Abstract
Background & Aims
In children with liver failure, as many as half remain of indeterminate etiology. This hinders timely consideration of optimal treatment options. We posit that a significant subset of these children harbor known inherited metabolic liver diseases with atypical presentation or novel inborn errors of metabolism. We investigated the utility of whole-exome sequencing in three children with advanced liver disease of indeterminate etiology.
Methods
Patient 1 was a 10 year-old female diagnosed with Wilson disease but no detectable ATP7B mutations, and decompensated liver cirrhosis who underwent liver transplant and subsequently developed onset of neurodegenerative disease. Patient 2 was a full-term 2 day-old male with fatal acute liver failure of indeterminate etiology. Patient 3 was an 8 year-old female with progressive syndromic cholestasis of unknown etiology since age 3 months.
Results
Unbiased whole-exome sequencing of germline DNA revealed homozygous mutations in MPV17 and SERAC1 as the disease causing genes in patient 1 and 2, respectively. This is the first demonstration of SERAC1 loss-of-function associated fatal acute liver failure. Patient 1 expands the phenotypic spectrum of MPV17-related hepatocerebral mitochondrial DNA depletion syndrome. Patient 3 was found to have syndromic cholestasis due to bi-allelic NOTCH2 mutations.
Conclusions
Our findings validate the application of whole-exome sequencing in the diagnosis and management of children with advanced liver disease of indeterminate etiology, with the potential to enhance optimal selection of treatment options and adequate counseling of families. Moreover, whole-exome sequencing revealed hitherto unrecognized phenotypic spectrum of inherited metabolic liver diseases.
Keywords: inherited metabolic liver diseases, whole-exome sequencing, genetic diagnosis, liver failure of indeterminate etiology, germline mutations
INTRODUCTION
Liver failure whether acute or chronic is a life-threatening event leading to multi-organ dysfunction, that requires rapid clinical decision about pursuing appropriate treatment including consideration of liver transplantation, which is the ultimate effective treatment option. Pediatric liver failure is a rare disease, but the precise frequency of liver failure in children is unknown. In 1999, the Pediatric Acute Liver Failure (PALF) Study Group was formed with the goal of developing a database of all individuals in the United States (US) younger than 18 years of age who present with new onset of severe liver-related coagulopathy with or without concomitant encephalopathy. According to PALF study group data, approximately half of the cases of acute liver failure in children in the US still remain of indeterminate etiology despite comprehensive evaluation [1].
We posit that a significant subset of these children suffer from known inborn metabolic liver disorders with atypical presentations (caused by inherited and/or de novo mutations) or have novel inborn errors of metabolism that lead to liver failure. With increasing access to affordable and rapid whole-exome capture and high throughput DNA sequencing, individual genomic analysis can be used in routine clinical practice [2–3]. The exome corresponds to 1% of the human genome that represent coding regions translated into proteins, and it harbors ~ 85% of all disease-causing variants [4]. Recent technological advances in high-throughput DNA sequencing allow deep coverage of coding DNA sequence from as little as 0.5 μg of genomic high-quality (non-degraded) DNA. Hence, whole-exome sequencing (WES) currently presents an unprecedented balance between length of analysis, cost and information collected, making it very attractive and suitable for clinical use [2–3]. However, its successful application mandates exquisite phenotype annotation and requires adequate bioinformatics analysis to identify causal gene variant(s).
Here, we describe three children with liver failure of indeterminate etiology despite standard evaluation, and whose diagnoses were uncovered using WES. We found rare homozygous mutations in two genes, MPV17 and SERAC1, both expressed in mitochondria and implicated in mitochondrial disorders. Patient 1 revealed an atypical presentation of MPV17-related hepatocerebral mitochondrial DNA depletion syndrome (MDS); whereas patient 2 is the first report of fatal acute liver failure related to homozygous splice site mutation in the SERAC1 gene, a genetic defect that has been predominantly associated with neurological phenotype and more recently with transient liver dysfunction [5–6]. Patient 3 who suffers from progressive syndromic cholestasis of unclear etiology, was found to harbor novel compound heterozygous mutations in NOTCH2 gene in conserved amino acids positions, establishing a presumed diagnosis of Notch2-related Alagille-like syndrome.
This study provides further evidence to recommend the clinical utility of WES in diagnosing selected infants and children with life-threatening liver diseases of indeterminate etiology, supporting its introduction into the clinical armamentarium in the field of hepatology [7–9].
PATIENTS and METHODS
Human Subjects
Study protocol was approved by the Yale Human Investigation Committee, and informed consent was obtained in accordance with institutional review board standards. Three children with advanced liver disease of indeterminate etiology despite comprehensive evaluation underwent further analysis using whole-exome sequencing.
Exome Capture and Sequencing
DNA was extracted from peripheral blood total leukocytes by standard procedures. We used 1 microgram of genomic DNA per patient for exome capture and sequencing. DNA fragments containing exonic sequence were captured using the Roche/Nimblegen SeqCap EZ Human Exome Library v1 (for patient 1) or v2 (for patients 2 and 3), and sequenced on the Illumina HiSeq platform. Mean coverage of the exome was greater than 100X with 96% of the exome covered at least 8 times and 90% covered at greater than 20X. The resulting sequence was analyzed for single nucleotide variants and small insertions and deletions (indels) differing from the reference genome (Human Genome 19, HG19).
At our Institution, we are able to identify a genetic liver disease using WES within 7 days of starting genetic analysis at a cost of $1700 per Exome, including DNA extraction, sequencing, quality control plus data analysis and report writing.
Exome Sequencing Analysis
We used whole-exome sequencing analysis pipeline described earlier in [4]. Variant filtering strategy for patient 1, 2 and 3 is outlined in supplementary figures 1, 2 and 3, respectively. In brief, patient's variants passing quality filters were filtered against public (NHLBI, 1000 Genome, dbSNP) and in-house (Yale 2500 exomes) databases for minor allele frequency (MAF) < 1%. Then, protein-altering variants were selected. For patients 1 and 2, which are offspring of consanguineous families, homozygous variants were selected. Additionally, for patient 1, since both parents' exomes were sequenced, a variant found in homozygous state from parents was excluded. For patient 3, the offspring of unrelated parents, the protein-altering variants were selected if they were related to a disease listed in Online Mendelian Inheritance in Man (OMIM) database. The variants (supplementary tables 1, 2 and 3 for patients 1, 2 and 3 respectively) derived from the variant filtering strategy were then prioritized based on their likelihood to have damaging functions to the protein using public algorithms such as Polyphen-2 and SIFT and/or matching totally or partially the patient's phenotype. Variants identified by exome sequencing that were potentially diagnostic for the patient's condition were confirmed by Sanger sequencing as depicted in the main figures.
Orthologues
Full-length orthologous protein sequences from both vertebrate and invertebrates were obtained from GenBank. Protein sequences were aligned using the ClustalW algorithm.
RESULTS
Patient 1: Recessive MPV17 mutation in a child with clinical diagnosis of Wilson disease
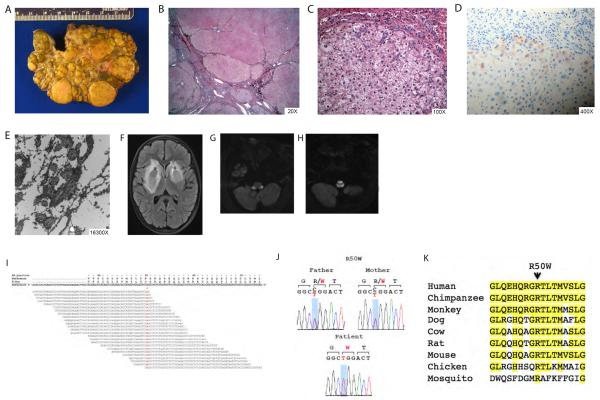
Patient 1 is the only child of consanguineous parents from India (table 1). She was in good state of health until age five years, when she developed incapacitating leg cramps, and was found to have abnormal liver function tests during work up. She was diagnosed with Wilson disease (WD) based on low serum ceruloplasmin (18mg/dL, normal range: 22–58mg/dL), elevated 24-hour urine copper excretion of 64 micrograms/24 hrs (normal range: 15–60 micrograms of urinary copper/24 hrs), and elevated liver copper content (468.1 mcg copper/gram liver dry weight (normal range = 10–35 mcg/g dry weight), which attributes a total score of 4 and establishes the clinical diagnosis of WD according to the scoring system adopted by the European Association for the Study of the Liver (EASL) clinical practice guidelines [10–11]. Pre-transplant liver biopsy showed mild micro and macrovesicular steatosis and copper stain shows hepatocellular accumulation (1–2/3+) predominantly in periportal areas with some staining within the lobules. However, sequencing of the coding regions of ATP7B gene did not reveal any disease causing mutations. She was started on copper-chelation therapy but progressed to end-stage liver disease (ESLD) with severe ascites, malnutrition and jaundice. During pre-transplant evaluation, the patient reported no new extra-hepatic signs or symptoms, and there were no ophthalmologic, renal or neurological findings. Baseline head computerized tomography (CT) was normal. She underwent uncomplicated deceased donor liver transplant for decompensated ESLD at nine years of age. Liver explant confirmed cirrhotic liver (figure 1A and 1B) and revealed two nodules of well-differentiated hepatocellular carcinoma of 1.9 and 1.5 cm of maximum dimension, which were incidentally found (figure 1C). In addition to multiple macronodules and dense fibrous septa, the explant showed focal steatosis, extensive oncocytic change, cholestasis, and focal areas of ballooning degeneration with Mallory's hyaline. There was heavy copper deposition in many of the hepatocytic nodules, predominantly in the periseptal hepatocytes (figure 1D). Electron microscopy revealed pleomorphic mitochondria with dilated cristae with occasional granular-dense deposits and crystalloid inclusion (figure 1E).
Table1.
Summary of the demographics, clinical features and genetic alterations of the three probands
| Patient Number |
Ethnicity (Consanguinity) |
Age of Onset |
Age at the time of transplant |
Age of Death/ Current Age |
Presenting Features |
Gene (NM_number) |
Mutation | Homozygous / Heterozygous |
Amino Acid or Nucleotide change (coding DNA nomenclature) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Indian (Yes) | 5 y | 9 y | 9 y / NA | bilateral lower extremities spasms and abnormal LFTs | MPV17 (NM_002437.4) | Missense | Homozygous | R50W (c.148T>C) |
| 2 | Pakistani (Yes) | 2 d | NA | 12 d / NA | shock, coagulopathy, lactic acidemia, hyperammonemia, elevated transaminases, hyperbilirubinemia and hypoalbuminemia | SERAC1 (NM_032861.3) | Splice site | Homozygous | IVS13+1OC (c.1403+1G>C) |
| 3 | Hispanic (No) | 3 m | NA | NA / 8 y | jaundice and pruritus | NOTCH2 (NM_024408.3) | Missense | Compound Heterozygous | S1741L (c.5222C>T) and H1882Y (c.5644C>T) |
y=years; d=days; m=months; NA=not applicable; LFTs=liver function tests.
Figure 1.
Liver histological, radiological and genetic findings in patient 1. (A and B) Liver explant with macroscopic nodular cirrhotic appearance at low and high (20x) power, respectively. (C) Well-differentiated hepatocellular carcinoma (100x). (D) Significant copper deposition in many of the hepatocytic nodules, predominantly in the periseptal hepatocytes (400x). (E) Pleomorphic mitochondria with dilated cristae with occasional granular-dense deposits and crystalloid inclusions. (F) Brain magnetic resonance imaging shows bilateral basal ganglia masses with surrounding edema as depicted by asterisks. (G and H) Representative images of brain magnetic resonance showing white matter abnormalities in the brain stem. (I) Missense homozygous mutation in MPV17 (R50W) detected by massively parallel sequencing. (J) Sanger sequence traces of codons 49–51 are shown. Patient 1 is homozygous for a single base change (CGG>TGG, in red) in MPV17, resulting in a R50W (c.148T>C) missense mutation. Proband's parents are heterozygous for the same mutation. (K) Conservation of R50 across species. The amino acid sequence of 42–58 of human MPV70 is shown and compared to the corresponding sequences of nine vertebrate and invertebrate orthologs and paralogs. Amino acid positions identical to human genome are highlighted in yellow.
Patient recovered rapidly after liver transplant and returned to school achieving high academic performance. However, nine months post-liver transplant, patient presented with new onset of dystonia and tremor. Brain magnetic resonance imaging (MRI) revealed new bilateral basal ganglia masses with surrounding edema (figure 1F). Initially, radiological findings were thought to be consistent with copper deposition in basal ganglia but her symptoms rapidly progressed to involve the white matter (figure 1G and 1H). She was found to have EBV viremia in blood and cerebral spinal fluid (CSF). Patient was treated for presumed central nervous system post-transplant lymphoproliferative disorder (not biopsy proven). Despite chemotherapy, patient developed progressive neurodegenerative disease, including seizures, which culminated with patient's death at ten-years of age. Autopsy was not performed in accordance with family wishes. WES of the proband and her parents revealed a homozygous missense mutation, R50W (c.148T>C), in MPV17 gene (NM_002437.4) (figure 1I), which was subsequently confirmed by Sanger sequencing (figure 1J). Both parents were heterozygous carriers for R50W variant (figure 1J). R50W occurs at an amino acid position that is 100% conserved between evolutionarily distant humans and flies (figure 1K). Upon review of the literature, the variant R50W has been previously reported in two patients, in one of the patients reported as a compound heterozygous [12] and in another patient in a homozygous state [13]. The R50W homozygous patient is a Hispanic girl who presented with failure to thrive at 8 months of age and died victim of liver failure with concomitant neurological symptoms when she was only 19 months old [13].
Patient 2: Neonatal Fatal Liver Failure in a patient with homozygous splice site mutation in SERAC1 gene
Patient 2 is the third child of consanguineous parents (first-degree cousins) born in Pakistan (table 1). The patient is a full term infant born via normal spontaneous vaginal delivery without any birth or immediate complications, with hospital discharge 32 hours after delivery. At birth, he weighed six pounds 10 ounces and pertinent birth history includes negative group B Streptococcus disease and negative serology. Few hours after discharge, patient was noted to be hypothermic, cyanotic and lethargic. In the Emergency Department, he was found to have a monomorphic wide complex tachycardia with weak pulses for which he required cardiac compressions, and ventilation and vasopressor support. Laboratory tests were remarkable for severe coagulopathy (INR of 7.37, normal range:0.8–1.15; and Partial Thromboplastin Time (PTT) > 120 sec, normal range: 23.2–31.6 sec), lactic acidemia (lactic acid of 7.0 mmol/L, normal range: 0.5–2.2 mmol/L), hyperammonemia (ammonia of 176 micromol/L, normal range: 11–35 micromol/L), elevated transaminases (AST of 322 U/L, normal range: 0–79 U/L; and ALT of 91 U/L, normal range: 0–34 U/L), hyperbilirrubinemia (total bilirubin of 6.08, normal is less than 1.20 mg/dL; and direct bilirubin of 0.80, normal range is less than 0.20 mg/dL) and hypoalbuminemia (albumin of 2.2 g/dL, normal range: 3.5–5.0 g/dL). Newborn screen revealed an elevated tyrosine level, but was thought not to be a primary disorder given the lack of reducing substances in the urine. At 3 days of age, qualitative urine analysis for organic acids was performed and revealed marked elevation of lactate, pyruvate and ketones. In addition, there was mild elevation of unrelated metabolites (3-hydroxyvaleric acid, 3-methyglucatonic acid, 2-ethy-3-hydroxypropionic acid and fumaric acid). The pattern was not diagnostic of any specific disorder. Genetic testing for known neonatal liver failure causing genes (FAH, ALDOB, ATP7B, GALT, GALE, GALK1, TRMU, SERPINA1, HPD) was unremarkable. Patient passed away on day 12 of life.
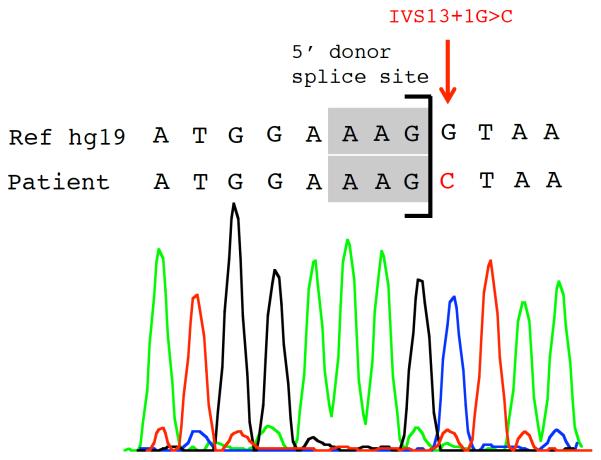
Post-mortem, high throughput WES revealed a homozygous splice site mutation (IVS13+1G>C, c.1403+1G>C) in SERAC1 (serine active site containing 1, NM_032861.3), which was confirmed by Sanger sequencing (figure 2). Interestingly, this patient had a two year-old brother who is alive, but had transient acute liver failure during his first few days of life that remained of unknown etiology. Applying WES, patient's brother was found to have the same homozygous splice site mutation (data not shown). SERAC1 encodes a phosphatidylglycerol remodeling protein that is essential for mitochondrial oxidative phosphorylation and intracellular cholesterol trafficking. This damaging mutation has been recently identified in two unrelated patients with type IV MEGDEL syndrome (Methylglutaconic Aciduria with sensori-neural Deafness, Encephalopathy and Leigh-like syndrome)[5] with predominant neurological phenotype and no reference of underlying liver dysfunction. One of the reported cases is a child of consanguineous parents originally from Pakistan, suggesting it may be a founder mutation.
Figure 2.
SERAC1 homozygous splice site mutation detected in patient 2 using WES. Sanger sequence trace of 5' donor splice site of codon 13 is shown. Patient 2 is homozygous for a base change (G>C, red arrow) in SERAC1, resulting in homozygous splice site mutation: IVS13+1G>C (c.1403+1G>C).
Patient 3: A case of chronic cholestasis and NOTCH2 gene mutation
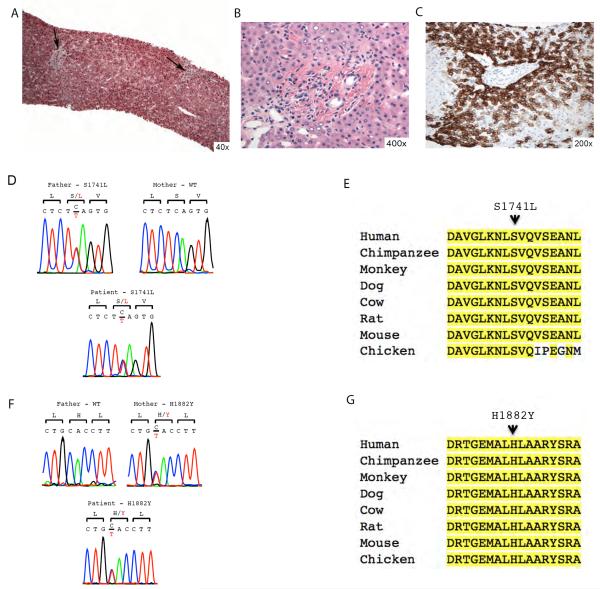
Patient 3 is the third child of non-consanguineous Puerto Rican parents. She is a full term infant, born from uneventful spontaneous vaginal delivery. She was healthy up to three months of age when she first presented with jaundice. Liver biopsy revealed bile duct paucity (figure 3B and 3C) in background of liver parenchyma with preserved lobular architecture and lack of significant fibrosis (figure 3A), suggestive of Alagille's syndrome. She is currently an 8 year-old girl with known bile duct paucity, progressive cholestasis, short stature (persistently on the third percentile for height), and dysmorphic facies. Evaluation for classic phenotypic features of Alagille syndrome was negative, namely cardiac, vertebral or ophthalmological abnormalities. Moreover, full JAG1 DNA sequencing was negative for disease-causing mutations. There is no family history of liver disease, including two healthy older siblings, a 15 year-old brother and a 10 year-old sister. She developed progressive hyperbilirubinemia and liver fibrosis, and she is currently awaiting liver transplant. WES revealed novel compound heterozygous variants in NOTCH2 (NM_024408.3), one inherited from her father (S1741L, c.5222C>T) and the other from her mother (H1882Y, c.5644C>T) (figure 3D and 3F). Both mutation variants are in the NOTCH2 intracellular domain, in amino acid residues that are conserved across several species (figure 3E and 3G), and probably damaging according to Polyphen-2 prediction scores. The clinical, histological and genetic findings together, suggest it is most likely that this patient has Notch2-related Alagille-like syndrome.
Figure 3.
Liver histological findings and NOTCH2 compound heterozygous mutations detected in patient 3. (A) Low magnification showing preserved lobular architecture of the liver parenchyma and lack of significant fibrosis (Trichrome stain, 40x). The portal tracts (arrows) seen at this magnification fail to show either significant inflammation or any obvious pathology. (B) Higher magnification (400x) of the portal tract to show absence of the bile duct and also any lack of ductular reaction. Occasional lymphocytes or histocytes can be seen in the portal tract. Few glycogenated hepatocytic nuclei can be seen in the periportal region (H&E stain). (C) Immunostain for CK7 highlights the absence of the native bile duct and shows intense staining of the periportal hepatocytes (200x). (D) Sanger sequence trace of codons 1720–1722 are shown. Patient 3 is heterozygous for a base change (TCA>TTA, in red) in NOTCH2, resulting in a S1741L (c. 5222C>T) missense heterozygous mutation. Proband's father is heterozygous for the same mutation whereas proband's mother is wild-type (WT). (E) Conservation of S1741 across species. The amino acid sequence of 1733–1749 of human NOTCH2 is shown and compared to the corresponding sequences of eight vertebrate orthologs and paralogs. Amino acid positions identical to human genome are highlighted in yellow. (F) Sanger sequence trace of codons 1881–1883 are shown. Patient 3 is heterozygous for a base change (CAC>TAC, in red) in NOTCH2, resulting in a H1882Y (c.5644C>T) missense heterozygous mutation, Proband's mother is heterozygous for the same mutation whereas proband's father is wild-type (WT). (G) Conservation of H1882Y across species. The amino acid sequence of 1874–1890 of human NOTCH2 is shown and compared to the corresponding sequences of eight vertebrate orthologs and paralogs. Amino acid positions identical to human genome are highlighted in yellow. H1882 is conserved among all species examined.
DISCUSSION
It was only in 2009, that the potential utility of WES in establishing a genetic diagnosis of an unanticipated congenital disease was demonstrated [4, 15]. In a short time, significant technological advances had occurred and WES is now available in the clinical setting at affordable cost and with the results available in a timely manner. Hence, WES is a powerful clinical diagnostic tool to investigate selected clinical conundrums that have remained unsolved, as exemplified by our cases. Children and young adults with advanced liver disease represent a population enriched for underlying inherited disorders. Several algorithms for biochemical testing to facilitate genetic diagnosis of common inherited diseases have been developed, including a `jaundice chip' that screens for five common genes and known mutations [16–17]. Thus, there is a significant unmet need in our diagnostic armamentarium that impacts directly on management of these patients. Our report highlights impressive potential of applying unbiased exomic analysis in evaluating these patients, predicting a new Era where hepatologists and the field of Pediatric Hepatology will fully embrace this technology.
Patient 1 highlights the limitations of current algorithms for diagnosis of Wilson disease. The patient fulfilled criteria for diagnosis of Wilson disease per EASL and American Association for the Study of Liver Diseases (AASLD) clinical practice guidelines [11, 18] but did not harbor ATP7B gene mutations. It is highly relevant in this regard that in several series, it has been reported that approximately 15% of patients with clinical and biochemical phenotype of WD do not have detectable ATP7B mutations [8]. The diagnosis of WD in patient 1 was questioned as her disease phenotype unfolded following successful liver transplant; in particular the finding of HCC in liver explant and neurodegenerative disease post-liver transplant. WES revealed a homozygous germline defect consistent with a diagnosis of MPV17-related hepatocerebral MDS. MPV17 encodes a mitochondrial inner-membrane protein of unknown function. Bi-allelic mutations in MPV17 underlie hepatocerebral MDS. To date, some 35 affected individuals have been described in the literature with MPV17-related hepatocerebral MDS. Combined data from these cases show that all had liver dysfunction, and 90% developed liver failure during infancy or early childhood. A quarter of the patients were found to have liver cirrhosis and two patients developed hepatocellular carcinoma [19]. Patient 1 did not present with lactic acidosis or early-onset hypoglycemia, which occur in approximately 75% and 50% of patients suffering from MPV17-related Hepatocerebral MDS, respectively. Approximately 90% of affected individuals have neurological symptoms, which includes developmental delay (83%), hypotonia and muscle weakness (66%), white matter abnormalities in brain MRI (34%), peripheral neuropathy (28%), and seizures, microcephaly and ataxia in less than 10% of the cases [19]. Most other multisystem mitochondrial disorders with prominent neuromuscular involvement, exhibit liver dysfunction as late feature in the natural history; in contrast, in MPV17-related hepatocerebral MDS, liver disease occurs early in the natural history. The mutation identified in our proband, R50W, has only been reported in two other affected individuals, one in an equivalent homozygous state [13] and the other as a compound heterozygous [12]. Interestingly, the only other case of R50W homozygosity in the literature had a much earlier clinical presentation with failure to thrive (at eight months of age) and rapid progression (died at 19 months of age from liver failure) compared to our patient. If the correct diagnosis was made prior to liver transplantation, the procedure would not have been undertaken due to high likelihood of development of neurological disease.
Interestingly, MPV17-related MDS phenocopies WD mitochondrial membrane crosslinking and mitochondrial destruction as well as oxidative-phosphorylation defects [20–21]; notably deletions of mitochondrial DNA have been described in ATP7B deficiency [22]. Thus, our findings suggest genetic heterogeneity in patients with phenotypic findings of WD, and the utility of WES to investigate patients with Wilsonian liver disease but no ATP7B mutations. Taken together, our findings suggest that MPV17 gene should be examined in patients with WD phenotypes and ATP7B mutation negative, and raises the possibility that MPV17 may be involved in regulation copper homeostasis.
Genetic analysis in patient 2 uncovered an uncharacterized clinical presentation of fatal liver failure in patients with homozygous splice site mutation in SERAC1 gene. Using an mRNA transcript analysis, Wortmann and colleagues demonstrated that exon 13 splice site mutations resulted in skipping of this exon, which encodes part of the lipase domain of SERAC1 protein [5]. While preparing this manuscript, Sarig and colleagues reported the first four cases of infantile hepatopathy in patients with decreased or absent expression of SERAC1, a gene which was previously thought to be exclusively associated with a neurological phenotype. Two novel homozygous mutations in SERAC1, distinct from the one detected in our patient, have been identified in two pairs of brothers of two consanguineous unions who presented with hypotonia and transient liver failure within the first 48 hours of life [14]. In contrast to our patient 2 who succumbed to liver failure, the four patients reported by Sarig et al had transient liver dysfunction in infancy that resolved spontaneously. Thus, our case broadens the genotype – phenotype correlation for patients with SERAC1 loss of function mutations and supports the suggestion recently made by Sarig and colleagues to rename this rare disorder to MEGDHEL (Methylglutaconic Aciduria with sensori-neural Deafness, Hepatopathy, Encephalopathy and Leigh-like) syndrome in order to reflect liver involvement [14].
Patients 1 and 2 had unsuspected mitochondrial hepatopathies which were only diagnosed using unbiased WES. Thus, the sequencing of the entire mitochondrial DNA, in addition to the exons of 1034 nuclear genes encoding mitochondrial proteins, also known as MitoExome [23], might also be considered in the investigation of children with liver failure of indeterminate etiology. However, WES has potential to reveal other etiologies of liver disease as illustrated by patient three. In contrast to patient 1 and 2, patient 3 was an offspring of non-consanguineous union and the case illustrates utility of WES, even in this more complex setting [24].
This proband has an atypical presentation of advanced liver disease with bile duct paucity, cholestasis, short stature and dysmorphic facies, suggestive of Alagille-like syndrome, but with negative JAG1 gene mutations. Alagille syndrome is thought to follow an autosomal dominant inheritance associated with haploinsufficiency, and with known incomplete penetrance [25]. Mutations in JAG1 gene are found in 94% of cases of Alagille syndrome. In <1% of cases, mutations in JAG1 receptor, NOTCH2 gene, had been described and associated with disease [26]. Interestingly, patient 3 was found to have novel NOTCH2 compound heterozygous mutations. Each parent carries one of the variants, in the absence of a liver phenotype. However, as the parents were evaluated for living liver donors, these genetic alterations in NOTCH2 will likely negatively impact their donor candidacy. Kamath and colleagues [26] also reports one patient with cholestasis, bile duct paucity and cataracts with two NOTCH2 missense variants. Otherwise, the remaining cases of Notch2-related Alagille syndrome reported in the literature show heterozygous NOTCH2 mutations. All NOTCH2 mutations related to Alagille syndrome are unique and are found across the entire gene. Previous study suggested that the spectrum of clinical features associated with NOTCH2 mutations differs from JAG1 mutations, with lower frequency of vertebral abnormalities and facial features [26], which is only partially concordant with our case. Similar to the study by Kamath et al. 2012, Marchetti and Iascone report two siblings affected by severe cholestasis who were found to have NOTCH2 frameshit mutations in exon 18, but who did not show any cardiac disease, skeletal, ocular abnormalities or facies typical of ALGS. Thus, in accordance to both studies from Kamath et al [26], and Marchetti and Iascone, our study provides further clinical and genetic evidence that classical clinical diagnostic criteria no longer capture the heterogeneity of this disorder, and a genetic and clinical redefinition of Alagille syndrome is warranted.
As WES is less expensive and data is generated and analyzed in a timely fashion, its use lends itself to further investigate cases who fulfill partial or full classical criteria of Alagille syndrome [27]. Identification of gene variants at the exome level has the potential to provide valuable information to improve our understanding of this heterogeneous disorder. For example, in murine models the same mutation may have distinct phenotypes, depending on genetic backgrounds [28]. Thus, similar findings likely occur in humans, and therefore a comprehensive characterization and integrative analysis of individual exomes has the potential to enhance our understanding of the impact of single nucleotide variants in clinical phenotype.
In summary, we provide evidence to recommend the utility of WES in diagnosing patients with severe liver disease of indeterminate etiology. This study is a proof of concept that inborn errors of metabolism will likely feature more prominently in clinical practice with greater access to WES in the clinic. Hence, WES may help improve diagnostic and management algorithms in rare instances of unexplained acute and chronic liver disease in children. Our data also generate new information on genotype-phenotype correlation, and it is expected that phenotypic spectrum of inherited metabolic liver diseases will expand beyond current understanding with this technology.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients and their families who contribute to the evolving understanding of liver disease. This study was funded by Yale Center for Mendelian Diseases, NHGRI/NHLBI U04HG.006504 (RPL) and K24 DK066306 (PKM).
List of Abbreviations
- WES
whole exome sequencing
- PALF
Pediatric Acute Liver Failure
- US
United States
- MDS
mitochondrial DNA depletion syndrome
- WD
Wilson Disease
- EASL
European Association for the Study of the Liver
- CT
computerized tomography
- MRI
magnetic resonance imaging
- PTT
Partial Thromboplastin Time
- MEGDEL syndrome
Methylglutaconic Aciduria with sensori-neural Deafness, Encephalopathy and Leigh-like syndrome
- AASLD
American Association for the Study of Liver Diseases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None.
REFERENCES
- [1].Squires RH, Jr., Shneider BL, Bucuvalas J, Alonso E, Sokol RJ, Narkewicz MR, et al. Acute liver failure in children: the first 348 patients in the pediatric acute liver failure study group. The Journal of pediatrics. 2006;148:652–658. doi: 10.1016/j.jpeds.2005.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical Whole-Exome Sequencing for the Diagnosis of Mendelian Disorders. The New England journal of medicine. 2013 doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jacob HJ, Abrams K, Bick DP, Brodie K, Dimmock DP, Farrell M, et al. Genomics in clinical practice: lessons from the front lines. Science translational medicine. 2013;5 doi: 10.1126/scitranslmed.3006468. 194cm195. [DOI] [PubMed] [Google Scholar]
- [4].Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wortmann SB, Vaz FM, Gardeitchik T, Vissers LE, Renkema GH, Schuurs-Hoeijmakers JH, et al. Mutations in the phospholipid remodeling gene SERAC1 impair mitochondrial function and intracellular cholesterol trafficking and cause dystonia and deafness. Nature genetics. 2012;44:797–802. doi: 10.1038/ng.2325. [DOI] [PubMed] [Google Scholar]
- [6].Wortmann S, Rodenburg RJ, Huizing M, Loupatty FJ, de Koning T, Kluijtmans LA, et al. Association of 3-methylglutaconic aciduria with sensori-neural deafness, encephalopathy, and Leigh-like syndrome (MEGDEL association) in four patients with a disorder of the oxidative phosphorylation. Molecular genetics and metabolism. 2006;88:47–52. doi: 10.1016/j.ymgme.2006.01.013. [DOI] [PubMed] [Google Scholar]
- [7].Sambrotta M, Strautnieks S, Papouli E, Rushton P, Clark BE, Parry DA, et al. Mutations in TJP2 cause progressive cholestatic liver disease. Nature genetics. 2014;46:326–328. doi: 10.1038/ng.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cnossen WR, te Morsche RH, Hoischen A, Gilissen C, Chrispijn M, Venselaar H, et al. Whole-exome sequencing reveals LRP5 mutations and canonical Wnt signaling associated with hepatic cystogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5343–5348. doi: 10.1073/pnas.1309438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Casey JP, McGettigan P, Lynam-Lennon N, McDermott M, Regan R, Conroy J, et al. Identification of a mutation in LARS as a novel cause of infantile hepatopathy. Molecular genetics and metabolism. 2012;106:351–358. doi: 10.1016/j.ymgme.2012.04.017. [DOI] [PubMed] [Google Scholar]
- [10].Ferenci P, Caca K, Loudianos G, Mieli-Vergani G, Tanner S, Sternlieb I, et al. Diagnosis and phenotypic classification of Wilson disease. Liver international : official journal of the International Association for the Study of the Liver. 2003;23:139–142. doi: 10.1034/j.1600-0676.2003.00824.x. [DOI] [PubMed] [Google Scholar]
- [11].EASL Clinical Practice Guidelines: Wilson's disease. Journal of hepatology. 2012;56:671–685. doi: 10.1016/j.jhep.2011.11.007. [DOI] [PubMed] [Google Scholar]
- [12].Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D'Adamo P, Calvo S, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nature genetics. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- [13].Wong LJ, Brunetti-Pierri N, Zhang Q, Yazigi N, Bove KE, Dahms BB, et al. Mutations in the MPV17 gene are responsible for rapidly progressive liver failure in infancy. Hepatology. 2007;46:1218–1227. doi: 10.1002/hep.21799. [DOI] [PubMed] [Google Scholar]
- [14].Sarig O, Goldsher D, Nousbeck J, Fuchs-Telem D, Cohen-Katsenelson K, Iancu TC, et al. Infantile mitochondrial hepatopathy is a cardinal feature of MEGDEL syndrome (3-Methylglutaconic aciduria type IV with sensorineural deafness, encephalopathy and leigh-Like Syndrome) caused by novel mutations in SERAC1. American journal of medical genetics Part A. 2013;161:2204–2215. doi: 10.1002/ajmg.a.36059. [DOI] [PubMed] [Google Scholar]
- [15].Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu C, Aronow BJ, Jegga AG, Wang N, Miethke A, Mourya R, et al. Novel resequencing chip customized to diagnose mutations in patients with inherited syndromes of intrahepatic cholestasis. Gastroenterology. 2007;132:119–126. doi: 10.1053/j.gastro.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Balistreri WF. Inherited disorders of bile Acid transport or synthesis. Gastroenterology & hepatology. 2007;3:343–345. [PMC free article] [PubMed] [Google Scholar]
- [18].Roberts EA, Schilsky ML. Diagnosis and treatment of Wilson disease: an update. Hepatology. 2008;47:2089–2111. doi: 10.1002/hep.22261. [DOI] [PubMed] [Google Scholar]
- [19].El-Hattab AW, Scaglia F, Craigen WJ, Wong LJC. MPV17-Related Hepatocerebral Mitochondrial DNA Depletion Syndrome. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews. Seattle (WA): 1993. [Google Scholar]
- [20].Zischka H, Lichtmannegger J, Schmitt S, Jagemann N, Schulz S, Wartini D, et al. Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease. The Journal of clinical investigation. 2011;121:1508–1518. doi: 10.1172/JCI45401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gu M, Cooper JM, Butler P, Walker AP, Mistry PK, Dooley JS, et al. Oxidative-phosphorylation defects in liver of patients with Wilson's disease. Lancet. 2000;356:469–474. doi: 10.1016/s0140-6736(00)02556-3. [DOI] [PubMed] [Google Scholar]
- [22].Mansouri A, Gaou I, Fromenty B, Berson A, Letteron P, Degott C, et al. Premature oxidative aging of hepatic mitochondrial DNA in Wilson's disease. Gastroenterology. 1997;113:599–605. doi: 10.1053/gast.1997.v113.pm9247482. [DOI] [PubMed] [Google Scholar]
- [23].Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Science translational medicine. 2012;4:118ra110. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, et al. Sequencing studies in human genetics: design and interpretation. Nature reviews Genetics. 2013;14:460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Turnpenny PD, Ellard S. Alagille syndrome: pathogenesis, diagnosis and management. European journal of human genetics : EJHG. 2012;20:251–257. doi: 10.1038/ejhg.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, et al. NOTCH2 mutations in Alagille syndrome. Journal of medical genetics. 2012;49:138–144. doi: 10.1136/jmedgenet-2011-100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vozzi D, Licastro D, Martelossi S, Athanasakis E, Gasparini P, Fabretto A. Alagille Syndrome: A New Missense Mutation Detected by Whole-Exome Sequencing in a Case Previously Found to Be Negative by DHPLC and MLPA. Molecular syndromology. 2013;4:207–210. doi: 10.1159/000347231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Erickson RP. Mouse models of human genetic disease: which mouse is more like a man? BioEssays : news and reviews in molecular, cellular and developmental biology. 1996;18:993–998. doi: 10.1002/bies.950181209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.