microarrays and protocols (array facility supported by NIH Grant No. 5 U24 DK58776 PI to Kenneth Williams). This work was supported by the Marcia Israel Early Cancer Detection Fund of the Yale Cancer Center, the NCI Early Detection Research Network Grant No. CA85065-03, and the NCI Innovative Technologies Grant No. CA81671-02. Work with BAC arrays was supported by NCI Grant No. CA83040.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL paul.lizardi@yale.edu; FAX (203) 785-7303.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.377203.
REFERENCES
- 1.Albertson D.G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S.H., Kowbel, D., Kuo, W.L., Gray, J.W., and Pinkel, D. 2000. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet. 25: 144-146. [DOI] [PubMed] [Google Scholar]
- 2.Aliotta J.M., Pelletier, J.J., Ware, J.L., Moran, L.S., Benner, J.S., and Kong, H. 1996. Thermostable Bst DNA polymerase I lacks a 3′ → 5′ proofreading exonuclease activity. Genet. Anal. 12: 185-195. [PubMed] [Google Scholar]
- 3.Ball S., Reeve, M.A., Robinson, P.S., Hill, F., Brown, D.M., and Loakes, D. 1998. The use of tailed octamer primers for cycle sequencing. Nucleic Acids Res. 26: 5225-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bebenek K., Joyce, C.M., Fitzgerald, M.P., and Kunkel, T.A. 1990. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 265: 13878-13887. [PubMed] [Google Scholar]
- 5.Bharathi A., Ghosh, A., Whalen, W.A., Yoon, J.H., Pu, R., Dasso, M., and Dhar, R. 1997. The human RAE1 gene is a functional homologue of Schizosaccharomyces pombe rae1 gene involved in nuclear export of poly(A)+ RNA. Gene 198: 251-258. [DOI] [PubMed] [Google Scholar]
- 6.Blanco L. and Salas, M. 1985. Characterization of a 3′ → 5′ exonuclease activity in the phage φ 29-encoded DNA polymerase. Nucleic Acids Res. 13: 1239-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkmann U., Gallo, M., Polymeropoulos, M.H., and Pastan, I. 1996. The human CAS (cellular apoptosis susceptibility) gene mapping on Chromosome 20q13 is amplified in BT474 breast cancer cells and part of aberrant chromosomes in breast and colon cancer cell lines. Genome Res. 6: 187-194. [DOI] [PubMed] [Google Scholar]
- 8.Bruder C.E., Hirvela, C., Tapia-Paez, I., Fransson, I., Segraves, R., Hamilton, G., Zhang, X.X., Evans, D.G., Wallace, A.J., Baser, M.E., et al. 2001. High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray–CGH. Hum. Mol. Genet. 10: 271-282. [DOI] [PubMed] [Google Scholar]
- 9.Buckley M.J., 2000. The Spot user's guide. CSIRO Mathematical and Informational Sciences, North Ryde, NSW, Australia (http://www.cmis.csiro.au/IAP/spotinfo.htm).
- 10.___,, 2001. Spot. CSIRO Mathematical and Informational Sciences, North Ryde, NSW, Australia.
- 11.Cheung V.G. and Nelson, S.F. 1996. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl. Acad. Sci. 93: 14676-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung V.G., Morley, M., Aguilar, F., Massimi, A., Kucherlapati, R., and Childs, G. 1999. Making and reading microarrays. Nat. Genet. 21 (1 Suppl): 15-19. [DOI] [PubMed] [Google Scholar]
- 13.Ciulla T.A., Sklar, R.M., and Hauser, S.L. 1988. A simple method for DNA purification from peripheral blood. Anal. Biochem. 174: 485-488. [DOI] [PubMed] [Google Scholar]
- 14.Dean F.B., Nelson, J.R., Giesler, T.L., and Lasken, R.S. 2001. Rapid amplification of plasmid and phage DNA using φ29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11: 1095-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean F.B., Hosono, S., Fang, L., Wu, X., Faruqi, A.F., Bray-Ward, P., Sun, Z., Zong, Q., Du, Y., Du, J., et al. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. 99: 5261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dooley W.C., Veronesi, U., Elledge, R., O'Shaughnessy, J., Ljung, B., and Arias, R. 2001. Detection of premalignant and malignant breast cells by ductal lavage. Obstet. Gynecol. 97 (4 Suppl 1): S2. [Google Scholar]
- 17.Dudoit S., Yang, Y.H., Callow, M.J., and Speed, T.P., 2000. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Technical report #578, pp. 1–38. Stanford University School of Medicine, Stanford, CA.
- 18.Esteban J.A., Salas, M., and Blanco, L. 1993. Fidelity of φ 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem. 268: 2719-2726. [PubMed] [Google Scholar]
- 19.Fiegl M., Tueni, C., Schenk, T., Jakesz, R., Gnant, M., Reiner, A., Rudas, M., Pirc-Danoewinata, H., Marosi, C., Huber, H., et al. 1995. Interphase cytogenetics reveals a high incidence of aneuploidy and intra-tumour heterogeneity in breast cancer. Br. J. Cancer 72: 51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Q., Schantz, S.P., Rao, P.H., Mo, J., McCormick, S.A., and Chaganti, R.S. 2000. Improving degenerate oligonucleotide primed PCR-comparative genomic hybridization for analysis of DNA copy number changes in tumors. Genes Chromosomes Cancer 28: 395-403. [PubMed] [Google Scholar]
- 21.Ihaka R. and Gentleman, R. 1996. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5: 299-314. [Google Scholar]
- 22.Kallioniemi A., Kallioniemi, O.P., Sudar, D., Rutovitz, D., Gray, J.W., Waldman, F., and Pinkel, D. 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258: 818-821. [DOI] [PubMed] [Google Scholar]
- 23.Kallioniemi A., Kallioniemi, O.P., Piper, J., Tanner, M., Stokke, T., Chen, L., Smith, H.S., Pinkel, D., Gray, J.W., and Waldman, F.M. 1994. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc. Natl. Acad. Sci. 91: 2156-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kittiniyom K., Gorse, K.M., Dalbegue, F., Lichy, J.H., Taubenberger, J.K., and Newsham, I.F. 2001. Allelic loss on Chromosome band 18p11.3 occurs early and reveals heterogeneity in breast cancer progression. Breast Cancer Res. 3: 192-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein C.A., Schmidt-Kittler, O., Schardt, J.A., Pantel, K., Speicher, M.R., and Riethmuller, G. 1999. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc. Natl. Acad. Sci. 96: 4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondoh S., Kaino, M., Okita, S., Ryozawa, S., Akiyama, T., and Okita, K. 1998. Detection of Ki-ras and p53 gene mutations in tissue and pancreatic juice from pancreatic adenocarcinomas. J. Gastroenterol. 33: 390-396. [DOI] [PubMed] [Google Scholar]
- 27.Lakhani S.R., Chaggar, R., Davies, S., Jones, C., Collins, N., Odel, C., Stratton, M.R., and O'Hare, M.J. 1999. Genetic alterations in ‘normal’ luminal and myoepithelial cells of the breast. J. Pathol. 189: 496-503. [DOI] [PubMed] [Google Scholar]
- 28.Lizardi, P.M. 2000. Multiple displacement amplification. United States Patent 6,124,120.
- 29.Lizardi P.M., Huang, X., Zhu, Z., Bray-Ward, P., Thomas, D.C., and Ward, D.C. 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19: 225-232. [DOI] [PubMed] [Google Scholar]
- 30.Loakes D. and Brown, D.M. 1994. 5-Nitroindole as an universal base analogue. Nucleic Acids Res. 22: 4039-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loakes D., Hill, F., Brown, D.M., and Salisbury, S.A. 1997. Stability and structure of DNA oligonucleotides containing non-specific base analogues. J. Mol. Biol. 270: 426-435. [DOI] [PubMed] [Google Scholar]
- 32.Lucito R., Nakimura, M., West, J.A., Han, Y., Chin, K., Jensen, K., McCombie, R., Gray, J.W., and Wigler, M. 1998. Genetic analysis using genomic representations. Proc. Natl. Acad. Sci. 95: 4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucito R., West, J., Reiner, A., Alexander, J., Esposito, D., Mishra, B., Powers, S., Norton, L., and Wigler, M. 2000. Detecting gene copy number fluctuations in tumor cells by microarray analysis of genomic representations. Genome Res. 10: 1726-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lüdecke H.J., Senger, G., Claussen, U., and Horsthemke, B. 1989. Cloning defined regions of the human genome by microdissection of banded chromosomes and enzymatic amplification. Nature 338: 348-350. [DOI] [PubMed] [Google Scholar]
- 35.Maruyama K., Tsukada, T., Bandoh, S., Sasaki, K., Ohkura, N., and Yamaguchi, K. 1995. Expression of NOR-1 and its closely related members of the steroid/thyroid hormone receptor superfamily in human neuroblastoma cell lines. Cancer Lett. 96: 117-122. [DOI] [PubMed] [Google Scholar]
- 36.Monni O., Barlund, M., Mousses, S., Kononen, J, Sauter, G., Heiskanen, M., Paavola, P., Avela, K., Chen, Y., Bittner, M.L., et al. 2001. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc. Natl. Acad. Sci. 98: 5711-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odelberg S.J., Weiss, R.B., Hata, A., and White, R. 1995. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase. Nucleic Acids Res. 23: 2049-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinkel D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W.L., Chen, C., Zhai, Y., et al. 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20: 207-211. [DOI] [PubMed] [Google Scholar]
- 39.Pollack J.R., Perou, C.M., Alizadeh, A.A., Eisen, M.B., Pergamenschikov, A., Williams, C.F., Jeffrey, S.S., Botstein, D., and Brown, P.O. 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23: 41-46. [DOI] [PubMed] [Google Scholar]
- 40.Pollack J.R., Sorlie, T., Perou, C.M., Rees, C.A., Jeffrey, S.S., Lonning, P.E., Tibshirani, R., Botstein, D., Borresen-Dale, A.L., and Brown, P.O. 2002. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. 99: 12963-12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riou G., Mathieu, M.C., Barrois, M., Le Bihan, M.L., Ahomadegbe, J.C., Benard, J., and Le, M.G. 2001. c-erbB-2 (HER-2/neu) gene amplification is a better indicator of poor prognosis than protein over-expression in operable breast-cancer patients. Intl. J. Cancer 95: 266-270. [DOI] [PubMed] [Google Scholar]
- 42.Saha S., Bardelli, A., Buckhaults, P., Velculescu, V.E., Rago, C., St Croix, B., Romans, K.E., Choti, M.A., Lengauer, C., Kinzler, K.W., et al. 2001. A phosphatase associated with metastasis of colorectal cancer. Science 294: 1343-1346. [DOI] [PubMed] [Google Scholar]
- 43.Saunders R.D., Glover, D.M., Ashburner, M., Siden-Kiamos, I., Louis, C., Monastirioti, M., Savakis, C., and Kafatos, F. 1989. PCR amplification of DNA microdissected from a single polytene chromosome band: A comparison with conventional microcloning. Nucleic Acids Res. 17: 9027-9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skerra A. 1992. Phosphorothioate primers improve the amplification of DNA sequences by DNA polymerases with proofreading activity. Nucleic Acids Res. 20: 3551-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snijders A.M., Nowak, N., Segraves, R., Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A.K., Huey, B., Kimura, K., et al. 2001. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 29: 263-264. [DOI] [PubMed] [Google Scholar]
- 46.Telenius H., Carter, N.P., Bebb, C.E., Nordenskjold, M., Ponder, B.A., and Tunnacliffe, A. 1992. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics 13: 718-725. [DOI] [PubMed] [Google Scholar]
- 47.Thompson C.T. and Gray, J.W. 1993. Cytogenetic profiling using fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH). J. Cell Biochem. Suppl. 17G: 139-143. [DOI] [PubMed] [Google Scholar]
- 48.Tomasetto C., Regnier, C., Moog-Lutz, C., Mattei, M.G., Chenard, M.P., Lidereau, R., Basset, P., and Rio, M.C. 1995. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11–q21.3 region of Chromosome 17. Genomics 28: 367-376. [DOI] [PubMed] [Google Scholar]
- 49.Voullaire L., Wilton, L., Slater, H., and Williamson, R. 1999. Detection of aneuploidy in single cells using comparative genomic hybridization. Prenat. Diagn. 19: 846-851. [PubMed] [Google Scholar]
- 50.Walker G.T., Fraiser, M.S., Schram, J.L., Little, M.C., Nadeau, J.G., and Malinowski, D.P. 1992. Strand displacement amplification—An isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 20: 1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson R.H., Neville, P.J., Roy, W.J., Jr., Hitchcock, A., and Campbell, I.G. 1998. Loss of heterozygosity on Chromosomes 7p, 7q, 9p and 11q is an early event in ovarian tumorigenesis. Oncogene 17: 207-212. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y.H., Buckley, M.J., Dudoit, S., and Speed, T.P. 2002. Comparison of methods for image analysis on cDNA microarray data. Journal of Computational and Graphical Statistics 11: 108-136. [Google Scholar]
- 53.Yang Y.H., Dudoit, S., Luu, P., Speed, T.P., et al. 2001. Normalization for cDNA microarray data. In Microarrays: Optical Technologies and Informatics (ed. M.L. Bittner), pp. 31 SPIE, Society for Optical Engineering, San Jose, CA.
- 54.Yuen T., Wurmbach, E., Pfeffer, R.L., Ebersole, B.J., and Sealfon, S.C. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L., Cui, X., Schmitt, K., Hubert, R., Navidi, W., and Arnheim, N. 1992. Whole genome amplification from a single cell: Implications for genetic analysis. Proc. Natl. Acad. Sci. 89: 5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]