Abstract
Strigolactones are phytohormones that stimulate seed germination of parasitic plants including Phelipanche aegyptiaca. Strigolactones are derived from carotenoids via a pathway involving the carotenoid cleavage dioxygenases CCD7 and CCD8. We report here identification of PaCCD7 and PaCCD8 orthologous genes from P. aegyptiaca. Expression analysis of PaCCD7 and PaCCD8 genes showed significant variation in their transcript levels in seeds and tubercles of P. aegyptiaca at different developmental stages. These two parasitic PaCCD7 and PaCCD8 genes were silenced in P. aegyptiaca using a trans-silencing approach in Nicotiana benthamiana. The transient knock-down of PaCCD7 and PaCCD8 inhibited tubercle development and the infestation process in host plants. Our results suggest an important role of the strigolactone associated genes (PaCCD7 and PaCCD8) in the parasite life cycle.
Keywords: Strigolactone, Phelipanche, root parasite, PaCCD7, PaCCD8, VIGS, trans-silencing
Phelipanche spp. and Orobanche spp. (Orobanchaceae) are economically destructive parasitic weeds, causing large crop losses in sunflower, tomato, faba bean, tobacco and many other field crops in the world.1Phelipanche plant development is divided into pre-parasitic and parasitic stages. The pre-parasitic stage starts with seed pre-conditioning followed by germination, which is induced by molecules secreted in the rhizosphere by the roots of host plants. Most of the known germination stimulants are strigolactones. Germination leads to the emergence of a radicle which attaches to the host root surface.2-4 The parasitic phase starts with the penetration of the parasite into the host root through a differentiating haustorium which connects to the host vascular tissues. The haustorium serves as both an attaching organ and a bridge for water and nutrient transfer from the host. The attached parasite first acts as a root, then diverts hormones and photoassimilates from the host plant. The parasite first develops a tubercle carrying numerous adventitious roots; the apical bud of this tubercle give rise to a subterranean shoot (spider) and then to a branched flowering spike after emergence from the soil (Fig. 1).2-4 The flower produces thousands of extremely small (0.15–0.5 mm long) tan-to-brown colored seeds, which blacken with age and can survive more than 15 years in a crop field until the favorable condition obtained.2-4
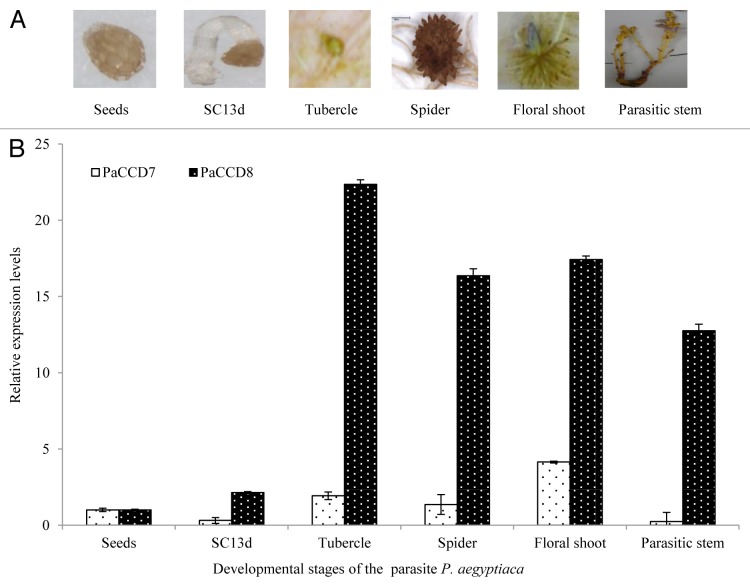
Figure 1. Expression patterns of P. aegyptiaca strigolactone biosynthetic genes during seeds and tubercles development. (A) Photography of different developmental stages of P. aegyptiaca including: dry seeds, seeds conditioned at 30ºC for 13 days (SC13d), tubercle, spider, floral shoot and parasitic stem. (B) Quantification of PaCCD7 and PaCCD8 transcript levels by real-time RT-PCR analysis normalized to equal levels of actin transcripts, in all different developmental stages of P. aegyptiaca. All analyses were performed using three biological replicates.
Strigolactones are plant hormones that were initially identified as signaling molecules in the rhizosphere. They are mainly produced in the roots and have been detected in the root exudates of a wide range of monocot and dicot plant species, supporting the ancient origin and importance in nature.5 In the rhizosphere, strigolactones act as host detection cues for symbiotic arbuscular mycorrhizal fungi and stimulate seed germination of parasitic plants.6 They are derived from carotenoids through sequential oxidative cleavage by carotenoid cleavage dioxygenases CCD7 and CCD8,7,8 thus belonging to the apocarotenoid class.7
Strigolactones occur in all green lineages of the plant kingdom.9-11 Although putative CCD7 was reported to exist in Phelipanche ramosa,12 endogenous strigolactone biosynthetic genes have not been reported in the parasitic plant Phelipanche aegyptiaca. Dependence of parasitic plant seed germination on exogenous strigolactones suggests the absence of endogenous strigolactones or a malfunctioning of strigolactone-related signaling in their seeds. Recently, Liu et al.13 provided the first evidence for strigolactones and strigolactone perception genes of the MAX2-type in the parasite Striga hermonthica. These authors also mentioned EST sequences of endogenous ShCCD7 and ShCCD8 genes but no further sequence information or accession numbers were published.
Here we present evidence of endogenous strigolactone biosynthetic genes CCD7 and CCD8 and their induced expression during development stages of Phelipanche tubercles. In our study, expression of dsRNA of Phelipanche CCD7 and CCD8 target genes in the host plants N. benthamiana by virus induced gene silencing (VIGS) strategy14 suppressed the candidate Phelipanche genes. VIGS is RNA silencing based technique used for the targeted downregulation of a host gene, allowing the analysis of the gene functions.15 The dsRNA replication intermediate would be processed so that the siRNA in the infected cell would correspond to parts of the viral vector genome, including any nonviral insert. Thus, if the insert is from a host gene, the siRNAs would target the RNase complex to the corresponding host mRNA and the symptoms in the infected plant would reflect the loss of the function in the encoded protein.15 VIGS has been used to silence a wide variety of genes in plants.16 The trans-specific RNAi silencing is a genetic interference phenomenon in which double-stranded RNA (dsRNA) induces individual sequence-specific posttranscriptional gene silencing.17 Trans-specific RNAi silencing was visualized first by transforming roots of the hemiparasite Triphysaria versicolor with the β-glucuronidase (GUS) reporter gene and then allowing the transgenic Triphysaria spp. to parasitize host roots expressing an RNAi hairpin construction of GUS (hpGUS).18 VIGS derived dsRNA can apparently transferred from host plant to either herbivores19 or parasitic plants and suppress the expression of target genes (Dubey et al., 2014, unpublished data). In the current study, silencing of Phelipanche CCD7 and CCD8 genes by TRV-VIGS system could lead to retardation of the parasite development.
Potential genes involved in the strigolactone biosynthesis (CCD7, CCD8) in Phelipanche were examined in the parasitic plant genome database (http://ppgp.huck.psu.edu/). Data mining of the parasitic plant genome resulted in identification of sequences PaCCD7 (NCBI Genbank accession number JN412814) and PaCCD8, displaying high sequence similarity to CCD7 and CCD8 from other species. PaCCD7 protein displays high sequence similarity to CCD7 (69% identity) from Nicotiana tabacum and Petunia x hybrida,20 and to CCD7 from Solanum lycopersicum (65% identity).21 PaCCD7 is also similar to CCD7 (59% identity) from Arabidopsis thaliana (Fig. S1). The PaCCD8 protein showed high amino acid similarity to Dad1/CCD8 (85% identity) from Petunia x hybrida,22 to CCD8 (84% identity) from Solanum lycopersicum,23 to CCD8 (84% identity) from Nicotiana tabacum, and also to CCD8 (66% identity) from Arabidopsis thaliana (Fig. S1).
In order to obtain further information about the presence of PaCCD7 and PaCCD8 in parasitic plant development, the expression pattern of P. aegyptiaca PaCCD7 and PaCCD8 genes was examined during different developmental stages of the parasite [dry seeds, seeds conditioned designated SC13d, and tubercle development] by quantitative RT-PCR (Fig. 1). PaCCD7 and PaCCD8 transcripts were detected in dry seeds, conditioned seeds, and in all P. aegyptiaca tubercle developmental stages. The abundance of PaCCD7 transcripts increased with the growth of parasite tubercle, reaching maximum levels at floral shoots followed by decreased levels in the parasite stems (Fig. 1B). Surprisingly, expression of PaCCD8 transcripts remained continuously high up to the last monitored stage, the fully grown parasite stems (Fig. 1B). To ensure that PaCCD7 and PaCCD8 primers are specific only for the parasite and do not amplify CCD7 and CCD8 from the Nicotiana benthamiana host plants, RT-PCR was performed on total RNA from the host roots. No amplification of these genes was detected when primers designed for the P. aegyptiaca genes were used. However, total RNA from VIGS-treated host plants revealed a strong amplification signal by the same primers (Fig. S2). Additionally, to ensure that the host tobacco plant CCDs (CCD7, CCD8) were not also suppressed by the VIGS gene silencing, qRT-PCR was performed on total root RNA from all N. benthamiana plant treatments. Transcripts of NbCCD7 and NbCCD8 in all four measurements, showing that the two investigated genes were expressed in all root of N. benthamiana plant treatments (control wild type plants, and plants infiltrated with TRV:PaCCD7, TRV:PaCCD8, or a mixture of TRV:PaCCD7 and TRV:PaCCD8) (Fig. S3). In general, no significant difference in the NbCCD7 and NbCCD8 transcripts expression levels were observed in all plant treatments (Fig. S3).
In conclusion, these data suggest presence of endogenous strigolactone biosynthesis genes and these genes were expressed in P. aegyptiaca seeds and tubercles. Those, suggest that might be parasitic plant produces its own strigolactones and those they are important for the development of the parasite.
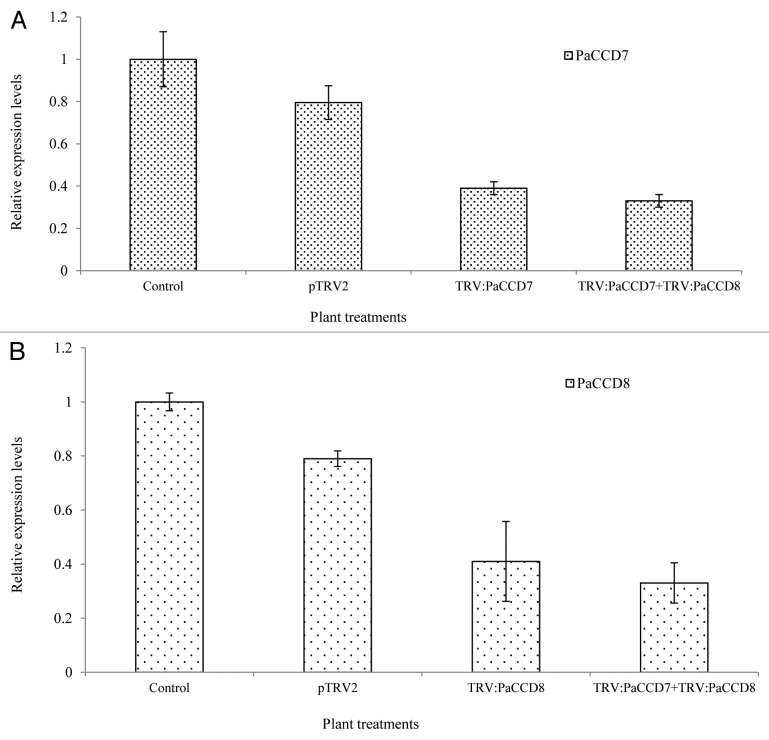
In order to determine whether PaCCD7 and PaCCD8 are functional in the parasite and whether they are indeed involved in the parasite strigolactone biosynthesis, the two genes were transiently knocked-down by the VIGS-mediated strategy.15 Systemic expression of VIGS vector in the host roots followed by Agro-infiltration was confirmed by RT-PCR (Fig. S4). These experiments demonstrated that endogenous PaCCD7 and PaCCD8 transcripts from P. aegyptiaca tubercles grown on N. benthamiana plants infiltrated with TRV:PaCCD7, TRV:PaCCD8 or a mixture of TRV:PaCCD7 and TRV:PaCCD8 were significantly reduced as compared to control plants (wild type plants or plants transformed with empty pTRV plasmids) (Fig. 2A and B).
Figure 2. Real-time RT-PCR analysis of PaCCD7 and PaCCD8 genes in P. aegyptiaca tubercles attached to control and VIGS-treated N. benthamiana plants. (A) Quantification of PaCCD7 transcript levels by qRT-PCR analysis normalized to equal levels of actin transcripts in the underground tubercles of P. aegyptiaca controls and VIGS-treated N. benthamiana plants. (B) Quantification of PaCCD8 transcript levels by qRT-PCR analysis normalized to equal levels of actin transcripts in the underground tubercles of P. aegyptiaca controls and VIGS-treated N. benthamiana plants. All analyses were performed using three biological replicates.
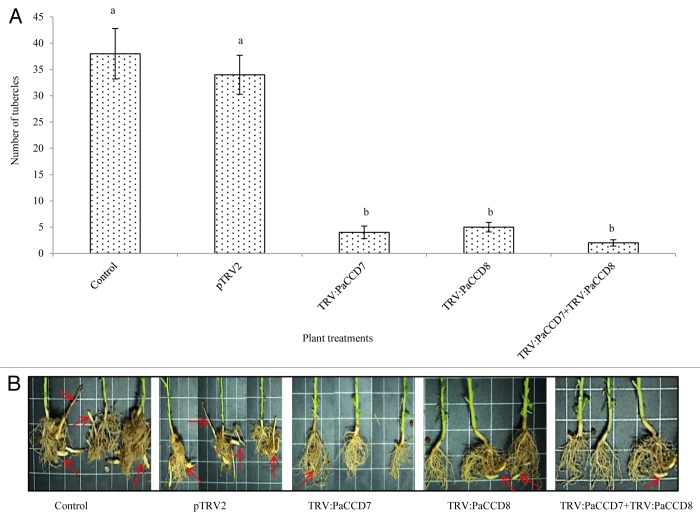
Additionally, the number of parasite tubercles attached to the roots of host plants treated with TRV:PaCCD7, TRV:PaCCD8, or a mixture of TRV:PaCCD7 and TRV:PaCCD8 was significantly reduced (by 95%) as compared to control plants (Fig. 3A and B).
Figure 3. Effect of P. aegyptiaca target genes (PaCCD7 and PaCCD8) silencing on development of the parasite in N. benthamiana host plants. (A) Effect of trans-specific gene silencing of the PaCCD7 and PaCCD8 on the tubercle numbers attached to N. benthamiana plants. (B)P. aegyptiaca tubercles and shoots attached to the controls and VIGS treated N. benthamiana plants indicated by red arrows. Mean ± SE were obtained from 20 independent plants. Bars labeled with different letters indicate the significant differences as determined by JMP statistic software (P ≤ 0.05).
The roles of CCD7 and CCD8 enzymes in the biosynthesis of strigolactones have been reported for several plant species.13,21,23 Recently, it has been shown that endogenous strigolactone biosynthesis and signaling are also present in the parasitic plant Striga hermonthica during their development.13 Specifically, partial sequences of two related CCD genes were identified in Striga EST database and designated as ShCCD7 and ShCCD8.13 Similarly, a full length parasite sequence for ShMAX2 was also reported, which is closely related to host plant MAX2-type genes.13 In this paper we describe two sequences of parasite strigolactone biosynthesis genes, PaCCD7 and PaCCD8 (Fig. S1) which display high sequence similarity to other plant CCD7 and CCD8.20,21,22,23
The PaCCD7 and PaCCD8 genes are expressed in P. aegyptiaca seeds and different development stages of P. aegyptiaca tubercles (Fig. 1), but they differ from the expression profiles of ShCCD7 and ShCCD8.13 Both ShCCD7 and ShCCD8 are expressed specifically in the pre-conditioned seeds of in Striga hermonthica.13 By contrast, in our study, expression of PaCCD8 was relatively high in developed tubercles, but low levels for PaCCD7 in parasitic stem stage of P. aegyptiaca (Fig. 1B). Our results showed that the transcript levels of PaCCD8 were more abundant than those of PaCCD7 in all examined tissues (Fig. 1B). Similar results were also reported by Liu et al.13
Vogel et al.21 showed that tomato SlCCD7 antisense lines display up to 90% reduction in strigolactone content. In this case the host SlCCD7 gene is expressed in all tissues, including roots, stems and fruits. Also, generating SlCCD8 knock-down tomato lines led to the reduction of strigolactone level.23 Based on the results of these antisense and knock-down analyses, we postulate that expression of CCD7 and CCD8 in the parasite is amenable to suppression approaches via VIGS-mediated and transgenic approaches. Interestingly, in our experiments using trans-silencing approaches by expressing parasite gene suppression in N. benthamiana, the infection by P. aegyptiaca was reduced by up to 95% in the VIGS-treated plants (Fig. 2). Reduction in the parasite seed germination and infection could be suggested due to the lack of the endogenous strigolactone of the parasite, which was inhibited following suppression of P. aegyptiaca endogenous strigolactone biosynthesis genes (PaCCD7 and PaCCD8). Liu et al.13 also suggested that Striga endogenous strigolactones possibly play a role in seed germination.
In conclusion, the trans-silencing of the parasitic P. aegyptiaca PaCCD7 and PaCCD8 genes significantly reduced the number of parasite tubercles attached to the host roots of N. benthamiana. To the best of our knowledge, this is the first report using the TRV-VIGS system for gene silencing of strigolactone biosynthetic genes from the parasitic weed P. aegyptiaca. The efficacy of the trans-silencing of PaCCD7 and PaCCD8 as a control strategy against parasitic plant management still needs to be determined in the field by using stable transgenic lines.
For the experiment setup, N. benthamiana plants were grown in polyethylene bags as described by Aly et al.24 Sterilized and conditioned seeds of P. aegyptiaca were spread into the properly established root. Tubercles of different stages (tubercle, spider, floral shoot, and parasitic stem) (Fig. 1) were isolated from the N. benthamiana roots. RNA from dry seeds, seeds conditioned at 30ºC for 13 days (SC13d), and tubercles of different stages was isolated using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). Five hundred ng of total RNA was reversed transcribed to cDNA by using reverse transcriptase (Verso cDNA kit, AB-1453/A, Thermo), followed by 5 fold dilution with molecular grade water. Quantitative RT-PCR (qRT-PCR) was performed by using the sequences of forward 5’-AAGACACCAC CACCATCAAT GC-3’ and reverse primer 5’-GGGACGGGTC TGTTGGTTTC-3’ in case of PaCCD7 and forward 5’-CACCCGATCG TCACGGATA-3’ and reverse primer 5’-CATCACCTTC CTCTCGTTCG TT-3’ in case of PaCCD8. Thermo-SYBR Green Master Mix (AB4162) was used in qRT-PCR according to manufacturer protocol, on the platform of ABI-Prism 7000 Real-Time PCR Detection System (Applied Biosystems). The expressions of target candidate gene were normalized with Actin gene of following forward and reverse primer sequences Actin-F: 5’-AATGATCGGA ATGGAAGCTG-3’, and Actin-R: 5’-TCCACTGAAG GACGATGTTT C-3’. The relative gene expression was calculated by using the 2-ΔΔCt method.25 The qRT-PCR experiment was performed in three biological replicates.
All sequences in the parasitic plant genome database (http://ppgp.huck.psu.edu/) that were similar to Arabidopsis thaliana CCD7 (accession number NM_130064) and CCD8 (accession number NM_119434) were translated using the bioinformatics resource “ExPASy” server (http://www.expasy.org) with several carotenoid cleavage dioxygenase 7 and 8 sequences being reassembled using the CAP3 sequence assembly program.
In this study, two carotenoid cleavage dioxygenases (CCD) genes were tentatively identified as P. aegyptiaca PaCCD7 and PaCCD8. For VIGS experiment, unique N-terminal regions about 300 bp of PaCCD7 and PaCCD8 (Fig. S5) were selected and amplified from the cDNA prepared from tubercles followed by cloning into pJET1.2 vector (Thermo Scientific). PaCCD7 was amplified by using the forward primer of 5’-GAGCTCATGC AGTCTGCCAC AGCTTGC-3’ sequence overhang with restriction site of Sac I and reverse primer of 5’-CTCGAGGGGA TGGACGGTAG ACCCG-3’ with the overhang of Xho I, respectively. Similarly, PaCCD8 was amplified by using the forward primer of sequence 5’-GAATTCTACA TTCAGCCCAC CCGATC-3’ with overhang of EcoR I and reverse primer with sequence of 5’-GGATCCCATG CATGTAACCT TTAGAGTTAG-3’ with BamH I restriction site. Both PaCCD7 and PaCCD8 genes were cloned separately into pTRV2 (Tobacco Rattle Virus vector 2) at the respective site and designated as TRV:PaCCD7 and TRV:PaCCD8, respectively. The VIGS assay was performed as described by the Bachan et al.26 The experiments were performed in N. benthamiana plants grown in the "Newe Yaar" Research Center in northern Israel, under standard field irrigation and fertilization conditions.4 Agrobacterium (strain EHA105) containing pTRV1 with TRV:PaCCD7 and/or TRV:PaCCD8 were mixed and infiltrated into the leaves of N. benthamiana. Non-infiltrated N. benthamiana plants, and plants infiltrated with mixtures of Agrobacterium transformed with empty plasmid of pTRV1 and pTRV2 were used as controls.
N. benthamiana plants were pre-challenged with parasitic seed about ten days before Agro-infiltration. After ten days of co-cultivation, RNA was isolated from leaves and roots of tobacco plants and systemic expression of TRV virus was analyzed. About 25 days after Agro-infiltration, tubercles attached to the VIGS-treated plants were isolated and the total number of tubercles attached to the VIGS-treated and control plants were counted.24 Additionally, the level of expression of the PaCCD7 and PaCCD8 genes was examined by qRT-PCR. All analyses were performed using three biological replicates.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Authors are very grateful to Professor Amit Gal-On for providing pTRV vectors. We also thank Anna Berim for valuable suggestions and scientific writing.
References
- 1.Parker C. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci. 2009;65:453–9. doi: 10.1002/ps.1713. [DOI] [PubMed] [Google Scholar]
- 2.Irving LJ, Cameron D. You are what you eat: interactions between root parasitic plants and their hosts. In: Kader JC, Delseny M, eds. Advances in Botanical Research, 2009:87-138. [Google Scholar]
- 3.Westwood JH, Roney JK, Khatibi PA, Stromberg VK. RNA translocation between parasitic plants and their hosts. Pest Manag Sci. 2009;65:533–9. doi: 10.1002/ps.1727. [DOI] [PubMed] [Google Scholar]
- 4.Aly R, Hamamouch N, Abu-Nassar J, Wolf S, Joel DM, Eizenberg H, Kaisler E, Cramer C, Gal-On A, Westwood JH. Movement of protein and macromolecules between host plants and the parasitic weed Phelipanche aegyptiaca Pers. Plant Cell Rep. 2011;30:2233–41. doi: 10.1007/s00299-011-1128-5. [DOI] [PubMed] [Google Scholar]
- 5.Xie X, Yoneyama K, Yoneyama K. The strigolactone story. In: VanAlfen NK, Bruening G, Leach JE, eds. Annu Rev Phytopathol, 2010:93-117. [DOI] [PubMed] [Google Scholar]
- 6.López-Ráez JA, Charnikhova T, Fernández I, Bouwmeester H, Pozo MJ. Arbuscular mycorrhizal symbiosis decreases strigolactone production in tomato. J Plant Physiol. 2011;168:294–7. doi: 10.1016/j.jplph.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Walter MH, Floss DS, Strack D. Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta. 2010;232:1–17. doi: 10.1007/s00425-010-1156-3. [DOI] [PubMed] [Google Scholar]
- 8.Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335:1348–51. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 9.Delaux P-M, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N. Origin of strigolactones in the green lineage. New Phytol. 2012;195:857–71. doi: 10.1111/j.1469-8137.2012.04209.x. [DOI] [PubMed] [Google Scholar]
- 10.Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 2012;159:1073–85. doi: 10.1104/pp.112.196253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brewer PB, Koltai H, Beveridge CA. Diverse roles of strigolactones in plant development. Mol Plant. 2013;6:18–28. doi: 10.1093/mp/sss130. [DOI] [PubMed] [Google Scholar]
- 12.Péron T, Véronési C, Mortreau E, Pouvreau J-B, Thoiron S, Leduc N, Delavault P, Simier P. Role of the sucrose synthase encoding PrSus1 gene in the development of the parasitic plant Phelipanche ramosa L. (Pomel) Mol Plant Microbe Interact. 2012;25:402–11. doi: 10.1094/MPMI-10-11-0260. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Zhang Y, Matusova R, Charnikhova T, Amini M, Jamil M, Fernandez-Aparicio M, Huang K, Timko MP, Westwood JH, et al. Striga hermonthica MAX2 restores branching but not the very low fluence response in the Arabidopsis thaliana max2 mutant. New Phytol. 2014;202:531–41. doi: 10.1111/nph.12692. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–29. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. Virus-induced gene silencing in plants. Methods. 2003;30:296–303. doi: 10.1016/S1046-2023(03)00037-9. [DOI] [PubMed] [Google Scholar]
- 16.Robertson D. VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol. 2004;55:495–519. doi: 10.1146/annurev.arplant.55.031903.141803. [DOI] [PubMed] [Google Scholar]
- 17.Bandaranayake PCG, Yoder JI. Trans-specific gene silencing of acetyl-CoA carboxylase in a root-parasitic plant. Mol Plant Microbe Interact. 2013;26:575–84. doi: 10.1094/MPMI-12-12-0297-R. [DOI] [PubMed] [Google Scholar]
- 18.Tomilov AA, Tomilova NB, Wroblewski T, Michelmore R, Yoder JI. Trans-specific gene silencing between host and parasitic plants. Plant J. 2008;56:389–97. doi: 10.1111/j.1365-313X.2008.03613.x. [DOI] [PubMed] [Google Scholar]
- 19.Kumar P, Pandit SS, Baldwin IT. Tobacco rattle virus vector: A rapid and transient means of silencing manduca sexta genes by plant mediated RNA interference. PLoS One. 2012;7:e31347. doi: 10.1371/journal.pone.0031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond RSM, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol. 2009;151:1867–77. doi: 10.1104/pp.109.146720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel JT, Walter MH, Giavalisco P, Lytovchenko A, Kohlen W, Charnikhova T, Simkin AJ, Goulet C, Strack D, Bouwmeester HJ, et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2010;61:300–11. doi: 10.1111/j.1365-313X.2009.04056.x. [DOI] [PubMed] [Google Scholar]
- 22.Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell. 2005;17:746–59. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohlen W, Charnikhova T, Lammers M, Pollina T, Tóth P, Haider I, Pozo MJ, de Maagd RA, Ruyter-Spira C, Bouwmeester HJ, et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012;196:535–47. doi: 10.1111/j.1469-8137.2012.04265.x. [DOI] [PubMed] [Google Scholar]
- 24.Aly R, Cholakh H, Joel DM, Leibman D, Steinitz B, Zelcer A, Naglis A, Yarden O, Gal-On A. Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol J. 2009;7:487–98. doi: 10.1111/j.1467-7652.2009.00418.x. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Bachan S, Dinesh-Kumar SP. Tobacco rattle virus (TRV)-based virus-induced gene silencing. In: Watson JM, Wang M-B, eds. Antiviral Resistance in Plants: Humana Press, 2012:83-92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.