Abstract
In order to shuttle substrates across the lipid bilayer, membrane proteins undergo a series of conformation changes that are influenced by protein structure, ligands, and the lipid environment. To test the effect of lipid on conformation change of the ABC transporter MolBC, EPR studies were conducted in lipids and detergents of variable composition. In both a detergent and lipid environment, MolBC underwent the same general conformation changes as detected by site-directed EPR spectroscopy. However, differences in activity and the details of the EPR analysis indicate conformational rigidity that is dependent on the lipid environment. From these observations, we conclude that native-like lipid mixtures provide the transporter with greater activity and conformational flexibility as well as technical advantages such as reconstitution efficiency and protein stability.
Keywords: ABC transporter, detergent, lipids, reconstitution, continuous wave-electron paramagnetic resonance, molybdate-transport system, Haemophilus influenza, conformational changes
Introduction
Integral membrane proteins play many essential roles from cellular signaling to the transport of molecules of various sizes and chemotypes.1 It is important to understand how these membrane-embedded proteins interact with the lipid molecules found in their natural environment.2 Transmembrane (TM) proteins insert into the lipid bilayer allowing for the non-polar regions to contact the hydrophobic lipid tails and polar regions to be exposed to the aqueous intra- or extracellular spaces.3 Detergents are often used to work with TM proteins in vitro because they mimic the lipid bilayer and prevent protein unfolding and aggregation.4,5 Though often effective in stabilizing a TM protein, the detergent micelle can alter the activity and regulation of a protein6-8 and may have effects on conformational shifts.9-12 This necessitates the study of TM proteins in a lipid bilayer.
Analysis of protein dynamics and conformation change has been greatly facilitated by site-directed spin-labeling electron paramagnetic resonance spectroscopy (SDSL-EPR).13,14 An EPR-sensitive spin-label is covalently attached to cysteine residues engineered at specific sites in the protein. Analysis of the EPR spectrum allows us to determine the mobility of the spin-label and the distance between spin-labels. EPR spectroscopy only detects lone-pair electrons such as those provided by the stable free-radical group of spin-label or certain oxidation states of transition metals. Since most lipids, detergents, and aqueous buffers do not contribute to the EPR spectrum, EPR spectroscopy is very advantageous for mechanistic studies in a lipid environment.15
The differences between a lipid bilayer and a detergent micelle begin with variances at the molecular scale. The detergents used for structural studies often have an uncharged head group that is large relative to the non-polar tails (Fig. 1A). This gives the molecule a cone shape, which forms micelles with a small radius.16 Commonly used lipids have a variety of head groups and two lipid tails that give the molecule a roughly cylindrical shape that enables for formation of lipid bilayers (Fig. 1B). The lipid composition of a bilayer has a well-documented effect on TM protein function,17,18 which can be roughly divided into direct and indirect interactions of lipid with protein. “Co-factor” lipid molecules can interact directly with a TM protein at specific sites with relatively high affinity,19,20 such interactions can be vital for protein function and stability.21,22 Other lipid-protein interactions are direct yet non-specific, involving the ring of annular lipids surrounding the TM protein. Often the hydrophobic region of a TM protein will have grooves or cavities where lipids or amphipathic molecules bind and affect the activity of the protein.23 Bulk properties of lipid bilayers such as thickness, lateral pressure, viscosity, and curvature may affect protein function and stability by altering the energy changes involved in conformation change.24 For example, if the thickness of the annular lipid bilayer does not match the thickness of the TM region, either the protein or lipid will alter its structure to minimize the mismatch. However, altering the bilayer thickness is a high energy solution, so hydrophobic mismatch is more likely to affect protein structure, especially for α-helical membrane proteins.24-26
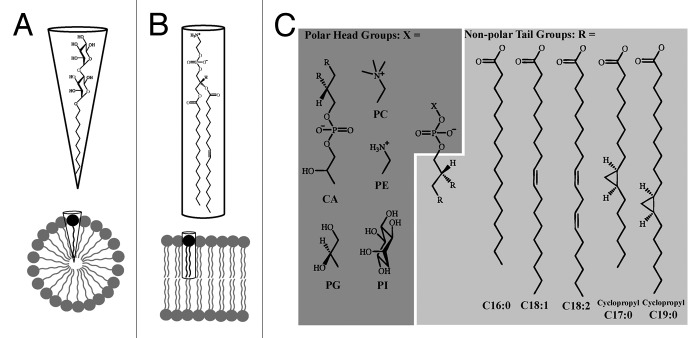
Figure 1. Chemical properties of detergents and lipids. (A) n-Decyl-β-D- maltopyranoside is a common non-ionic detergent used to stabilize TM proteins.5 The large polar head group and relatively short non-polar tail give the detergent a roughly conical shape, which facilitates the formation of detergent micelles. (B) Phosphatidylethanolamine can represent up to 70% of lipid in bacterial membranes;35,46 its roughly cylindrical shape enables the formation of lipid bilayers. (C) In addition to triglycerides and hopanoids, phospholipids are an important component of lipid bilayers. Phospholipids contain three components attached to a glycerol backbone: two non-polar tails (R, light gray) and one polar head group (X, dark gray). Here we show a selection of head groups including cardiolipin (CA), phosphatidylglycerol (PG), phosphatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylinositol (PI); and major tail groups from E. coli: palmitic acid (C16:0), oleic acid (C18:1), linoleic acid (C18:2), 9,10-methylene-hexadecanoic acid (cyclopropyl C17:0), and lactobacillic acid (cyclopropyl C19:0). Chemical structures were prepared using ChemDraw (CambridgeSoft Inc.).
Though studies in detergent are an appropriate starting point, the above effects of lipid on TM proteins require further study in a lipid environment. This is particularly true for mechanistic studies where a TM protein is being monitored for ligand-dependent conformation changes. Ideal targets of such studies are ABC (ATP-binding cassette) transporters, which undergo conformation change in the presence of ligands (e.g., substrate binding protein, ATP). These membrane pumps transport a variety of substrates across the lipid bilayer, utilizing ATP as an energy source.27 For Type II ABC importers, there is limited information on the mechanism, especially in a lipid environment.7,9,10,28,29 This family of membrane proteins includes many potential drug targets, which take up vital nutrients and cofactors and may be found in many pathogenic bacteria, so understanding the mechanism of transport is essential.
The mechanism of the Type II molybdate importer, MolBC-A,30 was recently elucidated using detergent-stabilized protein31 and reconstituted proteoliposomes.32 Two gates control the flux of substrate through the central translocation pathway. The periplasmic gate undergoes subtle conformation shifts, effectively opening in the presence of nucleotide. The cytoplasmic gate is formed by a loop between TM helices 2 and 3 and can be probed via site-directed CW-EPR at residue N93C. This cytoplasmic gate closes upon nucleotide binding, which could be monitored by an increase in coupling between EPR spin-labels attached to N93C.31,32 For this study, we also probed a third gate (“cytoplasmic gate I”) present in Type II large substrate transporters, which opens in the presence of nucleotides.9,29 In MolBC this site was probed by site directed EPR at I151C and was found to shift from a slightly open conformation to an open conformation.31 While this gate did not close, and thus was not considered a gate in a small substrate transporter like MolBC, conformational changes at this site provide valuable information on the effects of lipid composition on protein conformation.
To test the effects of lipid composition on the nucleotide triggered movement of sites along the transmembrane domain, we attached nitroxide based spin labels to two positions in the translocation channel, N93C and I151C. MolBC-A was then reconstituted into different lipid extracts and tested for activity and conformational shifts at spin-labeled N93C and I151C. EPR analysis of MolBC-A at both sites indicated that lipid restricted the conformational changes along the TMD, though some lipid types restricted movement more than others. Similarly, the type of lipid extract affected the ATP hydrolysis activity of the protein. Our results show that technical factors such as reconstitution efficiency and sample viscosity also varied in the different extracts. Our purpose in this work is to share observations from our studies and methodology, which may be applicable in studies to determine protein dynamics in a lipid environment.
Results
When choosing the lipid into which MolBC would be reconstituted, we had the option of lipids synthesized with a known head group and tail length or lipids extracted from cell membranes with different levels of purity (Fig. 1C). For example, phosphatidylcholine (PC) lipid could be extracted and purified from soybean or chicken egg, in which case PC is the most abundant head group and tail lengths vary in proportions. Alternatively, an extraction of all lipid species from an organism can be collected (e.g., E. coli total lipid extract) and optionally fractionated (E. coli polar lipid extract) eliminating some of the lipid species from the extract. The mixture of lipid chemistries found in the extracts confers some advantages in our mechanistic studies. First, if a TM protein required an unknown co-factor lipid for optimal activity, an extract has a greater chance of containing this potential co-factor. Second, the risk of hydrophobic mismatch is mitigated by the variety of tail lengths present in the extract; MolBC would pair with the appropriate length of lipid to form the lowest energy state. Third and most important, the mixture of lipid chemistries conferred by an extract better resembles the natural environment of the plasma membrane.33,34
Three lipid extracts were chosen for these studies due to their frequency of use in EPR membrane protein studies7,8 or their similarity to the native environment for MolBC. The first lipid selected, Soybean PC, is a lipid extract containing 14–23% PC in addition to phospholipids with other head groups such as phosphatidylethanolamine (PE) and phosphatidylinositol (PI). The major components of the hydrophobic tail region are 13% C16:0, 4% C18:0, 10% C18:1, 64% C18:2, and 6% C18:3 (Sigma: P5638, product information). We assumed that there would be variability between lots, so care was taken to choose a lot represented in the literature8 and to consistently use lipids from the selected lot. The second lipid used in this study, E. coli polar extract is acquired from E. coli (ATCC 11303) grown at 37 °C in Kornberg Minimal media (Avanti: 100600C, product information). The major head groups are 67% PE, 23% phosphatidylglycerol (PG), 9.8% cardiolipin (CA). The hydrophobic tails contain 1% C14:0, 36% C16:0, 2% C16:1, 20% cyclopropyl-C17:0, 1% C18:0, 28% 18:1, 12% cyclopropyl-C19:0.35 We mixed this lipid extract with Egg PC (99% PC; Avanti: 840051C, product information) in a 3:1 ratio following the lead of work done the lead of experiments conducted on another ABC transporter.7 The third lipid selected was E. coli total lipid extract, which did not undergo the additional fractionation of E. coli polar extract. The major head groups are 57.5% PE, 15.1% PG, 9.8% CA, and 17.6% of unclassified lipid content (Avanti: 100500C, product information). The tail content should be similar to the polar lipid extract described above. The E. coli total and polar extracts are closer to the native distribution lipids found in the organism of origin for MolBC-A, Hemophilus influenzae, which tend to have short tail lengths. For example, 35 tested strains of H. influenzae contained 12.7 ± 2.3% C14:0, 11.6 ± 1.08% 3-hydroxy-C14:0, 40.6 ± 2.41% C16:0, 31.4 ± 2.41% C16:1, 2.4 ± 1.13% C18:0, 0.5 ± 0.16% C18:1, and 0.2 ± 0.15% C18:2.34
For high signal to noise in an EPR spectrum, it is important to have a high concentration of spin-labeled protein. The reconstitution of TM protein into a lipid bilayer is a major factor that can limit the final protein concentration and required optimization to improve the quality of the final sample. A lipid to protein ratio of 10:1 (w/w) was chosen to maintain a stable reconstituted sample with enough excess lipid to mitigate the effect of residual detergent. This ratio was kept low relative to standard liposome ratios36 to maximize the final protein concentration in the sample. Reconstitution efficiencies into the E. coli polar lipid mix (77 ± 5%) and E. coli total lipid (72%) were often greater than the efficiency of Soybean PC (68 ± 7%).
Another factor that limits the final sample protein concentration is viscosity; a liposome sample can be easily concentrated by ultracentrifugation, but for EPR analysis the proteoliposome sample needs to be resuspended and fluid enough for loading and removal from a thin 50uL capillary. We found that the proteoliposome samples made with Soybean PC or E. coli polar lipid: Egg PC mixes were notably less viscous at 23 °C when compared with E. coli total lipid samples when resuspended to a similar volume.
For our studies on ligand-dependent conformation change of MolBC-A in a lipid environment, we were interested in the effect of lipid on protein activity. We found that reconstitution of MolBC from detergent to liposome decreased the basal (substrate-free) levels of ATP hydrolysis 3 to 24-fold (Table 1). Different lipid extracts had different effects on activity, with the two E. coli extracts showing higher specific activity than the Soybean PC proteoliposomes. However, all samples have a moderate capacity to bind and hydrolyze ATP. Therefore, incubation periods were timed so that a large majority of the transporter population should have bound ligand.
Table 1. ATP hydrolysis activity for MoIBC in liposomes.
| ATP hydrolysis activity for MolBC in liposomes | |||
|---|---|---|---|
| MolBC Mutant | Lipid | Specific activity (nmol Pi/mg enzyme/min) | Standard error of Activity (nmol Pi/mg enzyme/min) |
| N93C+MTSL | DM detergent | 1790 | 66 |
| N93C+MTSL | Soybean PC | 200 | 16 |
| N93C+MTSL | E. coli polar: Egg PC (3:1) | 415 | 6 |
| N93C+MTSL | E. coli Total lipid | 526 | 8 |
| I151C+MTSL | DM detergent | 1782 | 87 |
| I151C+MTSL | Soybean PC | 73 | 5 |
| I151C+MTSL | E. coli polar: Egg PC (3:1) | 288 | 7 |
Continuous wave-EPR spectroscopy indicates that reconstitution of MolBC from detergent to lipid affects the conformation cycle of the transmembrane domain (TMD), as reported by the spin labels at N93C and I151C. Spin-coupling is a measure of the proximity between spin-labels and can be qualitatively identified by broadening in the low-field region of the EPR spectrum.37 When the spin-labels assume a closer conformation the broadening shifts further down field; when broadening cannot be observed the spin-labels are beyond the detectable distance for CW-EPR spectroscopy (~1.8nm).
When the TMD was probed at N93C (Fig. 2A), we are able to observe spin-coupling in the EPR spectrum (Fig. 2B). Comparing detergent-solubilized MolBC with lipid reconstituted transporter, we see that the reconstituted sample shows less spin-coupling (Fig. 2B). When nucleotide-free (apo) MolBC_N93C spectra are simulated and quantified, we see that both detergent and lipid give a population of three distances 0.8, 0.95, and 1.2nm, though in lipid the larger distance is weighted more (21% in lipid vs 6% in detergent).31,32 This suggests that a wider spaced population of spin-label rotamers is stabilized by the lipid bilayer. In addition, we see a shift from predominately mobile spin-label in detergent to immobile spin-label in lipid, indicating that a lipid bilayer keeps the spin-label in a more restricted conformation. The presence of ATP did not alter the mobility in either sample, but spin-coupling was increased (Fig. 2C). When quantified, the majority of the spin for both detergent and lipid shifted to a close distance (at about 0.6nm), indicating a closure of the cytoplasmic gate. Residual spin at 0.9nm in the nucleotide-bound liposome sample indicates that the conformation shift in liposomes was not as complete as it had been in detergent. Addition of magnesium chloride allowed MolBC to hydrolyze the ATP and revert to an apo-like conformation with the spin-labels at N93C slightly separated (Fig. 2D).
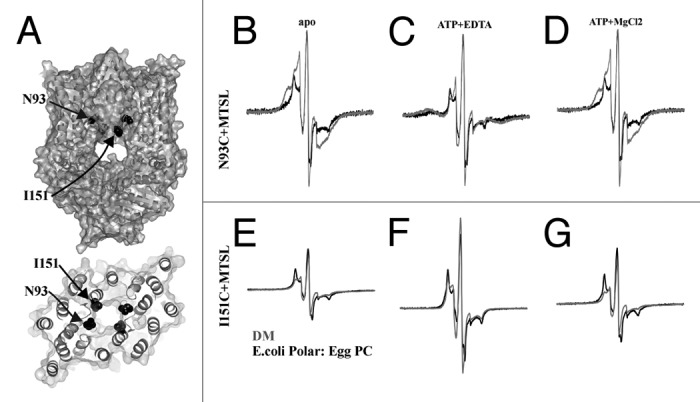
Figure 2. CW-EPR spectroscopy of MolBC_N93C+MTSL and I151C+MTSL showing the effect of lipid reconstitution on TMD conformation change. (A) Ribbon and space fill diagram of MolBC with N93 and I151 shown in black spheres. CW-EPR spectra of MolBC_N93C+MTSL (B-D) and I151C+MTSL (E-G) stabilized in E. coli polar lipid: Egg PC (3:1) liposomes or DM detergent micelles (black or gray respectively). Room temperature spectra (250 Gauss scan width) were recorded apo (B and E), ATP-bound (C and F), and post-hydrolysis (D and G). N93C spectra were normalized by the height of the central peak. I151C spectra were normalized for equal spin (normalized double integration values). MolBC surface and ribbon diagrams were prepared using PyMOL.47 Spectra were graphed using Grapher 9 (Golden Software Inc.).
For site I151C, we also see a decreased the mobility of spin-label when reconstitution from detergent to lipid (Fig. 2E). The change in mobility suggests that the lipid dependent conformation shift affects multiple residues along the translocation pathway. However, unlike N93C, liposome reconstitution does not affect the general conformation cycle at I151C, at least not with a mix of E. coli polar lipid and Egg PC. In both detergent and lipid, I151C assumes a close but not closed conformation apo and post-hydrolysis (Figs. 2E and 2G). And with ATP-bound, no broadening is observed, suggesting a widely separated conformation at I151C (Fig. 2F). The reconstitution of MolBC from a detergent micelle to a lipid bilayer appears to decrease spin mobility at both sites tested along the TMD and weaken conformation shifts at N93C.
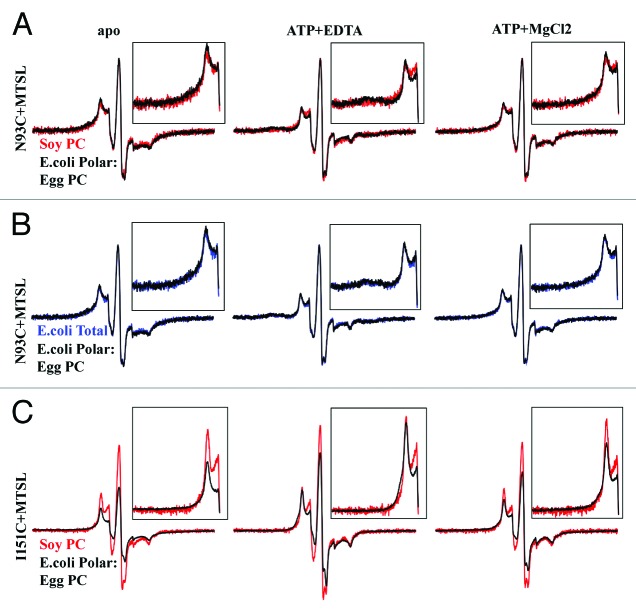
Continuing our EPR studies in liposomes of different lipid content, we see that the type of lipid does effect conformation change at both TMD sites. In the apo state, soybean PC and E. coli polar lipid mix appear to hold N93C in similar conformations (Fig. 3A). However, the nucleotide-dependent closure of the cytoplasmic gate at N93C is not observed in Soybean PC, while is it clearly observed in E. coli polar lipid mix. Alternatively, E. coli total lipid extract and E. coli polar lipid extract mix appear to give identical conformation shifts at N93C (Fig. 3B). At I151C, soybean PC does not appear to stabilize the close conformation of spin-label (Fig. 3C). A separated state is indicated by the greater intensity at the central peak and less broadening in the low-field spin. As stated above, in E. coli polar lipid mix, spin-label at I151C separates in the presence of ATP, but as there was no observable coupling in the apo state of soybean PC samples, we cannot confirm or rule out conformation change at I151C in soybean PC. Post-hydrolysis, we see again that soybean PC does not support a close conformation, though the difference between the two lipid types is less noticeable, which may be due to inefficient ATP hydrolysis in the reconstituted sample. As in the E. coli lipid mix, spin-label at I151C is mostly immobile in soybean PC, though there is a significant fraction of highly mobile spin indicative of denatured protein. This indicates that MolBC is less stable in soybean PC than the E. coli lipid extracts.
Figure 3. Effect of lipid identity on nucleotide-dependent conformation change, shown by CW-EPR. (A) MolB_N93C and MolBC_I151C were spin-labeled with MTSL then reconstituted into soybean PC liposomes (red), E. coli polar lipid: Egg PC (3:1) liposomes (black), or E. coli total lipid liposomes (blue). CW-EPR spectra (250G) were recorded apo, ATP-bound, and post-hydrolysis. For N93C, spectra from samples in E. coli polar: Egg PC are overlaid with soybean PC (A) or E. coli total lipid spectra (B) and were normalized by the height of the central peak. (C) For I151C, the E. coli polar: Egg PC spectra are overlaid with soybean PC and were normalized for equal spin (normalized double integration values).
Discussion
It is immediately clear that detergent and lipid provide different environments for MolBC. Reconstitution into proteoliposomes is known to eliminate basal levels of ATP hydrolysis for the Type I importer MalFGK assayed in the absence of substrate binding protein.8 And the basal activity of the Type II transporter BtuCD decreases from 980 to 200nmol Pi/mg enzyme/min upon reconstitution from LDAO detergent to liposomes.7 We observed a similar reduction for MolBC when reconstituting from DM to liposomes (Table 1). The most likely explanation for this effect on activity would be that a lipid environment slows or otherwise alters the conformation cycle required for ATP binding, hydrolysis, and release.
The difference between the detergent and lipid environments is also confirmed by CW-EPR spectroscopy. At the TMD cytoplasmic gate (N93C), a detergent micelle allows for a greater percentage of transporters with closed gates in the presence of ATP. Moreover, at both TMD sites and independent of nucleotide, the spin-labels shift from high mobility in detergent to low mobility in lipid. These data indicate that there are more rotameric sub-states available to spin-label when MolBC is embedded in a detergent micelle vs. a lipid bilayer. Possible explanations for this lipid effect can be found in the literature. Molecular crowding18 and lateral pressure38,39 applied by the lipid bilayer may be the source of this lipid-effect. Simulation studies with the α-helical protein GlpF40 and the β-barrel OmpA41 have indicated that a detergent micelle allows slightly greater flexibility to TM secondary structure relative to a simulated lipid bilayer. Similarly, the LCP structure of R. sphaeroides photosynthetic reaction center indicated a slight compression of TM helices relative to the in-detergent structure.42 Figure 4 illustrates the potential differences between the detergent and lipid environments, with MolBC surrounded by a thin detergent micelle (Fig. 4A; which may be better described as “a prolate ring” of detergent43) or a lipid bilayer (Fig. 4B). The detergent ring allows the protein to properly fold and undergo conformation change, but we can expect that it will enforce different restrictions on ATP-driven conformation change. Moreover, the lipid bilayer incurs a significant energetic cost upon deformation,24,26,44 which would resist conformation change in MolBC.
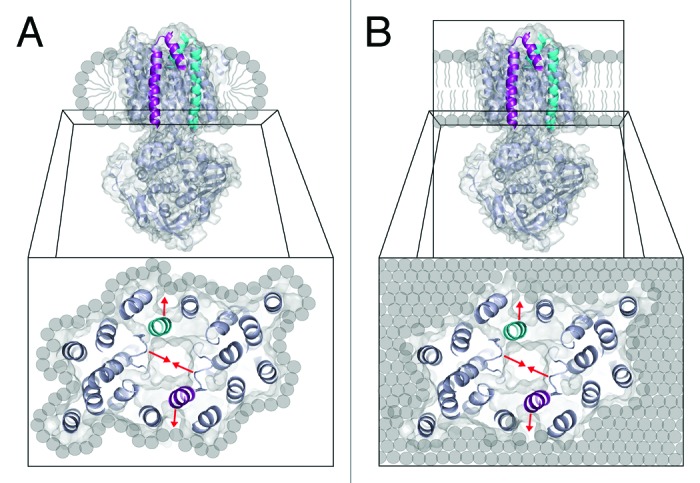
Figure 4. Schematic of MolBC in a detergent micelle and lipid bilayer. Ribbon and space fill diagram of MolBC with translocation pathway helices 5 and 5a colored cyan on one monomer and magenta on the other. (A) Detergent will form a band of hydrophobic tails that cover the hydrophobic region of MolB and stabilize the protein. When we analyze conformation change at the cytoplasmic region of the transporter, we see that the cytoplasmic gate closes in response to nucleotide binding (red arrows). (B) In the presence of lipid, the binding of ATP causes the NBDs and cytoplasmic gate to close. However, the lipid bilayer appears to resist conformation change of MolBC and allows the transporter less conformational flexibility.
The results we observed in different lipid extracts were more unexpected, indicating the significance of lipid composition. The E. coli lipid extracts allow similar levels of moderate ATP hydrolysis and conformation change at N93C. However, the Soybean PC extract diminished ATP hydrolysis to a greater extent and did not allow significant nucleotide-dependent conformation change at both TMD sites. It is reasonable to assume that transport-associated conformation change will be observed in more native-like lipids. Looking for a specific cause of the soybean PC effects, we can rule out the relatively high concentration of phoscholine head groups, since the E. coli polar: Egg PC mix had a similar percentage (25%). It is more likely that the shorter lipids present in the E. coli extracts supported the proper conformation changes of MolBC, accounting for the higher activity and greater conformational flexibility in these extracts relative to SoyPC. The heterogeneous nature of extracts limits our ability to pinpoint a particular cause of the different lipid effect. However, our findings highlight the importance of testing multiple lipid compositions when analyzing protein conformation change.
This study on MolBC-A highlights the importance of comparative analysis of EPR data in multiple environments. Combined, the results suggest that a lipid bilayer forms a more restrictive environment for the MolBC transporter relative to detergent micelles, where spin-label rotamer shifts and ligand-dependent conformation shifts are more limited. Moreover, we found that some lipid extracts were more restrictive than others. The more native-like mix of E. coli polar lipid and Egg PC offered our MolBC studies a good balance of reconstitution efficiency, sample viscosity, protein activity, and conformational freedom.
Materials and Methods
Mutagenesis, protein purification and spin-labeling
We have previously described the initial cloning, overexpression,30 mutagenesis, and purification of MolBC-A.31 Briefly describing our methods of purification and spin-labeling, cells with overexpressed MolBC_N93C were lysed in the presence of n-decyl-β-D-maltopyranoside (DM; Anatrace: D322) detergent and centrifuged. Lysate was loaded onto a Ni-NTA affinity column and washed. MTSL was added to a final concentration of 0.1mM and incubated with mixing at 4 °C for ~16 hours. Free-spin was removed via 80 column volumes of washing. Spin-labeled MolBC_N93C was eluted in 250mM imidazole, then dialyzed and concentrated to at least 120µM. A 27µL aliquot of concentrated, detergent-solublized, spin-labeled MolBC_N93C was used to quantify spin-labeling efficiency via CW-EPR.
Lipid preparation
Chloroform-dissolved E. coli Polar lipid (Avanti: 100600C) and Egg PC (Avanti: 840051C) were mixed in a 3:1 (w:w) ratio. The chloroform solvent dissolves both lipids and facilitates the formation of liposomes with uniform lipid content. A known amount of either E. coli Polar lipid: Egg PC lipid mixture or E. coli total lipid (Avanti: 100500C) was aliquoted into glass vials with Teflon coated caps. Lipids were dried under a stream of Argon while vortexing. Vortexing ensured that a thin layer of lipids was deposited on the walls of the vial. Residual chloroform was removed under vacuum for 16hrs and dry lipids stored at -20 °C for subsequent use. Soybean PC lipid extract (Sigma: P5638) was shipped dry and was resuspended directly in aqueous buffer.
The lipids were resuspended to 50mg/mL in 25mM Tris; pH 7.5; 100mM NaCl. To mitigate lipid oxidation, it was preferred to use degassed buffers and work under argon as much as possible. But working with the above lipid extracts, degassing did not affect the EPR results. A cloudy lipid suspension was produced by vortexing. The sample was sonciated 10 s on/ 10 s off for a total on time of 5min using a micro-tip probe sonicator (amplitude = 10–40 for samples ranging from 0.2 to 2mL respectively). Glass vials containing the lipid solution were clamped and incubated in room temperature water to disperse heat during sonication. A decrease in the optical density of the solution indicated that sonication successfully broke up the multi-laminar lipid sheets of the suspension and formed liposomes. Liposomes were kept covered, on ice, and used within a few hours of sonication.
Biobead preparation and use
Approximately 2mL biobeads (Bio-Rad: 152–8920) were washed with 2x10mL methanol, 3x10mL water, then 3x10mL 25mM Tris; pH 7.5; 100mM NaCl. To make the biobeads more effective at absorbing detergent, they were allowed to soak in buffer overnight before use. In order to measure the volume of dried biobeads, a clipped and calibrated Eppendorf pipette tip (P100 or larger) was used. Clipping the tip made the aperture large enough for biobeads to enter; the beads were allowed to settle in the tip forming a plug that reached the calibration mark. Excess liquid was then dispensed, leaving a plug of dry biobeads. Biobeads were then transferred to lipid protein solution as described below.
Protein reconstitution
The liposomes were dissolved with 40-80mM Sodium cholate. Different components of lipid extracts required more cholate than others to dissolve. An optical density measurement equal to buffer verified that 40mM cholate was the minimum concentration required to completely dissolve Soybean PC and E. coli Total extract liposomes (E. coli polar: Egg PC (3:1) liposomes required 80mM cholate ). Dissolved lipid and purified protein were mixed in a 10:1 (w:w) ratio, incubated on ice for 5min, then biobeads were added to the sample (2/3 of the sample volume in dry bio beads). Samples were incubated at 4 °C for ~16hrs with rocking, then at room temperature for 2–3hrs. The biobeads slowly removed the cholate (and residual DM from the protein purification), forming proteoliposomes. Samples were diluted with 25mM Tris; pH 7.5; 100mM NaCl to fill a Ti 45 rotor centrifuge tube (Beckman: 355622 ~73mL) and centrifuged at 150,000xg for 2hrs at 4 °C. The dilution and centrifugation steps aided liposome formation by diluting detergent and removing residual detergent-stabilized protein. Proteoliposomes were resuspended in a minimal volume of 25mM Tris; pH 7.5; 100mM NaCl. Reconstitution efficiency was calculated by determining the spin/protein ratio in the detergent-solubilized sample with the protein concentration determined via BCA protein assay (Thermo Scientific: 23250); the spin/protein ratio was used to calculate the amount of protein in a liposome sample, which was divided by the amount of protein reconstituted.
ATP hydrolysis Assay
An enzyme linked inorganic phosphate assay (Cytoskeleton Inc.) was used to assay rates of ATP hydrolysis. Assays were set-up in a UV transparent 96-well, half-area plate with 0.2mM 2-amino-6-mercapto-7-methylpurine riboside, 0.1 unit purine nucleoside phosphorylase, 1mM MgCl2, and MolBC. Each assay was 100μL, buffered with 25mM Tris pH 7.5; 100mM NaCl. 10–30μg/mL of MolBC (depending on activity of the mutant) was assayed in triplicate. The assay was started by addition of 2mM ATP. The initial rates of A360 increase for replicate samples were compared with determine standard error of the activity. The length of the initial region was varied to minimize standard error, while maintaining a high (> 0.98) r2 factor in the linear regression. A standard phosphate curve was used to quantify activity from the assay results. For specific activity calculations, the protein concentration was determined from the CW-EPR data by comparison of the spin concentration (from integrated absorbance peak compared with a 100µM MTSL standard) to the spin-labeling efficiency.
Continuous wave- electron paramagnetic resonance
The CW-EPR technique was described previously.31,45 Briefly, 25µL sample was aspirated into a 50µL glass capillary and nested in a quartz capillary. The following ligands were used: MolA at 2x molar excess over MolBC; 10mM ATP + 1.5mM EDTA; and 16.5mM MgCl2. Ligands were serially added to recovered protein samples, then incubated for at least 10min (60 min for MolA) before being re-scanned. X-band EPR spectra were collected at room temperature (296K) with a Bruker EMX-plus spectrometer with an ER 4119HS cavity, signal averaged 20 times in liposomes and 9 times in detergent. The scan width was set to 300Gauss and clipped to the values described in the figure legends. Distances were calculated using the Short Distances simulation software written by C. Altenbach.37
Acknowledgments
We thank Kari Tanaka for useful suggestions on the manuscript. This work was supported by National Science Foundation Grant MCB1121872.
Glossary
Abbreviations:
- ABC
ATP binding cassette
- CW-EPR
continuous wave-electron paramagnetic resonance
- MTSL
S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl) methylmethanesulfonothioate
- PC
phosphatidylcholine
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–38. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee AG. Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta. 2003;1612:1–40. doi: 10.1016/S0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 3.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–18. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 4.Newby ZE, O’Connell JD, 3rd, Gruswitz F, Hays FA, Harries WE, Harwood IM, Ho JD, Lee JK, Savage DF, Miercke LJ, et al. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nat Protoc. 2009;4:619–37. doi: 10.1038/nprot.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Privé GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–97. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Mao Q, Deeley RG, Cole SP. Functional reconstitution of substrate transport by purified multidrug resistance protein MRP1 (ABCC1) in phospholipid vesicles. J Biol Chem. 2000;275:34166–72. doi: 10.1074/jbc.M004584200. [DOI] [PubMed] [Google Scholar]
- 7.Borths EL, Poolman B, Hvorup RN, Locher KP, Rees DC. In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry. 2005;44:16301–9. doi: 10.1021/bi0513103. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez FJ, Orelle C, Davidson AL. Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J Am Chem Soc. 2010;132:9513–5. doi: 10.1021/ja104047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph B, Jeschke G, Goetz BA, Locher KP, Bordignon E. Transmembrane gate movements in the type II ATP-binding cassette (ABC) importer BtuCD-F during nucleotide cycle. J Biol Chem. 2011;286:41008–17. doi: 10.1074/jbc.M111.269472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joseph B, Korkhov VM, Yulikov M, Jeschke G, Bordignon E. Conformational cycle of the vitamin B12 ABC importer in liposomes detected by double electron-electron resonance (DEER) J Biol Chem. 2014;289:3176–85. doi: 10.1074/jbc.M113.512178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgieva ER, Borbat PP, Ginter C, Freed JH, Boudker O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat Struct Mol Biol. 2013;20:215–21. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hänelt I, Wunnicke D, Bordignon E, Steinhoff HJ, Slotboom DJ. Conformational heterogeneity of the aspartate transporter Glt(Ph) Nat Struct Mol Biol. 2013;20:210–4. doi: 10.1038/nsmb.2471. [DOI] [PubMed] [Google Scholar]
- 13.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–9. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 14.Klug CS, Feix JB. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol. 2008;84:617–58. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- 15.McHaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–61. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta. 2008;1778:1545–75. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta 2004; 1666:62-87. [DOI] [PubMed]
- 18.Lee AG. Biological membranes: the importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Powl AM, East JM, Lee AG. Heterogeneity in the binding of lipid molecules to the surface of a membrane protein: hot spots for anionic lipids on the mechanosensitive channel of large conductance MscL and effects on conformation. Biochemistry. 2005;44:5873–83. doi: 10.1021/bi047439e. [DOI] [PubMed] [Google Scholar]
- 20.Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–7. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 21.Powl AM, East JM, Lee AG. Anionic phospholipids affect the rate and extent of flux through the mechanosensitive channel of large conductance MscL. Biochemistry. 2008;47:4317–28. doi: 10.1021/bi702409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marius P, Zagnoni M, Sandison ME, East JM, Morgan H, Lee AG. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys J. 2008;94:1689–98. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossfield A, Feller SE, Pitman MC. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc Natl Acad Sci U S A. 2006;103:4888–93. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–85. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee AG. How lipids and proteins interact in a membrane: a molecular approach. Mol Biosyst. 2005;1:203–12. doi: 10.1039/b504527d. [DOI] [PubMed] [Google Scholar]
- 26.Andersen OS, Koeppe RE., 2nd Bilayer thickness and membrane protein function: an energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–30. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 27.Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Curr Opin Struct Biol. 2008;18:726–33. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locher KP. Review. Structure and mechanism of ATP-binding cassette transporters. Philos Trans R Soc Lond B Biol Sci. 2009;364:239–45. doi: 10.1098/rstb.2008.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korkhov VM, Mireku SA, Locher KP. Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature. 2012;490:367–72. doi: 10.1038/nature11442. [DOI] [PubMed] [Google Scholar]
- 30.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007;315:373–7. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 31.Rice AJ, Alvarez FJ, Schultz KM, Klug CS, Davidson AL, Pinkett HW. EPR spectroscopy of MolB2C2-a reveals mechanism of transport for a bacterial type II molybdate importer. J Biol Chem. 2013;288:21228–35. doi: 10.1074/jbc.M113.483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice AJ, Harrison A, Alvarez FJ, Davidson AL, Pinkett HW. Small Substrate Transport and Mechanism of a Molybdate ABC Transporter in a Lipid Environment. J Biol Chem. 2014 doi: 10.1074/jbc.M114.563783. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Law JH, Zalkin H, Kaneshiro T. Transmethylation Reactions in Bacterial Lipids. Biochim Biophys Acta. 1963;70:143. doi: 10.1016/0006-3002(63)90734-0. [DOI] [Google Scholar]
- 34.Jantzen E, Berdal BP, Omland T. Cellular fatty acid composition of Haemophilus species, Pasteurella multocida, Actinobacillus Actinomycetemcomitans and Haemophilus vaginalis (Corynebacterium vaginale) Acta Pathol Microbiol Scand B. 1980;88:89–93. doi: 10.1111/j.1699-0463.1980.tb02611.x. [DOI] [PubMed] [Google Scholar]
- 35.Stuart LJ, Buck JP, Tremblay AE, Buist PH. Configurational analysis of cyclopropyl fatty acids isolated from Escherichia coli. Org Lett. 2006;8:79–81. doi: 10.1021/ol052550d. [DOI] [PubMed] [Google Scholar]
- 36.Rigaud JL, Lévy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 37.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry. 2001;40:15471–82. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 38.Cantor RS. Lateral pressures in cell membranes: A mechanism for modulation of protein function. J Phys Chem B. 1997;101:1723–5. doi: 10.1021/jp963911x. [DOI] [Google Scholar]
- 39.Marsh D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys J. 2007;93:3884–99. doi: 10.1529/biophysj.107.107938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patargias G, Bond PJ, Deol SS, Sansom MSP. Molecular dynamics simulations of GlpF in a micelle vs in a bilayer: conformational dynamics of a membrane protein as a function of environment. J Phys Chem B. 2005;109:575–82. doi: 10.1021/jp046727h. [DOI] [PubMed] [Google Scholar]
- 41.Bond PJ, Sansom MSP. Membrane protein dynamics versus environment: simulations of OmpA in a micelle and in a bilayer. J Mol Biol. 2003;329:1035–53. doi: 10.1016/S0022-2836(03)00408-X. [DOI] [PubMed] [Google Scholar]
- 42.Katona G, Andréasson U, Landau EM, Andréasson LE, Neutze R. Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.35A resolution. J Mol Biol. 2003;331:681–92. doi: 10.1016/S0022-2836(03)00751-4. [DOI] [PubMed] [Google Scholar]
- 43.le Maire M, Champeil P, Moller JV. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim Biophys Acta. 2000;1508:86–111. doi: 10.1016/S0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 44.Lundbaek JA, Birn P, Hansen AJ, Søgaard R, Nielsen C, Girshman J, Bruno MJ, Tape SE, Egebjerg J, Greathouse DV, et al. Regulation of sodium channel function by bilayer elasticity: the importance of hydrophobic coupling. Effects of Micelle-forming amphiphiles and cholesterol. J Gen Physiol. 2004;123:599–621. doi: 10.1085/jgp.200308996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orelle C, Ayvaz T, Everly RM, Klug CS, Davidson AL. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc Natl Acad Sci U S A. 2008;105:12837–42. doi: 10.1073/pnas.0803799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sohlenkamp C, López-Lara IM, Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog Lipid Res. 2003;42:115–62. doi: 10.1016/S0163-7827(02)00050-4. [DOI] [PubMed] [Google Scholar]
- 47.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010.