Abstract
The rapid evolution of centromere sequences between species has led to a debate over whether centromere activity is sequence-dependent. The Arabidopsis thaliana centromere regions contain ∼20,000 copies of a 178-bp satellite repeat. Here, we analyzed satellites from 41 Arabidopsis ecotypes, providing the first broad population survey of satellite variation within a species. We found highly conserved segments and consistent sequence lengths in the Arabidopsis satellites and in the published collection of human α-satellites, supporting models for a functional role. Despite this conservation, polymorphisms are significantly enriched at some sites, yielding variation that could restrict binding proteins to a subset of repeat monomers. Some satellite regions vary considerably; at certain bases, consensus sequences derived from each ecotype diverge significantly from the Arabidopsis consensus, indicating substitutions sweep through a genome in less than 5 million years. Such rapid changes generate more variation within the set of Arabidopsis satellites than in genes from the chromosome arms or from the recombinationally suppressed centromere regions. These studies highlight a balance between the mechanisms that maintain particular satellite domains and the forces that disperse sequence changes throughout the satellite repeats in the genome.
[Supplemental material is available online at www.genome.org.]
The large heterochromatic constrictions known as centromeres play many roles in multicellular eukaryotes, holding sister chromatids together during the early stages of mitosis and, at later stages, assembling the kinetochores that bind to microtubules and mediate chromosome separation. These roles are conserved across eukaryotes, yet the DNA sequences that mediate centromere function have remained undefined in many cases. Overall, the sequence composition of the centromere region varies considerably across species, raising the possibility that epigenetic modifications, and not DNA sequence per se, govern centromere function (Choo 2000; Henikoff et al. 2001). Genetically, the DNA sequence composition of a centromere is defined as the portion of homologous chromosomes that segregate to opposite poles in meiosis I. The boundaries of such genetic intervals are defined by recombination events, and these intervals are often large, given the limited recombination in the centromere regions. In Arabidopsis, the genetically defined centromere regions contain large satellite arrays comprised of thousands of copies of ∼180-bp repeats (Martinez-Zapater et al. 1986; Copenhaver et al. 1999). We examined the patterns of satellite sequence evolution across Arabidopsis populations and found striking conservation of repeat sequence length as well as significantly conserved and variable regions within the repeats. We applied the same analysis to the previously published collections of human α-satellite DNA (Choo et al. 1991), finding similar patterns, albeit a higher degree of sequence variation. The presence of satellite domains with strikingly different rates of nucleotide substitution strongly indicates a sequence-dependent role for Arabidopsis centromere satellites.
Direct evidence of sequence-based centromere function comes from the budding yeast Saccharomyces cerevisiae, in which centromere function was first genetically defined by tetrad analysis, and subsequently reduced to a minimal functional region using minichromosomes (Clarke and Carbon 1980; Cottarel et al. 1989). In this species, the minimal DNA sequence necessary and sufficient to confer all centromere functions is only 125 bp in length. These centromeres are present on every chromosome and contain three conserved DNA elements (CDE): the 5′-CDEI (8 bp), a central, A + T-rich CDEII (78–86 bp), and a 3′-CDEIII (25 bp; Cottarel et al. 1989). Different protein complexes assemble on each of the CDE regions; mutations in either the CDE or various centromere-binding proteins reduce the efficiency of chromosome segregation (Clarke 1998). Although many of the proteins that assemble on the centromere DNA of yeast are well understood, the machinery that mediates attachment of these regions to microtubules for chromosome segregation remains unclear.
The centromere regions of budding yeast lack repetitive DNA, yet most other eukaryotes examined, including the fission yeast Schizosaccharomyces pombe, have numerous repeats at the centromere. Typically, eukaryotic centromere regions contain repeat units ranging in length from ∼150 to ∼210 bases, approximately the length required to form a single nucleosome (Henikoff et al. 2001). In humans, the centromere-specific α-satellites are AT-rich 171-bp repeats, tandemly arrayed in a head-to-tail arrangement. They are sufficiently variable to allow classification into distinct chromosome-specific subfamilies (Waye and Willard 1987; Choo et al. 1991). Synthetic minichromosomes that contain α-satellite arrays recruit essential centromere-binding proteins and are transmitted through mitosis, indicating that α-satellite arrays are sufficient to confer centromere function in human cell lines (Willard 1998, 2001; Yang et al. 2000; Schueler et al. 2001; Grimes et al. 2002). Neocentromeres that completely lack α-satellite DNA also have been characterized (DuSart et al. 1997; Barry et al. 2000). These centromeres form at a low frequency following disruption of a natural centromere, and indicate that, under some circumstances, α-satellite sequences are not necessary for centromere function.
Human α-satellite sequences share 90% identity with those from gorilla, chimpanzee, or orangutan (Durfy and Willard 1990; Baldini et al. 1991; Haaf and Willard 1998), and chimpanzee and gorilla satellites, like those of human, are organized into higher-order arrays (Durfy and Willard 1990; Baldini et al. 1991; Haaf and Willard 1998). Human satellites can acquire centromere activity when introduced into African green monkey cells (Haaf et al. 1992), indicating either that the features required for centromere function are conserved between the two species or that the satellite DNA sequence itself is unimportant. Despite this functional conservation, there is considerable divergence in array content between primate species (Waye and Willard 1989; Durfy and Willard 1990; Warburton and Willard 1990; Haaf and Willard 1997, 1998). For example, chromosome-specific α-satellites have been reported in humans, yet homologous primate chromosomes generally do not share the same satellite subfamilies (Haaf and Willard 1997, 1998).
Because satellites are present in thousands of copies, their divergence between species would require genome-wide homogenization, a process known as molecular drive (Dover 1982). Several mechanisms that could account for this homogenization have been postulated, including gene conversion and unequal crossing over (Smith 1976; Dover 1982; Stephan 1986; Charlesworth et al. 1994). In one model, the ancestor of closely related species contained a “library” of satellite variants within its genome, and as new species emerged, one satellite was predominately used as a template, resulting in the conversion of the other genomic copies (Mestrovic et al. 1998). Although it is attractive to postulate that selection could drive the choice of one satellite over others, chance could also account for biased amplification (Dover 1982; Nijman and Lenstra 2001). The continuous homogenization of satellite sequences within a genome can lead to a smaller within-species variation than between-species variation, an observation known as concerted evolution (Elder and Turner 1995).
To date, the diversification of satellites has been measured between closely related species, but not between the populations of an individual species. To provide a detailed understanding of the initial steps of satellite divergence, we characterized the satellites in geographically separated Arabidopsis thaliana populations (accessions or ecotypes). The genome sequencing project for the Columbia ecotype of Arabidopsis (The Arabidopsis Genome Initiative 2000) provided >5 Mb of assembled sequence from the genetically defined centromere intervals, and the unsequenced gaps within each centromere region are thought to be comprised primarily of satellite repeats (The Arabidopsis Genome Initiative 2000; Kumekawa et al. 2000, 2001; Hosouchi et al. 2002). The sequenced regions that flank these gaps contain numerous repetitive elements including retroelements, transposons, microsatellites, middle repetitive DNA, and tandemly organized satellites. The satellite repeats are not found elsewhere in the genome, but are restricted to the genetically defined centromere regions (Copenhaver et al. 1999; The Arabidopsis Genome Initiative 2000). Heslop-Harrison et al. (1999) analyzed 20 centromere satellite sequences from the Columbia ecotype and reported an ecotype-specific consensus, noting two regions of >99% conservation. By examining the sequence of satellites from other ecotypes, we explored whether the previously defined consensus could be extended to the species as a whole.
The Brassicaceae family, of which Arabidopsis is a member, has expanded from a common ancestor to 3350 species within a time frame of ∼40–50 million years (Al-Shehbaz 1984; Koch et al. 2001). Satellites homologous to the Arabidopsis 180-bp repeats have been discovered in other members of the Brassicaceae (Hallden et al. 1987), indicating that centromere satellites existed in a common ancestor. As described below, the satellite repeats from Arabidopsis are evolving rapidly among isolated populations, yet they contain highly conserved motifs. These studies set the stage for comparing satellite evolution patterns among the thousands of available species in the Brassicaceae family; identification of broadly conserved domains would imply possible selection for specific DNA sequence motifs.
RESULTS
Collection of Centromere Satellite Sequences From 41 Ecotypes
We used Polymerase Chain Reaction (PCR) to clone at least 10 satellite repeat sequences from each of 41 Arabidopsis ecotypes (Table 1); 457 clones were sequenced, resulting in 1029 whole or partial repeat sequences (GenBank accession nos. AF494837–AF495294). To reduce potential bias introduced by a particular PCR primer, two different PCR amplifications were performed for each ecotype using nonoverlapping primer sets (Fig. 1; see Methods). For most ecotypes, significant sequence differences were not detected between these amplifications; however in C24, Est-0, Mv-0, and Nok-0, the two primer sets amplified different repeat classes that varied at 4, 7, 12, and 9 sites, respectively. The satellite repeats were aligned using the location of the HindIII restriction site as the arbitrary beginning of the repeat. In total, the satellite repeats have an A + T content of 62.5%, similar to the genomic average of 65.1% (The Arabidopsis Genome Initiative 2000).
Table 1.
Arabidopsis Ecotypes Analyzed
| Name | Geographic origin | Name | Geographic origin |
| Aa-0 | Aua/Rhon, FRG. | Kil-0 | Killean, UK |
| Ba-1 | Blackmount, UK | Kin-0 | Kindalville, MI, USA |
| Bl-1 | Bologna, Italy | Kn-0 | Kaunas, Lithuania |
| Blh-1 | Bulhary, Czechoslovakia | Lc-0 | Loch Ness, GB. |
| C24 | common background line | Ler | Landsberg, FRG. |
| Can-0 | Canary Islands | Lip-0 | Lipowiec/Chrzanow, Poland |
| Chi-0 | Chisdra, Russai | Lu-1 | Lund, Sweden |
| Cl-0 | unknown | Ma-0 | Marburg (Lahn), FGR. |
| Col-0 | Columbia, MO, USA | Mt-0 | Martuba (Cyrenaika), Libya |
| Cvi-0 | Cape Verde Islands, Portugal | Mv-0 | Martha's Vineyard, MA, USA |
| Di-0 | Dijon, France | Nie-0 | Niederlauken/Ts., FRG. |
| Edi-0 | Edinburgh, GB. | Nok-0 | Noordwijk, Netherlands |
| Ei-5 | Eifel, Germany | Oy-0 | Oystese, Norway |
| En-2 | Enkheim/Frankfurt, FRG. | Pla-1 | Playa de Aro, Spain |
| Est-0 | Estland, Russia | Pog-0 | Point Grey, B.C., Canada |
| Go-2 | Gottingen, FRG. | Ri-0 | Richmond, B.C., Canada |
| Gr-1 | Graz, Austria | Ta-0 | Tabor, Czechoslovakia |
| Gre-0 | Greenville, MI, USA | Tsu-0 | Tsu, Japan |
| Hi-0 | Hilversum, Netherlands | Ws-0 | Wassilewskija/Dnjepr, USSR. |
| Kas-1 | Kashmir, India | Yo-0 | Yosemite National Park, USA |
| Kz-8 | Kazakhstan |
Figure 1.
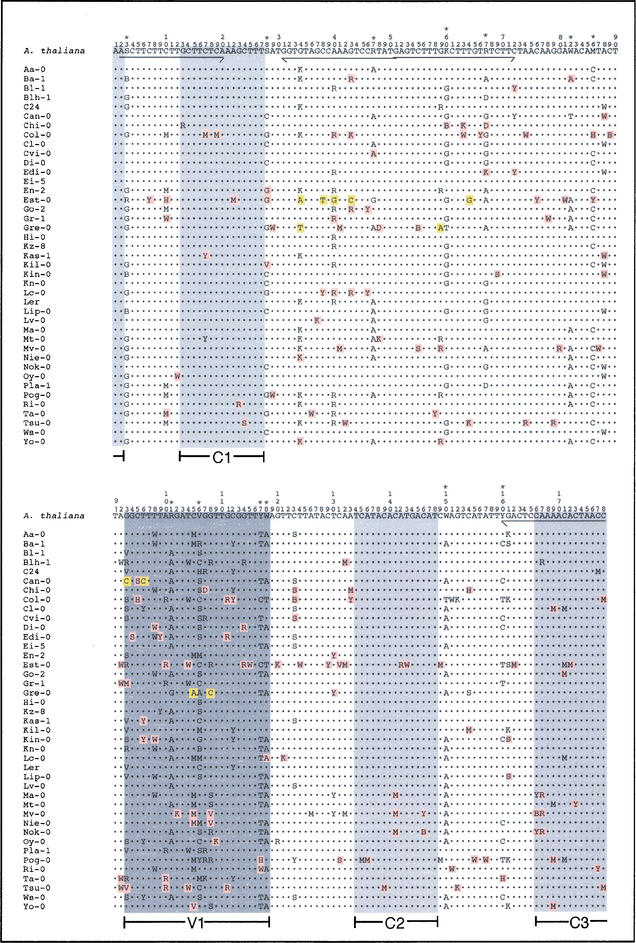
Satellite repeat consensus sequences from 41 Arabidopsis ecotypes. (Upper row) The overall consensus for the species; (asterisks) sites defined as polymorphic. Consensus sequences derived for each ecotype are indicated; (dots) identical nucleotides; (shading) nucleotide changes that are significantly different from the species consensus (χ2 test, P < 0.01); (pink) changes observed in multiple ecotype consensus sequences; (yellow) changes uniquely found in one ecotype consensus. Arrows below the species consensus indicate the primers (F2, R2 and F4, R4) used to amplify the repeats; (gray shading) conserved (C1, C2, C3) and variable (V1) regions (see also Fig. 3).
Based on migration through agarose gels, the Arabidopsis satellites were originally termed 180-bp repeats (Martinez-Zapater et al. 1986); sequence analysis of the Arabidopsis satellite repeats instead showed a mean length of 178 bp. Of the repeats we examined, 72% were 178 bp, 18% were 177 bp, and 8% were 179 bp. In addition, three outliers were observed at 176 bp, 182 bp, and 192 bp; the insertion and deletion events that gave rise to these variants differed in size and were scattered throughout the repeats. Consensus sequences derived for each ecotype were also 178 bp, demonstrating that repeat length is conserved across populations (Fig. 1). Similarly, analysis of α-satellite repeat length in primates has shown that α-satellite monomers are fairly constant in length, varying from 168 bp to 172 bp among species (Durfy and Willard 1990; Baldini et al. 1991; Fanning et al. 1993; Alves et al. 1994; Warburton et al. 1996; Haaf and Willard 1997, 1998).
Consensus Satellite Sequences From Individual Arabidopsis Ecotypes and the A. thaliana Species
We derived a consensus for the satellite repeat sequences from each Arabidopsis ecotype (Fig. 1), defining a consensus nucleotide as the base that occurs three times more often than any other at a given site; this definition was previously used to derive a consensus for α-satellite DNA (Waye and Willard 1987). In those cases in which these criteria were not met, the site was noted as polymorphic and the most predominant bases were indicated by the standard IUPAC symbols (Fig. 1). Next, the set of sequences from the 41 ecotypes was compiled to derive a consensus for the species; 13 of the 178 nucleotides comprising the repeat consensus were defined as polymorphic (Fig. 1, asterisks). These polymorphisms were also observed within individual ecotypes, indicating that they predate ecotype divergence. A consensus was previously reported from ∼20 satellite sequences from the Columbia ecotype (Heslop-Harrison et al. 1999); the consensus we defined for Arabidopsis differs at 15 sites, 13 of which reflect bases that are commonly polymorphic within the species.
Interestingly, there were notable sequence differences between the consensus derived for the species as a whole and the consensus of individual ecotypes, indicating rapid divergence within the past ∼5 million years (Koch et al. 2001). We used a χ2 test to identify those substitutions that are significantly different from the species consensus (Fig. 1, shading). In some cases, deviations were uniquely present in one ecotype consensus (Fig. 1, 11 cases, yellow); such substitutions were observed in Est-0, Gre-0, and Can-0. The substitutions seen in these ecotypes provide evidence for homogenization of new mutations across the genome in a short time frame. Termed molecular drive, such homogenization processes serve to increase the relative abundance of a particular variant; they can result from selective forces or as a consequence of random chance (Dover 1982). All of the substitutions observed in the consensus sequences for Est-0, Gre-0, and Can-0 were observed as minor sequence variants in some of the other ecotypes, indicating they were likely present in the ancestral population. Although the mechanisms behind this sequence divergence and subsequent amplification are unknown, it is of interest that in at least one case, Est-0, four unique substitutions are in close proximity, implying they originated from a single event (Fig. 1).
In addition to these ecotype-specific substitutions, we also identified 152 statistically significant deviations from the species consensus that correspond to nucleotide substitutions commonly found in multiple ecotypes (Fig. 1, pink). In many cases, these changes reflect sites that are more variable than in the species as a whole. Conversely, five changes correspond to the fixation of a single nucleotide at a site that is highly polymorphic in the population. Although broader sampling is required to interpret the significance of these events, they provide additional evidence that the Arabidopsis satellites are dynamic; new mutations are likely emerging continuously, replacing, by an undetermined mechanism, the predominant variants in the population (Nijman and Lenstra 2001).
Measuring Sequence Variation Across the Arabidopsis Centromere Satellite Repeats
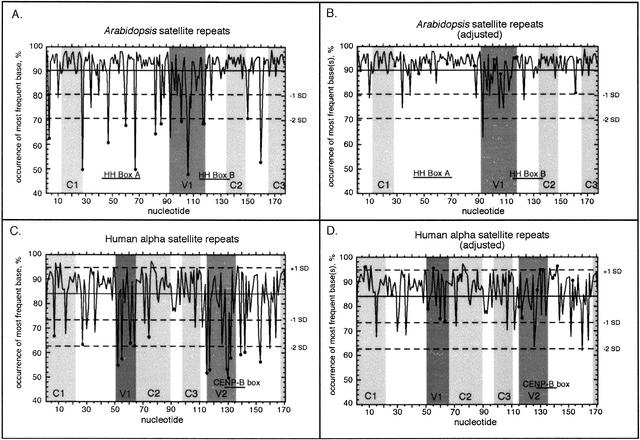
Despite mechanisms that homogenize repeat arrays across a genome, satellite repeats nonetheless accumulate variation at an appreciable rate. For example, human α-satellite repeats have a high degree of sequence heterogeneity, and variable sites are distributed among the satellite monomer classes in a nonrandom manner (Waye and Willard 1987; Choo et al. 1991). We used our entire data set of Arabidopsis satellite repeats to measure nucleotide variability, calculating the occurrence of the most frequent base as a percentage of all the nucleotides sequenced at each site (Fig. 2). Averaging these data across all of the sites in the repeat showed that most nucleotides are highly conserved, within 1 SD of a mean of 90.3 ± 9.8% (Fig. 2A). However, 21 sites showed more variation; 13 of these corresponded to polymorphisms identified previously (Fig. 2A, filled circles). We replotted these data (Fig. 2B), taking into account frequent polymorphisms (see Methods); this adjusted plot highlighted additional sites that exhibited unusually high variability.
Figure 2.
Variation across Arabidopsis and human satellite sequences. Percent occurrence of the most frequent base is plotted for each position in the Arabidopsis and α-satellite repeats, determined in the same manner for all nucleotides (A,C), or adjusted (B,D) by adding the frequencies of all the nucleotides that contributed to polymorphic sites (filled circles). (Solid line) The average percent occurrence of the most frequent base across all nucleotides; (dashed lines) SD from the average; (gray shading) cConserved (C1, C2, C3) and variable (V1, V2) regions (see also Fig. 3). (HH Boxes A and B) Conserved regions in the Columbia ecotype (Heslop-Harrison et al 1999); (CENP-B box) the binding site for the centromere-binding protein B (Muro et al. 1992).
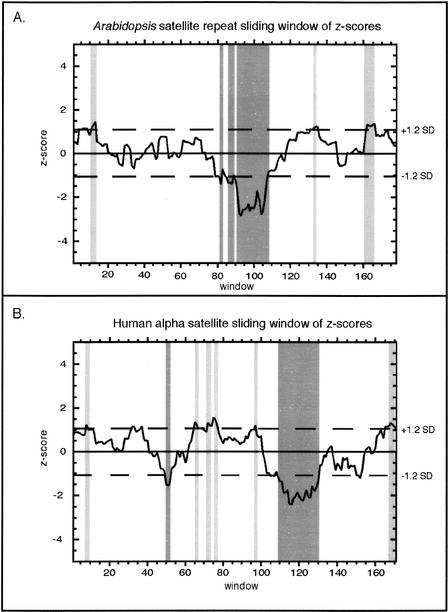
We identified conserved and variable segments within the satellite repeats by examining nucleotide occurrence frequencies over a sliding window of 15 bases. The 15-bp conserved domains C1, C2, and C3, and the 25-bp variable domain, V1, comprised of two overlapping windows, exhibited variation significantly different than the mean (Fig. 3A). Much of the variation within the satellite repeats is clustered near the V1 region, which contains 5 of the 8 highly variable sites (>2 SD from mean) and 3 of the 13 polymorphic sites. Strikingly, the same regions we identified as highly conserved or highly variable in the species as a whole showed similar patterns in the consensus sequences of individual ecotypes (Fig. 1). Thus, because these patterns occur repeatedly across Arabidopsis populations, they do not reflect chance variation within our sample of sequences. These nonrandom patterns of evolution within the Arabidopsis satellites strongly indicate biological constraints on satellite sequences. Whereas highly conserved domains may reflect important protein-binding sites, regions that exhibit extreme variation may point to areas where strict sequence consensus is not important. Alternatively, some sites may be under selection to remain polymorphic, creating a diversity of repeat monomers within arrays. In humans, such polymorphisms are organized into higher-order repeat units that might be important in the formation and structure of a centromere (Willard and Waye 1987; see Discussion).
Figure 3.
Identification of significantly conserved and variable domains. The percent occurrence of the most frequent base (Fig. 2A,C) was subjected to a z-score analysis, measured over a sliding window of 15 bp. This process sets the average at zero (solid line); dashed lines indicate ±1.2 SD. Significantly conserved windows (light gray) and significantly variable regions (dark gray) were merged when the sliding windows overlapped, and the entire window was represented as conserved (C1, C2, C3) and variable (V1, V2) regions (Figs. 1 and 2).
Lastly, we examined 950 repeats from the Columbia ecotype that were sequenced by the Arabidopsis Genome Project. These repeats are located on the edges of the satellite arrays; recent examinations of human α-satellites show that repeats on array edges are more variable than the repeats in the array core (Schueler et al. 2001). The Columbia sequences from the array edges differed from the species consensus at only 20 sites (Fig. 4), 18 of which were frequently polymorphic in the random sample of Arabidopsis populations (Fig. 1). Surprisingly, we found that the overall conservation of nucleotides within this large set of Columbia repeats was 89.4% ± 3.9%, similar to the Arabidopsis species average (90.3% ± 9.8%) and the Columbia consensus average derived from random sampling (91.3% ± 14.4%). We assessed monitored nucleotide conservation across these sequences, applying the same criteria used to generate Figure 3. The Columbia satellites from the array edges have an expanded C3 region and V1 region, and do not display any conservation above average in the C1 region, whereas the C2 region remains unchanged. Thus, in contrast to expectations based on human repeats, the edges of Arabidopsis satellite arrays are not more variable than sequences collected randomly from the genome. These observations may reflect a fundamental difference in the mechanisms that maintain human and Arabidopsis arrays.
Figure 4.
Comparison of Columbia ecotype satellite consensus sequences. The Col-0 consensus was derived from PCR-amplified sequences obtained in this study; the Col-edges consensus was derived using the 950 satellite sequences available from The Arabidopsis Genome Project (GenBank). The two consensus sequences were aligned with the species satellite consensus (Fig. 1); dots represent identity to the species consensus, and changes from the consensus are indicated.
Comparisons With Sequence Variation Across Human α-Satellite Repeats
To compare the composition of the Arabidopsis satellites to human α-satellite DNA, we reexamined the set of 293 human sequences compiled previously (Choo et al. 1991). In α-satellite DNA, 15 polymorphic sites have been identified, similar to the number in the Arabidopsis satellites. As with Arabidopsis, the percent occurrence of bases at these polymorphic sites is within 1 SD of the mean when the second-most-frequent base is considered (Fig. 2D). Interestingly, the average percent occurrence of the most abundant bases in the α-satellites was 84.0% ± 10.7% (Fig. 2C), indicating these repeats are significantly more variable (P < 0.0001) than the collection of Arabidopsis repeats, as determined by a univariate ANOVA test. This difference may reflect the dissimilarities in population structure and mating patterns between humans and Arabidopsis, the nearly fivefold difference in chromosome (and centromere) number (23 vs. 5, respectively), or a disparity in the functional roles of the repeats, accompanied by different selective pressures.
Using the same criteria as with the Arabidopsis satellites, we identified three regions of conservation (C1, C2, C3) and two regions of variability (V1, V2) in the α-satellite repeats (Fig. 3B). Whereas windows of significant conservation or variability in the Arabidopsis satellites tended to cluster, these windows were more scattered in α-satellites. Interestingly, the binding site for the 17-bp centromere protein B (CENP-B box), defined by DNA footprinting (Muro et al. 1992), resides in one of the variable regions, V2, which contains five polymorphic sites (Alexandrov et al. 1993; Rovanova et al. 1996). The average occurrence of the most frequent base across the entire CENP-B box is 78%, and when the nucleotides essential for CENP-B binding are considered (Tanaka et al. 2001), this percentage drops to only 68%, making this region notably more variable than the rest of the α-satellite repeat. This observation supports the model that many α-satellite repeats cannot tightly bind CENP-B, resulting in protein phasing and higher-order chromatin structure (Yoda et al. 1998). In fact, when we surveyed a set of 880 α-satellites from GenBank, only 23% had all of the nucleotides essential for CENP-B binding.
Variation of Single-Copy Sequences From the Recombinationally Suppressed Arabidopsis Centromeres
To better appreciate the diversity of the Arabidopsis centromere regions, we examined the sequence variation of three single-copy loci that are tightly linked to three different genetically defined centromeres (CEN2, CEN3, and CEN4), and DNA sequences from eight single-copy genes located in the chromosome arms (Supplemental Fig. 5, GenBank accession nos. AF494760–AF494836, AF495295–AF495335, AF495337–AF495375; available online at http://www.genome.org). For each intron and exon, we determined the sequence variability at each site by measuring the average occurrence of the most common nucleotide (Table 2). The exons from chromosome arms and from recombinationally supressed centromere regions displayed a similar rate of variation, with an average occurrence of the most frequent nucleotide ranging from 99.5% to 99.9% and 99.7% to 99.9%, respectively. Although two of the three centromeric introns had variation (95.6% and 95.8%) that was significantly different from that of introns from the chromosome arms (ANOVA, P < 0.0001), much of this variation is attributed to a single large deletion event, and therefore may not be representative of intron variation in the centromeric region.
Table 2.
Conservation of Satellites, Introns, and Exons
| Sequence type | Sequence | Average % occurrencea | Nucleotide diversitya |
| Satellites | Arabidopsis satellites | 90.8 ± 0.3 | 0.17 ± 0.08 |
| α-satellites | 83.9 ± 0.3 | n.d. | |
| Introns, | ACT8 | 99.1 ± 0.3 | 0.003 ± 0.004 |
| chromosome | ADH1 | 98.7 ± 0.1 | 0.007 ± 0.004 |
| arm | AP3 | 99.4 ± 0.1 | 0.008 ± 0.004 |
| CAL | 99.5 ± 0.1 | 0.006 ± 0.003 | |
| FAH1 | 99.2 ± 0.2 | 0.008 ± 0.005 | |
| PGI | 99.7 ± 0.1 | 0.059 ± 0.030 | |
| PI | 99.5 ± 0.1 | 0.006 ± 0.003 | |
| TT5 | 99.8 ± 0.2 | 0.003 ± 0.002 | |
| TT6 | 99.8 ± 0.3 | 0.003 ± 0.003 | |
| Introns, | ARP6 | 99.0 ± 0.4 | 0.006 ± 0.004 |
| centromere | MCM5-like | 95.6 ± 0.2 | 0.006 ± 0.005 |
| SL15-like | 95.8 ± 0.4 | 0.054 ± 0.031 | |
| Exons, | ACT8 | 99.5 ± 0.2 | 0.009 ± 0.006 |
| chromosome | ADH1 | 99.5 ± 0.1 | 0.007 ± 0.004 |
| arm | AP3 | 99.8 ± 0.1 | 0.005 ± 0.003 |
| CAL | 99.6 ± 0.1 | 0.006 ± 0.004 | |
| FAH1 | 99.8 ± 0.1 | 0.003 ± 0.002 | |
| PGI | 99.9 ± 0.1 | 0.074 ± 0.037 | |
| PI | 99.8 ± 0.1 | 0.003 ± 0.002 | |
| TT5 | 99.9 ± 0.1 | 0.001 ± 0.001 | |
| TT6 | 99.5 ± 0.1 | 0.007 ± 0.004 | |
| Exons, | ARP6 | 99.7 ± 0.1 | 0.001 ± 0.003 |
| centromere | MCM5-like | 99.9 ± 0.2 | 0.002 ± 0.002 |
| SL15-like | 99.9 ± 0.3 | 0.002 ± 0.002 |
±Standard error.
The variation of orthologous single-copy sequences cannot be directly compared with the variation among repetitive paralogous satellites; nonetheless, it is of interest that none of the intron or exon sequences showed as much variation as the collection of Arabidopsis satellite repeats (Table 2; ANOVA, P < 0.0001). Similarly, the nucleotide diversity of satellite repeats was substantially higher than that of genes (Table 2). Finally, we compared transition and transversion frequencies for the Arabidopsis and α-satellite repeats to the set of single-copy sequences (Table 3). Using the species consensus for each sequence, we tabulated the number of transitions and transversions for each individual sequence relative to the consensus. As expected, exons from both the chromosome arms and centromere regions showed more conservative changes than introns, having 68.3% and 75.0% transitions versus 56.6% and 60.0% transitions, respectively. In contrast, the Arabidopsis satellite repeats and α-satellite repeats had fewer transitions than either exons or introns (40.8% and 38.0% transitions, respectively) approaching the theoretical 33% value for a sequence that is mutating at random.
Table 3.
Percent Substitution Type in Satellites and Genes
| Sequence type | Transitionsa | Transversionsb | χ2c | Significance |
| α-Satellite repeats | 38.0 | 62.0 | 0.99 | P < 0.5 |
| Arabidopsis repeats | 40.8 | 59.2 | 2.5 | P < 0.5 |
| Introns | ||||
| Chromosome arm | 56.6 | 43.4 | 24 | P < 0.001 |
| Centromere | 60.0 | 40.0 | 32 | P < 0.001 |
| Exons | ||||
| Chromosome arm | 68.3 | 31.7 | 55 | P < 0.001 |
| Centromere | 75.0 | 25.0 | 78 | P < 0.001 |
Expected = 0.33.
Expected = 0.66.
χ2 test compared the expected frequency of transitions and transversion with the observed frequency; shading indicates significant deviations from expected values.
DISCUSSION
Satellite DNA and Centromere Functions
The development of human minichromosomes supports the idea of a functional role for satellite DNA in centromeres (Willard 2001). In contrast, the lack of sequence conservation across species and the prevalence of neocentromeres that lack satellite repeats have raised questions as to whether any specific satellite sequences are required for function (Choo 2000; Henikoff et al. 2001). In this study, we demonstrated that the centromeric satellite repeats of Arabidopsis have domains that are highly conserved, whereas other portions of these repeats vary considerably. Thus, the preservation of both conserved and variable domains across 41 different populations, along with a strict conservation of sequence length, strongly indicates that the evolution of the satellite repeats is constrained.
Conserved Domains in Satellite Repeats
We used a statistical test to define three regions (C1, C2, C3) of high average conservation (87.9% ± 1.5%, 88.4% ± 1.4%, 88.4% ± 1.8% conserved, respectively) and two variable regions (V1 and V2, 78.0% ± 3.3%, 76.1% ± 3.1% conserved, respectively) in human α-satellite DNA. None of these domains had been defined previously. Similarly, we defined three conserved regions (C1, 95.2% ± 0.9%; C2, 94.6% ± 0.7%; C3, 95.0% ± 0.9%) and one variable region (83.8% ± 2.4%) in the Arabidopsis satellite repeat consensus. These domains are distinct from the two conserved regions (Box A and Box B, 99% conservation) previously derived for 20 Columbia satellite sequences (Fig. 2A; Heslop-Harrison et al. 1999). Although Box A and Box B were not highly conserved across Arabidopsis or in the sample of Columbia satellites we obtained, some of these differences can be attributed to polymorphic sites, and others are more likely the result of the bias inherent in a smaller data set. The centromere satellite consensus sequence presented here was derived from 1029 repeats from 41 Arabidopsis ecotypes, and consequently more broadly reflects the species as a whole.
The presence of highly conserved domains within the satellites indicates that some repeat regions may be under selective pressure to maintain a particular DNA sequence, whereas other regions of the repeat evolve without constraint. One explanation for the differential rates of substitution in the Arabidopsis satellites could be the interaction of DNA-binding proteins with satellite DNA. In humans, centromere-binding proteins A, B, C, E, G, and H have been identified. Of those proteins, CENP-A, CENP-B, and CENP-C have been shown to have DNA-binding activity (Choo 2000). CENP-A is a histone H3-like protein that is found at active centromeres and is associated with α-satellite arrays in humans (Smith 2002). In addition, CENP-A homologs in Drosophila and Arabidopsis appear to be evolving adaptively, which could correlate with the sequence divergence of satellite arrays in the centromere (Malik and Henikoff 2001; Talbert et al. 2002). The association of CENP-A homologs with corresponding centromeric DNA could influence the maintenance of conserved sequence domains in the repeats.
Both CENP-B and CENP-C have been shown to associate with a subset of α-satellite repeats. However, the localizations of the two proteins on α-satellite arrays are distinct and nonoverlapping. CENP-C is found only at active centromeres, and the exact binding site of CENP-C within the α-satellite is still unknown (Politi et al. 2002). CENP-B is found associated with α-satellite arrays at both active and inactive centromeres; it binds to α-satellite monomers at a specific 17-bp sequence named the CENP-B box (Muro et al. 1992). Interestingly, the CENP-B box in the α-satellite repeats overlaps with the highly variable V2 region, and contains five polymorphic sites in its consensus. Combining the insights from the recently solved CENP-B/CENP-B box cocrystal (Tanaka et al. 2001) with the survey of published α-satellite sequences, we found four of the nine bases essential for CENP-B binding are also polymorphic; CENP-B would be unable to interact with a highly common base at each of these four sites. Ikeno et al. (1994) analyzed a higher order α-satellite array comprised of 11 repeats, and found that CENP-B-binding sites are located in alternating repeat monomers. Taken together, these results raise the possibility that polymorphisms serve to phase CENP-B binding within the satellite arrays, potentially aiding in the assembly of the α-satellite DNA into a higher-order structure recognized by other centromere-binding proteins (Yoda et al. 1998; Choo 2000). Although centromere-binding proteins from plants are less well characterized, it is possible that a similar phasing mechanism could be operating, given the patterns of nonrandom variation that we observed within the Arabidopsis satellite repeats.
Conservation of Satellite Sequence Length
A requirement for uniform nucleosome phasing and the subsequent propagation of centromeric heterochromatin has often been ascribed as the source of the uniform satellite length observed within a species and between closely related species (Henikoff et al. 2001). In primates, satellite monomers vary from 168 to 172 bp (Durfy and Willard 1990; Baldini et al. 1991; Fanning et al. 1993; Alves et al. 1994; Warburton et al. 1996; Haaf and Willard 1997, 1998). Average centromere satellite lengths have also been determined for a wide range of other species, including maize (156 bp; Ananiev et al. 1998), rice (159 bp; Dong et al. 1998), and insects in the genus Palorus (143 bp; Mestrovic et al. 1998). We found that Arabidopsis centromere satellites were remarkably conserved within all 41 ecotypes (178 ± 0.1 bp). The highly invariant length of Arabidopsis satellite repeats indicates a rigid length requirement. Because nucleosome arrays can accommodate insertions of several base pairs without a dramatic alteration in phasing patterns (Simpson 1991), other explanations, such as length requirements that modulate higher-order structures across entire arrays, may be more appropriate. CENP-B, known to bind as a dimer, may require rigid monomer length so that CENP-B boxes are in appropriate locations within a centromere structure for protein binding (Yoda et al. 1998). Alternatively, the length requirement could be a result of the satellite array interaction with specialized centromere histones, such as CENP-A (Talbert et al. 2002). If nucleosome phasing is involved, then the diversity of satellite lengths among Arabidopsis, maize, and rice would require invoking a species-specific nucleosome length restriction.
The Diversity of Satellites Across Populations
Despite the presence of conserved domains, many portions of the satellite repeats exhibit notable variation. Considered as a whole, the centromere satellite repeats are more variable across the Arabidopsis population than any other single-copy sequence examined, including noncoding DNA. Interestingly, the Arabidopsis satellite repeats were significantly less variable than α-satellite repeats. Reproductive strategies may explain some of this difference; because Arabidopsis is self-pollinating, it is expected to have less heterozygosity and less genetic diversity than individuals in an outcrossing population (Charlesworth and Wright 2001).
The vast number of satellite copies in the genome provides tremendous redundancy and an enhanced opportunity for divergence; they can undergo various mechanisms of evolution, homogenizing new changes through gene conversion, unequal exchange, and transposition (Dover 1982). Moreover, recombination and repair enzymes may have a limited access to heterochromatic satellites, increasing the rate of nucleotide substitution relative to the rest of the genome. If satellite sequences indeed provide critical functions, this redundancy and high rate of change could allow organisms to sample substitutions, even in functional domains, without deleterious effects.
Evolution of Satellite DNA
Although the function of satellite DNA remains questionable (Csink and Henikoff 1998), satellite evolution has attracted much attention. Many studies have compared the satellites from closely related species (Waye and Willard 1989; Grebenstein et al. 1996; Alix et al. 1998; Mestrovic et al. 1998; Rajagopal et al. 1999; Landais et al. 2000; Nijman and Lenstra 2001). In these analyses, homogenization of satellite repeats within the genome has typically occurred, resulting in less variation within a species than between closely related species. This type of change, termed concerted evolution (Elder and Turner 1995), likely relies on mechanisms of molecular drive: unequal exchange, gene conversion, or transposition (Smith 1976; Dover 1982; Stephan 1986; Charlesworth et al. 1994).
The results presented here indicate a balance between the stochastic and selective pressures that drive satellite diversity. Our finding of significantly conserved and variable regions across ecotypes indicates a strong bias in the turnover of satellite sequences (Mestrovic et al. 1998). Molecular drive may account for the homogenization of 11 substitutions observed in individual Arabidopsis ecotypes that differ from the species consensus (Fig. 1, yellow shading). Because these 11 substitutions also occur at a low frequency in other ecotypes, they were likely present in the ancestral parent, and homogenization of the variant occurred since the ecotype populations diverged (Nijman and Lenstra 2001). In addition, the precise conservation of satellite length is particularly striking. Taken together, these observations indicate a model in which higher-order structures have a strict requirement for sequence length and conservation of particular repeat regions. Satellite evolution may progress in a manner that retains all of these features, maintaining essential protein-binding sites, structural domains, and sites for epigenetic modification.
METHODS
Source of DNA Sequences Analyzed
Arabidopsis centromere satellite repeat sequences were from ecotypes obtained primarily from the Arabidopsis Biological Resource Center (ABRC), Ohio State University; Kz-8 was obtained from Joy Bergelson, University of Chicago. DNA was extracted from a rosette leaf of an individual plant as described (McKinney et al. 1995). Two sets of primers (F2: 5′-AGCTTCTTCTTGCTTCTCA; R2: 5′-CCAATCACAAAACCT CAGC; and F4: 5′-GAGTCTTTGGCTTTGTATCTTC; R4: 5′-GTATACCTGAAACCGATGTGG; Fig. 1) were used to amplify satellite repeats; PCR was performed as recommended (PanVera Corporation). Amplification products were separated by gel electrophoresis, and the ladder of repeats was visualized after ethidium bromide staining. Bands measuring ∼180 bp, 360 bp, and 540 bp were purified, cloned (TOPO TA kit, Invitrogen), and sequenced using the M13 forward and reverse primers. A minimum of 10 clones was sequenced for each of the ecotypes. The resulting 457 clones sequenced gave 1029 whole or partial repeat sequences. For analysis of repeat length, 176 internal repeats derived from 360-bp or 540-bp amplified bands were considered. For this study, we did not include the 950 satellite repeat sequences from the Columbia ecotype that were deposited in GenBank by the Arabidopsis Genome Sequencing Project; many of these sequences reside at the borders of satellite arrays and consequently contain biases not representative of the genome. Furthermore, to consider all ecotypes equally, we also excluded a set of 624 satellite repeat sequences that were obtained on random clones from the Landsberg ecotype (http://www.tigr.org/tdb/e2k1/ath1/atgenome/Ler.shtml).
The analysis of human α-satellite DNA described here relied on data compiled previously by Choo et al. (1991). This earlier study derived a consensus of available human satellite sequences, and tabulated the variation at each site.
The sequence variation among Arabidopsis ecotypes was analyzed for 12 genes (Supplemental Fig. 5, available online at http://www.genome.org; Table 2). These genes are all expressed, with known EST or cDNA counterparts (The Arabidopsis Genome Initiative 2000). We performed PCR and DNA sequencing in four cases (ARP6, MCM5-like, SL15-like, and ACTIN8; GenBank accession nos. AF494760–AF494836, AF495295–AF495335, AF495337–AF495375); for the remainder, we analyzed sequence available in GenBank (for references, see Supplemental Fig. 5, available online at http://www.genome.org).
Sequence Analysis
Prior to analysis, primer and vector sequences were trimmed, concatenated satellite repeat arrays were separated, and sequences were aligned using Seqman (DNAStar). Polymorphic sites within the consensus are indicated (Fig. 1) by IUPAC symbols: (B) C or G or T; (D) A or G or T; (H) A or C or T; (K) G or T; (M) A or C; (R) A or G; (S) C or G; (V) A or C or G; (W) A or T; (Y) C or T. A χ2 test determined the significance of nucleotide differences observed in individual ecotypes. For the expected nucleotide occurrence at a given site, we used the overall nucleotide frequencies, as determined for the 41 ecotypes combined, at each position. The data were divided into two classes (consensus and nonconsensus) for a single degree of freedom; differences from the consensus were considered significant when P ≤ 0.0001. Nucleotides within a given ecotype that showed a significant deviation in frequency from the overall species consensus are indicated in Figure 1 (shading). Some substitutions in ecotype consensus sequences are not shaded, as the nucleotide frequency does not significantly differ from the overall consensus.
The percent occurrence of the most frequent base at each site was calculated for Arabidopsis satellite repeats, α-satellite repeats, and gene sequences; for the satellites, this is plotted in Figure 2. At polymorphic sites (i.e., sites where the most common base is not three times more frequent than any other, filled circles in Fig. 2) either the percent occurrence of the most frequent base was calculated (Fig. 2A,C), or the percent occurrence of the polymorphic nucleotides, considered as a group, was considered (Fig. 2B,D). The average percent occurrence and standard deviations are also depicted in Figure 2; for these calculations, polymorphic sites were not treated differently from other nucleotides. A univariate ANOVA test (α = 0.05) with a Bonferroni adjustment (Sokal and Rohlf 1997) was used to determine if the average values for the percent occurrence differed when Arabidopsis satellites, α-satellites, and genes were considered.
Conserved and variable regions within the Arabidopsis satellite repeats and α-satellites were defined by a sliding-window analysis of the percent occurrence data; z-scores were used to define windows of significantly higher or lower variation than the average. Windows of 5 bp, 10 bp, 15 bp, and 20 bp were initially analyzed, and results from a 15-bp window analysis are presented. The average percent occurrence for each window was tabulated, and an overall average and standard deviation for these window data points were used to produce a z-score (z = [x − μ]/ς, where x is each window data point, μ is the average of all windows, and ς is the standard deviation). Windows that had a z-score of ±1.2 SD from the mean (∼20% of all windows) were considered significant (Fig. 3). For clusters of windows with significant deviations from the mean, the window with the largest departure from the mean was used as the center for the conserved or variable regions; the Arabidopsis satellite repeat variable region V1 consists of two independent overlapping windows (depicted in Figs. 1 and 2). This analysis made it possible to use the same criteria to define conserved and variable regions in the Arabidopsis and α-satellite repeats.
Nucleotide diversity was calculated for Arabidopsis satellite repeats and centromere and arm genes using ARLEQUIN software (Schneider et al. 2000). Insertions and deletions were not considered in the calculations; the Tajima and Nei method was used by the software.
WEB SITE REFERENCES
http://www.arabidopsis.org; The Arabidopsis Information Resource.
http://www.tigr.org/tdb/e2k1/ath1/atgenome/Ler.shtml; The Institute for Genomic Research, Landsberg erecta random sequence Database (Ler).
Acknowledgments
We thank S. Duffy and K. Thornton for helpful discussions; and members of the Preuss laboratory, M. Sharp, A. Hall, K. von Besser, and K. Keith, for critical reading of the manuscript. This work was supported in part by an NIH Training Grant in Genetics and Regulation (S.E.H.), and by grants from the National Science Foundation, the David and Lucile Packard Fellows Program, and the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL dpreuss@midway.uchicago.edu; FAX (773) 702-6648.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.593403. Article published online before print in January 2003.
REFERENCES
- 1.Alexandrov I.A., Medvedev, L.I., Mashkova, T.D., Kisselev, L.L., Romanova, L.Y., and Yurov, Y.B. 1993. Definition of a new α satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 21: 2209-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alix K., Baurens, F.-C., Paulet, F., Glaszmann, J.-C., and D'Hont, A. 1998. Isolation and characterization of a satellite DNA family in the Saccharum complex. Genome 41: 854-864. [PubMed] [Google Scholar]
- 3.Al-Shehbaz I.A. 1984. The tribes of Cruciferae (Brassicaceae) in the southeastern United States. J. Arnold Arbor 65: 343-373. [Google Scholar]
- 4.Alves G., Seuanez, H.N., and Fanning, T. 1994. α Satellite DNA in neotropical primates (Platyrrhini). Chromosoma 103: 262-267. [DOI] [PubMed] [Google Scholar]
- 5.Ananiev E.V., Phillips, R.L., and Rines, H.W. 1998. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. 95: 13073-13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Arabidopsis Genome Initiative 2000. Analysis of the genome sequencing of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- 7.Baldini A., Miller, D.A., Miller, O.J., Ryder, O.A., and Mitchell, A.R. 1991. A chimpanzee-derived chromosome-specific α-satellite DNA sequence conserved between chimpanzee and human. Chromosoma 100: 156-161. [DOI] [PubMed] [Google Scholar]
- 8.Barry A.E., Bateman, M., Howman, E.V., Cancilla, M.R., Tainton, K.M., Irvine, D.V., Saffery, R., and Choo, K.H. 2000. The 10q25 neocentromere and its inactive progenitor have identical primary nucleotide sequence: Further evidence for epigenetic modification. Genome Res. 10: 832-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlesworth B., Sniegowski, P., and Stephan, W. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215-220. [DOI] [PubMed] [Google Scholar]
- 10.Charlesworth D. and Wright, S.I. 2001. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 11: 685-690. [DOI] [PubMed] [Google Scholar]
- 11.Choo K.H. 2000. Centromerization. Trends Cell Biol. 10: 182-188. [DOI] [PubMed] [Google Scholar]
- 12.Choo K.H., Vissel, B., Nagy, A., Earle, E., and Kalitsis, P. 1991. A survey of the genomic distribution of α satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 19: 1179-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke L. 1998. Centromeres: Proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev. 8: 212-218. [DOI] [PubMed] [Google Scholar]
- 14.Clarke L. and Carbon, J. 1980. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504-509. [DOI] [PubMed] [Google Scholar]
- 15.Copenhaver G.P., Nickel, K., Kuromori, T., Benito, M., Kaul, S., Lin, X., Bevan, M., Murphy, G., Harris, B., Parnell, L.D., et al. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468-2474. [DOI] [PubMed] [Google Scholar]
- 16.Cottarel G., Shero, J.H., Hieter, P., and Hegemann, J.H. 1989. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 3342-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csink A. K. and Henikoff, S. 1998. Something from nothing: The evolution and utility of satellite repeats. Trends Genet. 14: 200-204. [DOI] [PubMed] [Google Scholar]
- 18.Dong F., Miller, J.T., Jackson, S.A., Wang, G.-L., Ronald, P.C., and Jiang, J. 1998. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. 95: 8135-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dover G. 1982. Molecular drive: A cohesive mode of species evolution. Nature 299: 111-117. [DOI] [PubMed] [Google Scholar]
- 20.Durfy S.J. and Willard, H.F. 1990. Concerted evolution of primate α satellite DNA: Evidence for an ancestral sequence shared by gorilla and human X chromosome α satellite. J. Mol. Biol. 216: 555-566. [DOI] [PubMed] [Google Scholar]
- 21.DuSart D., Cancilla, M.R., Earle, E., Mao, J.-I, Saffery, R., Tainton, K.M., Kalitsis, P., Martyn, J., Barry, A.E., and Choo, K.H.A. 1997. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat. Genet. 16: 144-153. [DOI] [PubMed] [Google Scholar]
- 22.Elder J.F. and Turner, B.J. 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Quart. Rev. Biol. 70: 297-320. [DOI] [PubMed] [Google Scholar]
- 23.Fanning T.G., Seuanez, H.N., and Forman, L. 1993. Satellite DNA sequences in the New World primate Cebus apella (Platyrrhini, Primates). Chromosoma 102: 306-311. [DOI] [PubMed] [Google Scholar]
- 24.Grebenstein B., Grebenstein, O., Sauer, W., and Hemleben, V. 1996. Distribution and complex organization of satellite DNA sequences in Aveneae species. Genome 39: 1045-1050. [DOI] [PubMed] [Google Scholar]
- 25.Grimes B.R., Rhoades, A.A., and Willard, H.F. 2002. α-Satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol. Therapy 5: 798-805. [DOI] [PubMed] [Google Scholar]
- 26.Haaf T. and Willard, H.F. 1997. Chromosome-specific α-satellite DNA from the centromere of chimpanzee Chromosome 4. Chromosoma 106: 226-232. [DOI] [PubMed] [Google Scholar]
- 27.___, 1998. Orangutan α-satellite monomers are closely related to the human consensus sequence. Mam. Genome 9: 440-447. [DOI] [PubMed] [Google Scholar]
- 28.Haaf T., Warburton, P.E., and Willard, H.F. 1992. Integration of human α-satellite DNA into simian chromosomes: Centromere protein binding and disruption of normal chromosome segregation. Cell 70: 681-696. [DOI] [PubMed] [Google Scholar]
- 29.Hallden C., Bryngelsson, T., Sall, T., and Gustafsson, M. 1987. Distribution and evolution of a tandemly repeated DNA sequence in the family Brassicaceae. J. Mol. Evol. 25: 318-323. [Google Scholar]
- 30.Henikoff S., Ahmad, K., and Malik, H.S. 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098-1102. [DOI] [PubMed] [Google Scholar]
- 31.Heslop-Harrison J.S., Murata, M., Ogura, Y., Schwarzacher, T., and Motoyoshi, F. 1999. Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11: 31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosouchi T., Kumekawa, N., Tsuruoka, H., and Kotani, H. 2002. Physical map-based sizes of the centromeric regions of Arabidopsis thaliana Chromosomes 1, 2, and 3. DNA Res. 9: 117-121. [DOI] [PubMed] [Google Scholar]
- 33.Ikeno M., Masumoto, H., and Okazaki, T. 1994. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range α-satellite DNA arrays of human Chromosome 21. Hum. Mol. Genet. 3: 1245-1257. [DOI] [PubMed] [Google Scholar]
- 34.Koch M., Haubold, B., and Mitchell-Olds, T. 2001. Molecular systematics of the Brassicaceae: Evidence from coding plastidic MATK and nuclear CHS sequences. Am. J. Botany 88: 534-544. [PubMed] [Google Scholar]
- 35.Kumekawa N., Hosouchi, T., Tsuruoka, H., and Kotami, H. 2000. The size and sequence organization of the centromeric region of Arabidopsis thaliana Chromosome 5. DNA Res. 7: 315-321. [DOI] [PubMed] [Google Scholar]
- 36.___, 2001. The size and sequence organization of the centromeric region of Arabidopsis thaliana Chromosome 4. DNA Res. 8: 285-290. [DOI] [PubMed] [Google Scholar]
- 37.Landais I., Chavigny, P., Castagnone, C., Pizzol, J., Abad, P., and Vanlerberghe-Masutti, F. 2000. Characterization of a highly conserved satellite DNA from the parasitoid wasp Trichogramma brassicae. Gene 255: 65-73. [DOI] [PubMed] [Google Scholar]
- 38.Malik H.S. and Henikoff, S. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157: 1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Zapater J.M., Estelle, M.A., and Somerville, C.R. 1986. A highly repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204: 417-423. [Google Scholar]
- 40.McKinney E.C., Ali, N., Traut, A., Feldmann, K.A., Belostotsky, D.A., McDowell, J.M., and Meagher, R.B. 1995. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: Actin mutants act2-1 and act4-1. Plant J. 8: 613-622. [DOI] [PubMed] [Google Scholar]
- 41.Mestrovic N., Plohl, M., Mravinac, B., and Ugarkovic, D. 1998. Evolution of satellite DNAs from the genus Palorus—Experimental evidence for the “library” hypothesis. Mol. Biol. Evol. 15: 1062-1068. [DOI] [PubMed] [Google Scholar]
- 42.Muro Y., Masumoto, H., Yoda, K., Nozaki, N., Ohashi, M., and Okazaki, T. 1992. Centromere protein B assembles human centromeric α-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol. 116: 585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nijman I.J. and Lenstra, J.A. 2001. Mutation and recombination in cattle satellite DNA: A feedback model for the evolution of satellite DNA repeats. J. Mol. Evol. 52: 361-371. [DOI] [PubMed] [Google Scholar]
- 44.Politi V., Perini, G., Trazzi, S., Pliss, A., Raska, I., Earnshaw, W.C., and Della Valle, G. 2002. CENP-C binds the α-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 115: 2317-2327. [DOI] [PubMed] [Google Scholar]
- 45.Rajagopal J., Das, S., Khurana, D.K., Srivastava, P.S., and Lakshmikumaran, M. 1999. Molecular characterization and distribution of a 145-bp tandem repeat family in the genus Populus. Genome 42: 909-918. [DOI] [PubMed] [Google Scholar]
- 46.Rovanova L.Y., Deriagin, G.V., Mashkova, T.D., Tumeneva, I.G., Mushegian, A.R., Kisselev, L.L., and Alexandrov, I.A. 1996. Evidence for selection in evolution of α-satellite DNA: The central role of CENP-B/pJα binding region. J. Mol. Biol. 261: 334-340. [DOI] [PubMed] [Google Scholar]
- 47.Schneider S., Roessli, D., and Excoffier, L., 2000. Arlequin ver. 2.000: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- 48.Schueler M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109-115. [DOI] [PubMed] [Google Scholar]
- 49.Simpson R.T. 1991. Nucleosome positioning: Occurrence, mechanisms, and functional consequences. Prog. Nucleic Acid Res. Mol. Biol. 40: 143-184. [DOI] [PubMed] [Google Scholar]
- 50.Smith G.P. 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191: 528-535. [DOI] [PubMed] [Google Scholar]
- 51.Smith M.M. 2002. Centromeres and variant histones: What, where, and why? Curr. Opin. Cell Biol. 14: 279-285. [DOI] [PubMed] [Google Scholar]
- 52.Sokal R.R. and Rohlf, F.J., 1997. Biometry W.H. Freeman and Company, New York, NY.
- 53.Stephan W. 1986. Recombination and the evolution of satellite DNA. Genet. Res. 47: 167-174. [DOI] [PubMed] [Google Scholar]
- 54.Talbert P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14: 1053-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka Y., Nureki, O., Kurumizaka, H., Fukai, S., Kawaguchi, S., Ikuta, M., Iwahara, J., Okazaki, T., and Yokoyama, S. 2001. Crystal structure of the CENP-B protein–DNA complex: The DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 20: 6612-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warburton P.E. and Willard, H.F. 1990. Genomic analysis of sequence variation in tandemly repeated DNA. J. Mol. Biol. 216: 3-16. [DOI] [PubMed] [Google Scholar]
- 57.Warburton P.E., Haaf, T., Gosden, J., Lawson, D., and Willard, H.F. 1996. Characterization of a chromosome-specific chimpanzee α-satellite subset: Evolutionary relationship to subsets on human chromosomes. Genomics 33: 220-228. [DOI] [PubMed] [Google Scholar]
- 58.Waye J.S. and Willard, H.F. 1987. Nucleotide sequence heterogeneity of α satellite repetitive DNA: A survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 15: 7549-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.___, 1989. Concerted evolution of α satellite DNA: Evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma 98: 273-279. [DOI] [PubMed] [Google Scholar]
- 60.Willard H.F. 1998. Human artificial chromosomes coming into focus. Nat. Biotech. 16: 415-416. [DOI] [PubMed] [Google Scholar]
- 61.___, 2001. Genomics and gene therapy: Artificial chromosomes coming to life. Science 290: 1308. [DOI] [PubMed] [Google Scholar]
- 62.Willard H.F. and Waye, J.S. 1987. Hierarchical order in chromosome-specific human α satellite DNA. Trends Genet. 3: 192-198. [Google Scholar]
- 63.Yang J.W., Pendon, C., Yang, J., Haywood, N., Chand, A., and Brown, W.R.A. 2000. Human mini-chromosomes with minimal centromeres. Hum. Mol. Genet. 9: 1891-1902. [DOI] [PubMed] [Google Scholar]
- 64.Yoda K., Ando, S., Okuda, A., Kikuchi, A., and Okazaki, T. 1998. In vitro assembly of the CENP-B/α-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes to Cells 3: 533-548. [DOI] [PubMed] [Google Scholar]