Abstract
We compared the Y-chromosome linkage maps for four salmonid species (Arctic charr, Salvelinus alpinus; Atlantic salmon, Salmo salar; brown trout, Salmo trutta; and rainbow trout, Oncorhynchus mykiss) and a putative Y-linked marker from lake trout (Salvelinus namaycush). These species represent the three major genera within the subfamily Salmoninae of the Salmonidae. The data clearly demonstrate that different Y-chromosomes have evolved in each of the species. Arrangements of markers proximal to the sex-determining locus are preserved on homologous, but different, autosomal linkage groups across the four species studied in detail. This indicates that a small region of DNA has been involved in the rearrangement of the sex-determining region. Placement of the sex-determining region appears telomeric in brown trout, Atlantic salmon, and Arctic charr, whereas an intercalary location for SEX may exist in rainbow trout. Three hypotheses are proposed to account for the relocation: translocation of a small chromosome arm; transposition of the sex-determining gene; or differential activation of a primary sex-determining gene region among the species.
The important developmental process of sex determination in vertebrates involves considerable evolutionary plasticity (for review, see Bull 1983). Accordingly, sex-determination pathways harbor substantial differences both between and within classes (for review, see Marshall-Graves and Shetty 2001). For example, the male development switch in placental mammals is controlled by SRY, a single dominant gene on the Y-chromosome (Gubbay et al. 1990; Sinclair et al. 1990), with no sex-specific orthologs in monotremes, birds, reptiles, amphibians, and fish (for review, see Baroiller and Guiguen 2001; Clinton and Haines 2001; Marshall-Graves and Shetty 2001; Pieau et al. 2001; Schmid and Steinlein 2001). Although various environmental and genetic signals may initiate the regulatory cascade, vertebrate sex-determining pathways may converge to one ancestral biochemical pathway (Koopman 2001; Marshall-Graves and Shetty 2001). Current data indicate that certain sex-determination-regulatory genes have evolved rapidly, yet others are fairly conserved (Marin and Baker 1998). An illustrative example is the recent recruitment of SRY as a sex-determination switch in placental mammals. SRY is a member of a group of High Mobility Group (HMG) Box bearing genes (SOX genes) that have major regulatory roles in transcription. SRY is likely derived from the SOX3 gene (Bowles et al. 2000) found on the X-chromosome of therian mammals. It has been proposed that SOX3 and SRY interact with SOX9 for testis differentiation (Graves 1998).
Fish as a group encompass a wide range of sex-determination patterns from environmental mechanisms (e.g., temperature or group dynamics), through primary genetic sex determination modulated by environmental factors, to strict genetic sex determination (for review, see Chourrout 1988; Baroiller and Guiguen 2001). Genetic sex determination may involve a variety of mechanisms including polygenic inheritance, male or female heterogamety, multiple sex chromosomes, and autosomal factors (for reviews, see Price 1984; Devlin and Nagahama 2002). Alternative mechanisms (e.g., male and female heterogamety) may occur among closely related species (e.g., tilapia, Oreochromis spp.) or even among populations of the same species (e.g., platyfish, Xiphophorus maculatus), reflecting recent changes in sex determination. Also in contrast to higher vertebrates, only a few species have evolved morphologically distinguishable sex chromosomes (for review, see Beçak 1983). Surprisingly, we have yet to witness elucidation of the sex-determination mechanisms involved in genome model species (i.e., zebrafish, Danio rerio; and pufferfish, Fugu rubripes). Furthermore, aside from the medaka, (Oryzias latipes), there has been no characterization of a major sex-determining gene in teleost fish. In medaka, the sex-determining gene DMY shares phylogenetic affinities to DMRT1 and may have arisen from an ancestral duplication event with this gene (Matsuda et al. 2002).
Male heterogamety has long been accepted as a general rule in salmonid fish, although sex chromosomes still await identification in most species (for review, see Phillips and Ráb 2001). Primary evidence for male heterogamety is derived from analysis of sex ratios in the progeny of hormonally sex-reversed individuals. For example, sex-reversed females of rainbow trout (Oncorhynchus mykiss), chinook salmon (Oncorhynchus tshawytscha), coho salmon (Oncorhynchus kisutch), and Atlantic salmon (Salmo salar) produce all female progeny when crossed with normal females, indicating that females are homogametic XX (Johnstone et al. 1979; Hunter et al. 1982, 1983; Johnstone and Youngson 1984). Subsequent characterization of sex-linked markers has also provided support for male heterogamety in Arctic charr (Salvelinus alpinus), lake trout (Salvelinus namaycush), masu salmon (Oncorhynchus masou), pink salmon (Oncorhynchus gorbuscha), chum salmon (Oncorhynchus keta), brown trout (Salmo trutta), rainbow trout (O. mykiss), coho salmon (O. kisutch), and chinook salmon (O. tshawytscha; May et al. 1989; Du et al. 1993; Forbes et al. 1994; Prodöhl et al. 1994; Young et al. 1998; Nakayama et al. 1999; Sakamoto et al. 2000; Devlin et al. 2001; Zhang et al. 2001; Stein et al. 2002).
Heteromorphic sex chromosomes have been detected in only a handful of species (for review, see Hartley 1987; Phillips and Ráb 2001). The largest pair of submetacentrics have been identified as the sex chromosomes in lake trout and brook trout (Salvelinus fontinalis) based on an X-specific heterochromatic block at the end of the short arm (Phillips and Ihssen 1985; Phillips et al. 2002). In sockeye salmon (Oncorhynchus nerka), a male-specific Robertsonian translocation has been reported, presumably resulting from a Y–autosome fusion (Fukuoka 1972; Thorgaard 1978). Size differences in a homologous pair of chromosomes between the sexes have also been observed in the short arm of a small subtelocentric pair in rainbow trout (Thorgaard 1977). Interestingly, males lacking this heteromorphic condition have been observed in rainbow trout (Thorgaard 1983) and sockeye salmon (Fukuoka 1972), which indicates that chromosome rearrangements differentiating the sex chromosomes are still in the process of fixation. Sex-specific probes now facilitate identification of sex chromosomes using fluorescent in situ hybridization (FISH) techniques (Reed et al. 1995; Moran et al. 1996; Iturra et al. 1998, 2001; Phillips 2001; Phillips et al. 2001, 2002; Stein et al. 2001). Altogether, current cytogenetic data support the view that salmonid species represent early stages of sex chromosome differentiation (Phillips et al. 2001). Consistent with this is the viability and fertility of YY males (Chevassus 1988; Onozato 1989), suggesting that X- and Y-chromosomes still share a similar repertoire of functional genes.
Linkage data indicate that there is a lack of conservation regarding the sex-determining locus across salmonids. An early study conducted by May et al. (1989) in the genus Salvelinus found that sex-linked allozyme markers in Arctic charr were not linked to the phenotypic sex-determining locus (thereafter denoted as SEX) in lake trout and brook trout. Similar results were subsequently reported in other genera: A growth hormone marker was shown to be sex-linked in coho salmon, chinook salmon, and masu salmon, but sex linkage was not conserved in amago salmon (Oncorhynchus rhodurus) and rainbow trout (Forbes et al. 1994; Nakayama et al. 1999; Zhang et al. 2001). Similarly, a minisatellite locus shown to be in tight linkage with SEX in brown trout by Prodöhl et al. (1994) was mapped to an autosomal pair in Atlantic salmon (Taggart et al. 1995). Interspecific disruption of sex linkage is surprising because extensive conservation of linkage arrangements has been observed for biochemical (Johnson et al. 1987) and microsatellite (Gharbi 2001) loci. We report the comparative mapping of sex-linkage groups in rainbow trout, Atlantic salmon, brown trout, Arctic charr, and lake trout using molecular markers. Our results confirm and extend preliminary observations indicating that the sex-determining locus is not conserved with respect to synteny with identified homologous chromosome sets among these various species.
RESULTS
Sex Linkage of Molecular Markers
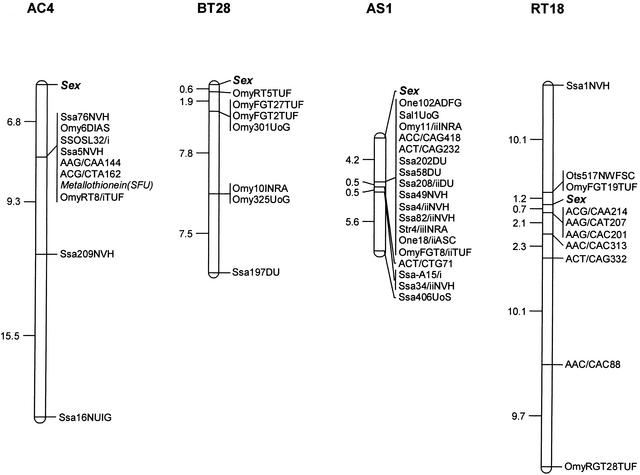
Gene mapping in Arctic charr (data not shown) identified 12 sex-linked markers including two amplified fragment length polymorphisms (AFLP), nine microsatellite loci, and one functional gene, with SEX located at the distal end of linkage group AC4 (Fig. 1). The microsatellite markers Ssa72NVH and Ots500NWFSC were not unequivocally localized to the map because genotypic data were only obtained for approximately half of the mapping progeny. Both parents were heterozygous for the same pair of alleles in the mapping families used. Consequently, these markers are not shown in the figure. The brown trout linkage map (data not shown) includes seven sex-linked microsatellite markers on linkage group BT28 (Fig. 1). The relative position of SEX and OmyRT5TUF at the distal end of the sex-linkage group could not be determined unambiguously because markers were informative in different families. Therefore, the terminal location of SEX remains tentative in this species. SEX was located distally on linkage group AS1 in Atlantic salmon using 3 and 15 AFLP and VNTR markers, respectively (Fig. 1). Sakamoto et al. (2000) previously reported two microsatellite markers (OmyFGT19TUF and OmyRGT28TUF) linked to SEX in rainbow trout. We identified two additional sex-linked microsatellite markers (Ots517NWFSC and Ssa1NVH) and six AFLP markers. Recombination among the microsatellite and AFLP markers in this linkage group strongly supported a more intercalary location for the SEX locus in this species (Fig. 1) compared with the other three species.
Figure 1.
Genetic map of the sex linkage groups in Arctic charr (AC4), brown trout (BT28), Atlantic salmon (AS1), and rainbow trout (RT18) generated from male parents. Estimates of map distances between markers are indicated in centiMorgans.
Microsatellite Comparative Mapping of Sex-Linkage Groups
A microsatellite marker (Yp136) isolated from the lake trout Y-chromosome (Stein et al. 2002) was tested for sex linkage in Arctic charr. Yp136 showed no evidence of linkage with SEX and mapped to linkage group AC18 (data not shown). Two of the microsatellite markers (Omy6DIAS and Ssa209NVH) located on AC4 were polymorphic in the male parent of lake trout cross 2. Neither marker was linked to the sex-determining locus. These markers were also unlinked to each other at an LOD = 3.0 threshold. Omy6DIAS was also polymorphic for the male parent in lake trout cross 1, but the female parent was also heterozygous for the same two alleles. Consequently, the informative number of meioses scored in the mapping progeny was too small to accurately assess linkage affinities.
The putative Y-chromosome of Arctic charr (AC4) incorporated markers from linkage groups BT2, BT7, and BT17 in brown trout; AS2, AS10, and AS25 in Atlantic salmon; and RTK, RTE, and RT15 in rainbow trout (Table 1). The rainbow trout sex-linkage chromosome also demonstrated mosaic affinities to linkage groups in the other species, possessing markers that mapped to AC5 and AC27 in Arctic charr, and BT13 and BT22 in brown trout (Table 1). However, the available evidence indicates that RT18 only localizes to AS9 in Atlantic salmon based on homologies detected with Ssa1NVH in males (Table 1) and Ssa96NVH in female mapping parents (data not shown). In contrast, the sex chromosome of brown trout was completely syntenic with RTB markers in rainbow trout, and possibly AS8 in Atlantic salmon, although more cross-priming markers need to be examined to confirm this. In addition, the distal region of BT28 from SEX appears syntenic with AC7 (based on shared affinities with Omy301UoG, Omy10INRA, and Ssa197DU in Atlantic salmon and rainbow trout and the conserved marker order detected among these species; data not shown), whereas the proximal region of the linkage group may be syntenic with either AC1 or AC13 because OmyFGT27TUF is duplicated in Arctic charr (Table 1). The sex-linkage group of Atlantic salmon also demonstrated a high degree of synteny with linkage groups in brown trout (syntenic with BT1 and BT11 markers) and rainbow trout (syntenic with RT2 and RT5 markers; Table 1). In rainbow trout, the localization of syntenic blocks was confounded by the fact that 4 of the 7 cross-priming markers (i.e., Omy11INRA, Str4INRA, One18ASC, and OmyFGT8TUF) showed duplicate expression in rainbow trout. However, the fact that the three single-copy markers (i.e., Sal1UoG, One102ADFG, and Ssa406UoS) examined mapped to either RT5 or RT2 allowed us to tentatively assign the syntenic blocks to these two rainbow trout linkage groups. Also several markers are syntenic between BT1 and RT2 (data not shown), supporting the mosaic arrangements detected in the AS1 linkage group. Unfortunately, none of the polymorphic markers detected in the male Atlantic salmon mapping parents were informative in the male Arctic charr mapping parents.
Table 1.
Homologous Marker Locations Among Four Different Species of Salmonid Fishes
| Sex chromosome | Marker | Homologous chromosome location | |||
| AC | BT | AS | RT | ||
| AC4 | Ssa72NVH | — | AS10 | RTK | |
| SSOSL32/i | — | BT2 | RTE | ||
| Omy6DIAS | — | RT15 | |||
| Ssa16NUIG | — | BT7 | AS2 | RT15 | |
| Ssa76NVH | — | AS25 | |||
| Ssa5NVH | — | BT17 | AS10 | ||
| BT28 | OmyRT5TUF | — | RTB | ||
| OmyFGT27TUF | AC1, AC13 | — | RTB | ||
| OmyFGT2TUF | — | RTB | |||
| Omy301UoG | AC7 | — | AS8 | RTB | |
| Omy10INRA | AC7, AC27 | — | RTB | ||
| Omy325UoG | AC7 | — | RTB | ||
| Ssa197DU | — | AS8 | RTB | ||
| AS1 | Omy11/iiINRA | BT1a | — | RT2a | |
| Sal1UoG | — | RT2 | |||
| Ssa4/iiNVH | BT11a | — | |||
| One102ADFG | — | RT5 | |||
| Ssa406UoS | BT11 | — | RT5 | ||
| Str4/iiINRA | BT11a | — | RT5a | ||
| One18/iiASC | BT11a | — | RT5a | ||
| OmyFGT8/iiTUF | BT11a | — | RT5a | ||
| RT18 | Ots517NWFSC | AC5 | — | ||
| OmyFGT19TUF | AC27 | BT13 | — | ||
| OmyRGT28TUF | — | ||||
| Ssa1NVH | UNA | BT22 | AS9 | — | |
(AC) Arctic charr, (BT) brown trout, (AS) Atlantic salmon, (RT) rainbow trout, (UNA) unassigned marker. Missing marker assignments indicate that the marker either did not amplify in the species indicated or was monomorphic in the mapping parents tested.
Duplicated markers were assigned based on single-copy markers shared between homologous linkage groups.
DISCUSSION
Position of the Sex-Determining Locus on the Y-Chromosome
With the exception of rainbow trout, genetic maps of the other salmonid species studied indicate that SEX occurs at the end of the Y linkage group (Fig. 1). Although distal ends of linkage groups do not necessarily coincide with telomeric regions of chromosomes, recombination between SEX and other markers (Fig. 1) may be indicative of a more terminal placement. According to Wright Jr. et al.'s (1983) model of chromosome pairing in male salmonids, only the telomeric regions of homologously paired multivalents may experience recombination. Furthermore, increased male recombination distances toward the end of several linkage groups in rainbow trout (Sakamoto et al. 2000) and brown trout (Gharbi 2001) indicate that higher recombination in telomeric regions may represent a common trend in male meiosis (Gharbi 2001). In rainbow trout, the sex-determining factor has been mapped close to putative centromeric markers (Allendorf et al. 1994; Sakamoto et al. 2000). For example, SEX maps close to OmyFGT19TUF (∼1%–2% recombination across several rainbow trout families descended from two males in each of two unrelated hatchery strains), and gynogenetic gene–centromere distances with this marker are ∼2 cM (Sakamoto et al. 2000). Thus, given the close proximity of OmyFGT19TUF to the centromere, the data may be supportive of an intercalary location for SEX on the chromosomes of rainbow trout, at least in the strains surveyed by Sakamoto et al. (2000). Given the lack of recombination across male chromosomes, however, further characterization of the sex linkage groups by other methods is required. Direct evidence for the location of SEX from fluorescent in situ hybridization of DNA probes has presently been inconclusive because none of the published studies used probes shown to contain the sex-determining factor (Iturra et al. 1998, 2001; Phillips 2001; Phillips et al. 2001, 2002; Stein et al. 2001).
Homologies Among Y-Chromosomes
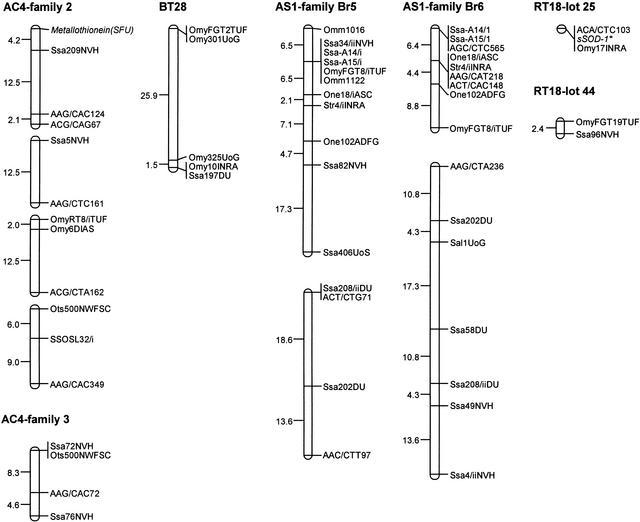
The Arctic charr sex-linkage group demonstrates the greatest variability in its affinities to the linkage groupings found in other species examined. This may be indicative of a greater phylogenetic divergence of this species compared with the two Salmo species and rainbow trout. Unfortunately, too few homologous markers were examined in lake trout to permit even a cursory assessment with this species. Also, caution must still be exercised in the interpretation of the observed linkage group affinities because the linkage maps in all these species are still largely incomplete. In addition, because male salmonids demonstrate the phenomenon of pseudolinkage (Wright Jr. et al. 1983), it is possible for markers from two separate linkage groups to appear physically linked as a consequence of the ancient homologous chromosome pairings that can occur in male salmonids. This phenomenon often results in an apparent linkage of telomeric markers from different linkage groups. Because many of the markers demonstrating tight linkage in the male Arctic charr mapping parents show large recombination distances in the female parents (Fig. 2), AC4 may represent a pseudolinked group. Similarly, in Atlantic salmon, the female parents from families Br5 and Br6 show separate linkage groupings for the same syntenic markers that are tightly linked in the male parents (Figs. 1 and 2). In Atlantic salmon, however, all the markers in the male map with the exception of Ssa406UoS and SEX show a low level of recombination with one another, indicating an intercalary location of these markers on one linkage group. Although gene–centromere distances have not been assessed for this linkage group it is likely that these separate linkage groupings in the female parents may represent different metacentric chromosome arms. It is also possible that they represent separate chromosome segment domains spanning a region of chromosome arm fusions, because Atlantic salmon are known to have undergone multiple arm fusions that have reduced the fundamental number of chromosome arms in their karyotype (Hartley 1987). Consistent with either interpretation is the fact that the separate linkage group segments detected in the Atlantic salmon females appear to be homologous to two separate linkage groups in rainbow trout (i.e., RT2 and RT5) and brown trout (i.e., BT1 and BT11; Table 1; Fig. 2).
Figure 2.
Genetic map of the sex-linkage groups in Arctic charr (AC4), brown trout (BT28), Atlantic salmon (AS1), and rainbow trout (RT18) generated from female parents. Vertically aligned linkage groups represent chromosome segments linked in the male map (see text). Estimates of map distances between markers are indicated in centiMorgans.
Mechanisms for the Disruption of Sex Linkage
Comparative mapping of sex-linked microsatellite markers clearly demonstrates that Arctic charr, brown trout, Atlantic salmon, and rainbow trout have evolved different sex chromosomes. In addition, limited linkage data indicate that sex chromosomes are not conserved between Arctic charr and lake trout. Occurrence of alternative Y-chromosomes among salmonid species is in accordance with emerging patterns from chromosome painting showing that sex chromosome probes generally cross-hybridize to autosomes (Phillips et al. 2001). However, a few species may still share a common Y-chromosome, as recently evidenced from fluorescent in situ hybridization in the closely related lake trout and brook trout (Phillips et al. 2002). Although we confirm previous findings of a general lack of conservation for sex linkage among salmonid species (see above), our results extend knowledge to include the fact that arrangements of markers proximal to the sex-determining locus are preserved on homologous, yet autosomal, linkage groups (Table 1). Therefore, a small segment of DNA must be involved in relocation of the sex-determining region.
Differential inactivation of a duplicated SEX locus on salmonid homologs may be a possible explanation for our results; however, the present evidence does not support this interpretation. Although the homologies for RT18 and BT28 are unknown, AS1 shows homology to AS6 and AS12, whereas AC4 shows homology to AC25 and AC28 (data not shown). None of the markers on these ancestrally duplicated linkage groups show homology to the sex-linkage groups of the other species studied. This hypothesis cannot be entirely discounted, however, as RT5 and RT15 are homologous in rainbow trout (Sakamoto et al. 2000), which may imply a potential homologous affinity between AC4 and AS1. This interpretation is confounded by the fact that we cannot be certain at present which copy of an ancestrally duplicated pair is being expressed in another species.
We believe that there are three possible models that may explain the observed differences in Y linkage, and that these mechanisms are not necessarily mutually exclusive. First, the chromosomes of salmonid species are believed to have undergone a series of Robertsonian translocations throughout evolutionary time, resulting in a fairly constant number of chromosome arms and a wide variety of diploid numbers across the family (for review, see Phillips and Ráb 2001). Robertsonian rearrangement involving the sex chromosome has been reported in at least one species (Thorgaard 1978). If the sex-determining region is being rearranged during a whole arm translocation during or after speciation, we would expect the chromosome arm bearing the sex locus to be conserved, whereas the region across the centromere would be different (May et al. 1989). This indicates that the sex-determining region must fall on a relatively small chromosome arm within the genome (i.e., present lack of detectable conserved markers). Although rainbow trout, coho salmon, and chinook salmon do exhibit sex chromosomes with a short chromosome arm (Thorgaard 1977; Iturra et al. 2001; Stein et al. 2001), chromosome arms carrying the sex-determining factor in lake trout (Phillips and Ihssen 1985), brook trout (Phillips et al. 2002), and sockeye salmon (Thorgaard 1978) are probably too large to fit into this model. Translocation of a shorter chromosome segment, however, may still account for the lack of common sex-linked markers.
Second, it is also possible that the sex-determining region has been transposing throughout the genome without relocating adjacent markers, thus causing disruption of sex linkage across species. A similar mechanism of transposition has been postulated in the black fly (Megaselia scalaris), in which a Maleness factor is moved at a low rate from one chromosome to another while closely linked markers remain in their original position (Traut and Willhoeft 1990; Traut 1994), and transposition of active Y genes from autosomal sources has been reported in humans (Lahn and Page 1999).
Third, it is also possible that salmonids have evolved different sex-determining genes. Given the multiplicity of different sex-determining or sex-associated genes that have been identified in vertebrates, it possible that differential mutations to ancestral genes within the sex-determining suite of genes (e.g., SOX-family, DMRT1, or TDF gene mutations) may result in the acquisition of new locations for the primary sex-determining region among species (Marshall-Graves 2002). Unless common sex-linked markers are identified between species with divergent sex chromosomes, the question may remain open until sequence characterization of the sex-determining genes.
METHODS
Mapping Families
Source material for this study was reference families used for genome-wide mapping projects in our laboratories. Rainbow trout families were backcross pedigrees between phenotypically divergent strains previously referred to as lot 25 (Jackson et al. 1998) and lot 44 (Sakamoto et al. 1999). Details for Arctic charr, brown trout, and Atlantic salmon pedigrees will be provided in forthcoming reports on the present map status in these species. Briefly, Arctic charr families were backcross material between two Canadian aquaculture strains (Nauyuk and Fraser) originating from the Rockwood Aquaculture Research Station near Gunton, Manitoba. Two crosses were performed: a Nauyuk × Fraser F1 female was crossed to a Fraser strain male to produce family 2 (n = 48); conversely, a Nauyuk × Fraser F1 male was crossed to a Fraser strain female to produce family 3 (n = 48). Brown trout families were backcrosses between evolutionary lineages (for review, see Bernatchez 2001). Two Mediterranean (Reverotte stream, France) × Atlantic (Gournay hatchery, France) F1 males were crossed to Atlantic dams to produce families 12 (n = 45) and 15 (n = 45). Two additional pedigrees, 14 (n = 48) and 17 (n = 48), were generated by mating marmoratus (Pellice stream, Italy) × Atlantic F1 males into Atlantic females. The two Atlantic salmon families (Br5 and Br6; n = 48 in both cases) were outcrosses involving four parents sampled from a large natural population (River Tay, Scotland). Two lake trout crosses were made between unrelated fish from the Manitou strain that were maintained at the former Maple Research Station (Ontario Ministry of Natural Resources). The offspring were reared at this facility until they could be sexed. Sex was unambiguously determined by internal examination of ovary or testis development in Arctic charr family 3, brown trout families 14 and 17, rainbow trout lot 44, Atlantic salmon families Br5 and Br6, and both lake trout families. Phenotypic sex was used as the marker for SEX in the linkage analysis.
Marker Analysis
Because of the collaborative nature of this study, different protocols were used to analyze microsatellite polymorphism. Genomic DNA was phenol-extracted from fin, gill, liver, or muscle tissue as outlined in Taggart et al. (1992), Estoup et al. (1993), and Bardakci and Skibinski (1994). Polymerase chain reaction, electrophoresis, and DNA fragment visualization of microsatellite markers were performed using radioactive or fluorescent end-labeled primers as described in Sakamoto et al. (1996), Estoup et al. (1998), and Sakamoto et al. (1999). Alternatively, PCR was carried out in 11-μL reaction volumes with direct incorporation of fluorescently labeled deoxynucleotide triphosphate. The PCR reaction mixture contained 30 ng of genomic DNA, 1× PCR buffer (20 mM Tris-HCl, 50 mM KCl at pH 8.4; GIBCO BRL), 136 μM each dNTP (Roche Diagnostics), 0.9 μM Tamra-dCTP (Applied Biosystems), 1.4 mM MgCl2 (GIBCO BRL), 0.09 mg/mL BSA (GIBCO BRL), 0.04 μM each primer, and 0.2 U of Taq DNA polymerase (GIBCO BRL). The PCR temperature profile consisted of an initial denaturing step of 95°C for 5 min; 36 cycles of 95°C for 30 sec, 1 min at a specific annealing temperature, and 72°C for 1 min; and a final extension step of 72°C for 10 min. PCR products were subsequently separated on a 6% polyacrylamide denaturing gel (7 M urea), and DNA fragments were visualized by scanning with a fluorescent imaging system (Hitachi FMBIOII).
AFLP analysis was performed as described by Vos et al. (1995) with some modifications. The two restriction enzymes used were EcoRI and MseI. Fragments were amplified by PCR in a MJ Research PTC-100 or PTC-200 with the following temperature profile: 94°C for 2 min; 9 cycles of 94°C for 20 sec, 66°C for 20 sec (0.5°C per cycle), 72°C for 2 min; 19 cycles of 94°C for 20 sec, 56°C for 30 sec, 72°C for 2 min; and 60°C for 30 min. EcoRI primers were labeled with the single-isomer fluorescein dye TET (Applied Biosystems), and amplified fragments were separated on a 6% polyacrylamide denaturing gel (7 M urea) using a Model SA gel electrophoresis unit (GIBCO BRL), and visualized with a fluorescent imaging system (Hitachi FMBIO). The lane standard 350-TAMRA (Applied Biosystems) was included on each gel, and AFLP band size in base pairs was determined by comparison with the lane standard using FMBIO Analysis 8.0. Minisatellite DNA polymorphisms were detected on Southern blots of Hae III-digested genomic DNA using isotopically labeled (32P) single-locus minisatellite probes as described by Taggart et al. (1995).
A single nucleotide polymorphism (SNP) was detected in the second intron of the metallothionein B gene using heteroduplex analysis (HA) as described by White et al. (1992). The Primers for PCR amplification were 5′-GCATGCACCAGT TGTAAGAAA-3′ and 5′-TCACTGACAACAGCTGGTATC-3′. Amplification was carried out using a temperature profile of one cycle at 95°C for 5 min followed by 30 cycles at 95°C for 45 sec, 55°C for 45 sec, and 72°C for 45 sec. PCR products were labeled by incorporation of [32P]dCTP during amplification. To drive the formation of heteroduplexes, PCR products were heated to 95°C for 5 min then cooled to 20°C over a period of 1 h. Products were separated according to size and conformation by subjecting them to electrophoresis through a 10% low cross-link (37.5:1) native polyacrylamide gel in TBE buffer and 10% glycerol for 16 h at a constant power of 3 W. Gels were 30 cm long and 0.4 mm thick. They were dried onto filter paper, and products were visualized using autoradiography.
Linkage Analysis
Linkage analysis was primarily conducted using LINKMFEX version 1.5 (see http://www.uoguelph.ca/∼rdanzman/software/LINKMFEX). All linkage maps reported here have been constructed using sex-specific data (i.e., data generated from the female or male parent) and a minimum LOD score of 4.0 to assign markers to linkage groups. The linear order of markers within linkage groups was assisted with both LINKMFEX (module MAPORD) and CARTHAGENE version 0.5 (see http://www-bia.inra.fr/T/CarthaGene). Because recombination is largely repressed along salmonid chromosomes during male meiosis (e.g., Sakamoto et al. 2000), linkage maps derived from the male parent are inherently error-prone. Admittedly, a few genotyping errors may alter the order of closely spaced markers in such a way that linkage-group architecture should be regarded as tentative unless female data provide better support. Tentatively ordered linkage groups were printed out with MAPCHART (Voorrips 2002) using recombination fractions as estimates of map distances to account for the high level of interference in salmonid species (e.g., Thorgaard et al. 1983; Guyomard 1986).
Genetic Nomenclature
Naming of microsatellite markers follows the convention outlined by Jackson et al. (1998). Species abbreviations, common names, and lab affiliations are outlined in Sakamoto et al. (2000) with the following additions: NWFSC (Northwest Fisheries Science Center) and UoS (University of Stirling). Atlantic salmon single-locus minisatellite nomenclature follows Taggart et al. (1995). The naming of AFLP loci follows the convention in which the three-base selective primer extensions used to produce the loci are listed first, followed by the base pair size of the locus (Young et al. 1998). For example, AAG/CAA334 indicates the three nucleotides (AAG) for the EcoRI primer and the three nucleotides (CAA) for the MseI primer amplified a product at 334 bp. Genes are identified with an italicized code referring to the gene name. The institution where the gene polymorphism was identified is listed in parentheses following the gene code.
Linkage groups have been identified in rainbow trout with either a number or a letter (Sakamoto et al. 2000). Linkage groups for Arctic charr, Atlantic salmon, and brown trout have been numbered arbitrarily. In this report, linkage groups are identified by a two-letter code referring to the species (AC, Arctic charr; AS, Atlantic salmon; BT, brown trout; RT, rainbow trout), followed by their present numerical or alphabetical code.
WEB SITE REFERENCES
http://www-bia.inra.fr/T/CarthaGene; CARTHAGENE version 0.5.
http://www.uoguelph.ca/∼rdanzman/software/LINKMFEX; LINKMFEX version 1.5.
Acknowledgments
The authors thank the following individuals for their contribution: Martine Andriamanga, Angélique Gautier, Sonia Gharbi, and Arnaud Estoup (INRA). This research was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada, the European Union FAIR program (SALMAP project contract CT96-1591), INRA (Hydrobiology and Wildlife Department), Natural Environment Research Council (NERC), and the United States Department of Agriculture (USDA). This work is dedicated to the memory of Richard Powell, a respected friend and colleague from the SALMAP project.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL rdanzman@uoguelph.ca; FAX (519) 767-1656.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.578503.
REFERENCES
- 1.Allendorf F.W., Gellman, W.A., and Thorgaard, G.H. 1994. Sex linkage of two enzyme loci in Oncorhyncus mykiss (rainbow trout). Heredity 72: 498-507. [DOI] [PubMed] [Google Scholar]
- 2.Bardakci F. and Skibinski, D.O.F. 1994. Application of the RAPD technique in tilapia fish: Species and subspecies identification. Heredity 73: 117-123. [DOI] [PubMed] [Google Scholar]
- 3.Baroiller J.F. and Guiguen, Y. 2001. Endocrine and environmental aspects of sex differentiation in gonochoristic fish. EXS 91: 177-201. [DOI] [PubMed] [Google Scholar]
- 4.Beçak W. 1983. Evolution and differentiation of sex chromosomes in lower vertebrates. Differentiation 23 (Suppl.): S3-S12. [Google Scholar]
- 5.Bernatchez L. 2001. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 55: 351-379. [DOI] [PubMed] [Google Scholar]
- 6.Bowles J., Schepers, G., and Koopman, P. 2000. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227: 239-255. [DOI] [PubMed] [Google Scholar]
- 7.Bull J.J., 1983. Evolution of sex determining mechanisms. Benjamin/Cummings, Don Mills, Ontario, Canada.
- 8.Chevassus B., 1988. “Modification du phénotype sexuel et du mode de reproduction chez les poissons salmonidés: Inversion sexuelle, gynogénèse, hybridation interspécifique et polyploïdisation.” Ph.D. dissertation Université Paris XI, Orsay, France.
- 9.Chourrout D. 1988. Revue sur le déterminisme génétique du sexe des poissons téléostéens. Bull. Soc. Zool. Fr. 113: 123-144. [Google Scholar]
- 10.Clinton M. and Haines, L.C. 2001. An overview of factors influencing sex determination and gonadal development in birds. EXS 91: 97-115. [DOI] [PubMed] [Google Scholar]
- 11.Devlin R.H. and Nagahama, Y. 2002. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 208: 191-364. [Google Scholar]
- 12.Devlin R.H., Biagi, C.A., and Smailus, D.E. 2001. Genetic mapping of Y-chromosomal DNA markers in Pacific salmon. Genetica 111: 43-58. [DOI] [PubMed] [Google Scholar]
- 13.Du S.J., Devlin, R.H., and Hew, C.L. 1993. Genomic structure of growth hormone genes in Chinook salmon (Oncorhynchus tshawytscha): Presence of two functional genes, GH-I and GH-II, and a male specific pseudogene, GH-ψ. DNA Cell Biol. 12: 739-751. [DOI] [PubMed] [Google Scholar]
- 14.Estoup A., Presa, P., Krieg, F., Vaiman, D., and Guyomard, R. 1993. (CT)n and (GT)n microsatellites: A new class of genetic markers for Salmo trutta L. (Brown trout). Heredity 71: 488-496. [DOI] [PubMed] [Google Scholar]
- 15.Estoup A., Gharbi, K., San Cristobal, M., Chevalet, C., Haffray, P., and Guyomard, R. 1998. Parentage assignment using microsatellites in turbot (Scophtalmus maximus) and rainbow trout (Oncorhynchus mykiss) hatchery populations. Can. J. Fish. Aquat. Sci. 57: 715-725. [Google Scholar]
- 16.Forbes S.H., Knudsen, K.L., North, T.W., and Allendorf, F.W. 1994. One of two growth hormone genes in coho salmon is sex-linked. Proc. Natl. Acad. Sci. 91: 1628-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuoka H. 1972. Chromosomes of the sockeye salmon (Oncorhynchus nerka). Jpn. J. Genet. 47: 459-464. [Google Scholar]
- 18.Gharbi K., 2001. “Construction of a microsatellite linkage map in the tetraploid derivative brown trout (Salmo trutta): Comparative mapping of paralogous regions, map alignment with orthologous segments in rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar).” Ph.D. dissertation Institut National Agronomique Paris-Grignon, Paris, France.
- 19.Graves J.A. 1998. Interactions between SRY and SOX genes in mammalian sex determination. BioEssays 20: 264-269. [DOI] [PubMed] [Google Scholar]
- 20.Gubbay J., Collignon, J., Koopman, P., Capel, B., Economou, A., Munsterberg, A., Vivian, N., Goodfellow, P., and Lovell-Badge, R. 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346: 245-250. [DOI] [PubMed] [Google Scholar]
- 21.Guyomard R. 1986. Gene segregation in gynogenetic brown trout (Salmo trutta L.): Systematically high frequencies of post-reduction. Genet. Sel. Evol. 18: 385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartley S.E. 1987. The chromosomes of salmonid fishes. Biol. Rev. 62: 197-214. [Google Scholar]
- 23.Hunter G.A., Donaldson, E.M., Goetz, F.W., and Edgell, P.R. 1982. Production of all female and sterile Coho salmon, and experimental evidence for male heterogamety. Trans. Am. Fish. Soc. 111: 367-372. [Google Scholar]
- 24.Hunter G.A., Donaldson, E.M., Stoss, J., and Baker, I.I. 1983. Production of monosex female groups of chinook salmon (Oncorhynchus tshawytscha) by the fertilization of normal ova with sperm from sex-reversed females. Aquaculture 33: 355-364. [Google Scholar]
- 25.Iturra P., Medrano, J.F., Bagley, M., Lam, N., Vergara, N., and Marin, J.C. 1998. Identification of sex chromosome molecular markers using RAPDs and fluorescent in situ hybridization in rainbow trout. Genetica 101: 209-213. [DOI] [PubMed] [Google Scholar]
- 26.Iturra P., Lam, N., de la Fuente, M., Vergara, N., and Medrano, J.F. 2001. Characterization of sex chromosomes in rainbow trout and coho salmon using fluorescence in situ hybridization (FISH). Genetica 111: 125-131. [DOI] [PubMed] [Google Scholar]
- 27.Jackson T.R., Ferguson, M.M., Danzmann, R.G., Fishback, A.G., Ihssen, P., O'Connell, M., and Crease, T.J. 1998. Identification of two QTL influencing upper temperature tolerance in three rainbow trout (Oncorhynchus mykiss) half-sib families. Heredity 80: 143-151. [Google Scholar]
- 28.Johnson K.R., Wright, J.E., Jr., and May, B. 1987. Linkage relationships reflecting ancestral tetraploidy in salmonid fish. Genetics 116: 579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnstone R. and Youngson, A.F. 1984. The progeny of sex-inverted female Atlantic salmon (Salmo salar L.). Aquaculture 37: 179-182. [Google Scholar]
- 30.Johnstone R., Simpsons, T.H., Youngson, A.F., and Whitehead, C. 1979. Sex reversal in salmonid culture. Part II. The progeny of sex reversed rainbow trout. Aquaculture 18: 13-19. [Google Scholar]
- 31.Koopman P. 2001. Sry, Sox9 and mammalian sex determination. EXS 91: 25-56. [DOI] [PubMed] [Google Scholar]
- 32.Lahn B.T. and Page, D.C. 1999. Retroposition of autosomal mRNA yielded testis specific gene family on human Y chromosome. Nat. Genet. 21: 429-433. [DOI] [PubMed] [Google Scholar]
- 33.Marin I. and Baker, B.S. 1998. The evolutionary dynamics of sex determination. Science 281: 1990-1994. [DOI] [PubMed] [Google Scholar]
- 34.Marshall-Graves J.A. 2002. The rise and fall of SRY. Trends Genet. 18: 259-264. [DOI] [PubMed] [Google Scholar]
- 35.Marshall-Graves J.A. and Shetty, S. 2001. Sex from W to Z: Evolution of vertebrate sex chromosomes and sex determining genes. J. Exp. Zool. 290: 449-462. [DOI] [PubMed] [Google Scholar]
- 36.Matsuda M., Nagahama, Y., Shinomlya, A., Sato, T., Matsuda, C., Kobayashi, T., Morrey, C.E., Shibata, N., Asakawa, S., Shimizu, N., et al. 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559-563. [DOI] [PubMed] [Google Scholar]
- 37.May B., Johnson, K.R., and Wright, J.E., Jr. 1989. Sex linkage in salmonids: Evidence from a hybridized genome of brook trout and Arctic charr. Biochem. Genet. 27: 291-301. [DOI] [PubMed] [Google Scholar]
- 38.Moran P., Martinez, J.L., Garcia-Vazquez, E., and Pendas, A.M. 1996. Sex chromosome linkage of 5S rDNA in rainbow trout (Oncorhynchus mykiss). Cytogenet. Cell Genet. 75: 145-150. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama I., Biagi, C.A., Koide, N., and Devlin, R.H. 1999. Identification of a sex-linked GH pseudogene in one of two species of Japanese salmon (Oncorhynchus masou and O. rhodurus). Aquaculture 173: 65-72. [Google Scholar]
- 40.Onozato H. 1989. Androgenesis in masu salmon. Physiol. Ecol. Japan Spec. 1: 543. [Google Scholar]
- 41.Phillips R.B. 2001. Application of fluorescence in situ hybridization (FISH) to fish genetics and genome mapping. Marine Biotechnol. 3: 5145-5152. [DOI] [PubMed] [Google Scholar]
- 42.Phillips R.B. and Ihssen, P. 1985. Identification of sex chromosomes in lake trout (Salvelinus namaycush). Cytogenet. Cell Genet. 39: 14-18. [DOI] [PubMed] [Google Scholar]
- 43.Phillips R. and Ráb, P. 2001. Chromosome evolution in the Salmonidae (Pisces): An update. Biol. Rev. 76: 1-25. [DOI] [PubMed] [Google Scholar]
- 44.Phillips R.B., Konkol, N.R., Reed, K.M., and Stein, J.D. 2001. Chromosome painting supports lack of homology among sex chromosomes in Oncorhynchus, Salmo, and Salvelinus (Salmonidae). Genetica 111: 119-123. [DOI] [PubMed] [Google Scholar]
- 45.Phillips R.B., Matsuoka, M.P., and Reed, K.M. 2002. Characterization of charr chromosomes using fluorescence in situ hybridization. Environ. Biol. Fish. 64: 223-228. [Google Scholar]
- 46.Pieau C., Dorizzi, M., and Richard-Mercier, N. 2001. Temperature-dependent sex determination and gonadal differentiation in reptiles. EXS 91: 117-141. [DOI] [PubMed] [Google Scholar]
- 47.Price D.J. 1984. Genetics of sex determination in fishes: A brief review. In Fish reproduction: Strategies and tactics (eds. G.W. Pottsand & R.J. Wooton), pp. 77–89. Academic Press, London, UK.
- 48.Prodöhl P.A., Taggart, J.B., and Ferguson, A. 1994. Single locus inheritance and joint segregation analysis of minisatellite (VNTR) DNA loci in brown trout (Salmo trutta L.). Heredity 73: 556-566. [Google Scholar]
- 49.Reed K.M., Bohlander, S.K., and Phillips, R.B. 1995. Microdissection of the Y chromosome and fluorescence in situ hybridization analysis of the sex chromosomes of lake trout, Salvelinus namaycush. Chromosome Res. 3: 221-226. [DOI] [PubMed] [Google Scholar]
- 50.Sakamoto T., Okamoto, N., and Ikeda, Y. 1996. Application of PCR primer pairs from rainbow trout to detect polymorphism of CA repeat DNA loci in five confamilial species. Fish Sci. 62: 552-555. [Google Scholar]
- 51.Sakamoto T., Danzmann, R.G., Okamoto, N., Ferguson, M.M., and Ihssen, P. 1999. Linkage analysis of quantitative trait loci associated with spawning time in rainbow trout (Oncorhynchus mykiss). Aquaculture 173: 33-43. [Google Scholar]
- 52.Sakamoto T., Danzmann, R.G., Gharbi, K., Howard, P., Ozaki, A., Khoo, S.K., Woram, R.A., Okamoto, N., Ferguson, M.M., Holm, L-E., et al. 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmid M. and Steinlein, C. 2001. Sex chromosomes, sex-linked genes, and sex determination in the vertebrate class amphibia. EXS 91: 143-176. [DOI] [PubMed] [Google Scholar]
- 54.Sinclair A.H., Berta, P., Palmer, M.S., Hawkins, J.R., Griffiths, B.L., Smith, M.J., Foster, J.W., Frischauf, A-M., Lovell-Badge, R., and Goodfellow, P.N. 1990. A gene from the human sex determining region encodes a protein with homology to a conserved DNA binding motif. Nature 346: 240-244. [DOI] [PubMed] [Google Scholar]
- 55.Stein J., Phillips, R.B., and Devlin, R.H. 2001. Identification of the Y chromosome in chinook salmon (Oncorhynchus tshawystcha). Cytogenet. Cell Genet. 92: 108-110. [DOI] [PubMed] [Google Scholar]
- 56.Stein J.D., Reed, K.M., and Phillips, R.B. 2002. Isolation and characterization of a sex-linked microsatellite locus from the lake trout sex chromosome. Environ. Biol. Fish. 64: 211-216. [Google Scholar]
- 57.Taggart J.B., Hynes, R.A., Prodöhl, P.A., and Ferguson, A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J. Fish Biol. 40: 963-965. [Google Scholar]
- 58.Taggart J.B., Prodöhl, P.A., and Ferguson, A. 1995. Genetic markers for Atlantic salmon (Salmo salar L.): Single locus inheritance and joint segregation analyses of minisatellite (VNTR) DNA loci. Anim. Genet. 26: 13-20. [Google Scholar]
- 59.Thorgaard G.H. 1977. Heteromorphic sex chromosomes in male rainbow trout. Science 196: 900-902. [DOI] [PubMed] [Google Scholar]
- 60.___, 1978. Sex chromosomes in the sockeye salmon: A Y–autosome fusion. Can. J. Genet. Cytol. 20: 349-354. [DOI] [PubMed] [Google Scholar]
- 61.___, 1983. Chromosomal differences among rainbow trout populations. Copeia 3: 650-662. [Google Scholar]
- 62.Thorgaard G.H., Allendorf, F.W., and Knudsen, K.L. 1983. Gene–centromere mapping in rainbow trout: High interference over long map distances. Genetics 103: 771-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Traut W. 1994. Sex determination in the fly Megaselia scalaris, a model system for primary steps of sex chromosome evolution. Genetics 136: 1097-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Traut W. and Willhoeft, U. 1990. A jumping sex determining factor in the fly Megaselia scalaris. Chromosoma 99: 407-412. [Google Scholar]
- 65.Voorrips R.E. 2002. Mapchart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77-78. [DOI] [PubMed] [Google Scholar]
- 66.Vos P., Hoger, M., Bleeker, M., Rijans, M., Van Der Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, M., Kuiper, M., et al. 1995. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White M.B., Carvalho, M., Derse, D., O'Brien, S.J., and Dean, M. 1992. Detecting single base substitutions as heteroduplex polymorphisms. Genomics 12: 301-306. [DOI] [PubMed] [Google Scholar]
- 68.Wright J.E., Johnson, K., Hollister, A., and May, B. 1983. Meiotic models to explain classical linkage, pseudolinkage, and chromosome pairing in tetraploid derivative salmonid genomes. Isozymes: Curr. Top. Biol. Med. Res. 10: 239-260. [PubMed] [Google Scholar]
- 69.Young W.P., Wheeler, P.A., Coryell, V.H., Keim, P., and Thorgaard, G.H. 1998. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics 148: 839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Q., Nakayama, I., Fujiwara, A., Kobayashi, T., Oohara, I., Masaoka, T., Kitamura, S., and Devlin, R.H. 2001. Sex identification by male-specific growth hormone pseudogene (GH-ψ) in Oncorhynchus masou complex and related hybrid. Genetica 111: 111-118. [DOI] [PubMed] [Google Scholar]