Abstract
Dual channel imaging and warping of two-dimensional (2D) protein gels were used to visualize global changes of the gene expression patterns in growing Bacillus subtilis cells during entry into the stationary phase as triggered by glucose exhaustion. The 2D gels only depict single moments during the cells' growth cycle, but a sequential series of overlays obtained at specific points of the growth curve facilitates visualization of the developmental processes at the proteomics scale. During glucose starvation a substantial reprogramming of the protein synthesis pattern was found, with 150 proteins synthesized de novo and cessation of the synthesis of almost 400 proteins. Proteins induced following glucose starvation belong to two main regulation groups: general stress/starvation responses induced by different stresses or starvation stimuli (ςB-dependent general stress regulon, stringent response, sporulation), and glucose-starvation-specific responses (drop in glycolysis, utilization of alternative carbon sources, gluconeogenesis). Using the dual channel approach, it was not only possible to identify those regulons or stimulons, but also to follow the fate of each single protein by the three-color code: red, newly induced but not yet accumulated; yellow, synthesized and accumulated; and green, still present, but no longer being synthesized. These green proteins, which represent a substantial part of the protein pool in the nongrowing cell, are not accessible by using DNA arrays. The combination of 2D gel electrophoresis and MALDI TOF mass spectrometry with the dual channel imaging technique provides a new and comprehensive view of the physiology of growing or starving bacterial cell populations, here for the case of the glucose-starvation response.
[This is presented as a movie of B. subtilis's growth/glucose-starvation response, available at www.genome.org and also at http://microbio1.biologie.uni-greifswald.de/starv/movie.htm.]
In 1975 O'Farrell (1975) and Klose (1975) independently described a highly sensitive protein separation technique that enabled the simultaneous analysis of a very large number of proteins. The introduction of this technique into bacterial physiology by F. Neidhardt and R. vanBogelen more than 20 years ago (Herendeen et al. 1979), and decorated with the catchy term “proteomics” in the mid-90s (Wilkins et al. 1996), opened a new era in this field of research. Recent gene sequencing efforts dramatically stimulated progress and interest in proteomics because it is still rather difficult to identify protein spots without knowledge about genome sequences. The genome of Bacillus subtilis, the model organism for Gram-positive bacteria, comprises more than 4100 genes, among them 1700 whose function is still unknown (Kunst et al. 1997). There is an enormous increase in new information simply derived from the genome sequence. The discovery of a large number of putative and still unknown alternative ς factors, two-component systems, and probably many more global regulators in this intensively studied bacterial species was somewhat surprising. However, the genome sequence provides only the blueprint for a living cell. On the other hand, the proteome can be considered as an image taken from a living cell at the molecular level, and could be used to bring the blueprint to life. Up to 10,000 proteins (Klose and Kobalz 1995) can be separated on one single 2D gel. Even if alkaline or extracellular proteins are included in the study, the majority of all proteins synthesized in bacterial cells can be visualized simultaneously. If the genome sequence of the organism is known, the identification of proteins by mass spectrometry (MALDI TOF MS, ESI MS, etc.) supported by N-terminal sequencing no longer represents a problem.
At present our B. subtilis master gel contains ∼600 entries organized in a B. subtilis 2D protein database named Sub2D (Büttner et al. 2001; Werner and Bernhardt 1998; accessible via http://microbio2.biologie.uni-greifswald.de:8880/sub2d.htm). However, this master gel only provides the experimental tool for physiological proteomics visualizing the physiological state of a cell at the level of proteins. The next step in proteomics is to analyze the kinetics of the protein pattern in response to the changes in environmental conditions that are typical of the natural habitat.
Growth of B. subtilis in its natural environment, the upper layers of soil, is characterized by alternation of short periods allowing growth and long nongrowth periods caused by stress and starvation. Consequently, cellular adaptation strategies were optimized by evolution of a highly sophisticated and very complex adaptational genetic and regulatory network that represents one of the most essential features of cell physiology ensuring bacterial survival. This network consists of single regulons, which are groups of genes distributed over the whole genome but with a unique adaptive function and controlled by one global regulator. The analysis of this network, its dissection into single regulons, and a definition of the adaptive function of all proteins within the regulons is crucial for understanding bacterial physiology (Msadek 1999; Sonenshein 2000; Hecker and Völker 2001).
Extracellular stimuli frequently induce more than one regulon. The entire set of proteins induced by one stimulus has been called a stimulon (vanBogelen and Neidhardt 1990). All proteins induced by a specific stimulus contribute to stress adaptation, and therefore defining the size and structure of a stimulon represents the first step in elucidating adaptation to the stimulus. The next step in analyzing adaptational networks is the dissection of stimulons into single regulons. Proteins/genes belonging to regulons can be identified if mutants in global regulators are available. The structure of presently known as well as still unknown regulons can be analyzed by comparing wild-type protein expression patterns to deregulated mutant strains carrying inactivated global regulatory genes. This has been demonstrated in the case of the ςB-dependent general stress regulon of B. subtilis as a model (for reviews, see Hecker et al. 1996; Hecker and Völker 1998; Price 2000). Dissection of the entire genome into single regulons is not yet sufficient for understanding global gene regulation because single regulons do not exist independently from one another but are tightly connected, forming a complex adaptational network (Hecker and Völker 2001). Finally, by using this comprehensive computer-aided inspection and matching of various 2D gels loaded with radioactively labeled proteins from growing, starved, or stressed B. subtilis cells, it is possible to proceed from a 2D protein index to a more global analysis and description of the gene regulation map of a cell (Antelmann et al. 1997, 2000). The most comprehensive results are provided by DNA array techniques, which, however, generate a huge quantity of data that is difficult to interpret. A fast but nevertheless sufficient overview on cell physiology can be obtained by proteomics when the dual channel imaging of 2D protein gels is used (Bernhardt et al. 1999).
The dual channel imaging technique for 2D protein gel analysis was developed to make the search for proteins belonging to stimulons or regulons more feasible. This simple technique allows a rapid allocation of proteins to stimulons or regulons simply by looking for red (newly induced) proteins (see Methods) or green (repressed) proteins. The advantage of this technique becomes apparent in the matching of protein patterns on a single gel. This means that the matching of different gel images, one of the bottlenecks in data analysis, is no longer necessary (Bernhardt et al. 1999). Dual channel imaging provides a large quantity of information about the relative amount and synthesis rate of each single protein. However, one gel visualizes only a single moment (snapshot) in the growth cycle of a bacterial cell population, presenting a nondynamic gene expression pattern. But if one places these composite images in a sequential order, the growth and development of a bacterial cell can be depicted at the molecular level as a movie of life. The combination of 2D protein gel electrophoresis and MALDI TOF mass spectrometry with this dual channel imaging technique can provide a comprehensive overview of the physiology of a bacterial cell population entering the stationary growth phase.
The main goal of the present study is to provide experimental evidence for this technique using glucose-starved cells as a model system, a quite normal environmental situation for B. subtilis cells. This semiquantitative approach can be complemented by quantitative evaluation of the synthesis pattern for single proteins or protein groups, and by DNA array techniques for the entire genome.
RESULTS
The adaptation of B. subtilis to glucose as a growth-dictating parameter was investigated by embedding composite images of the entire proteome into the time scale of the growth curve. Cultivation of B. subtilis was done in synthetic minimal medium with 0.05% glucose as the sole carbon and energy source. Samples were taken at several time points of the growth curve, pulse-labeled, and separated by 2D PAGE. Furthermore, the redistribution of the overall protein pattern of starved cells into that of growing cells after readdition of glucose was analyzed.
For this purpose the dual channel imaging technique for 2D protein gels was used (Bernhardt et al. 1999). This technique combines staining techniques to visualize accumulated proteins and autoradiography to uncover proteins that were synthesized at defined states. Two digitized images of 2D gels have to be generated and combined in using alternate additive color channels. One of them—the densitogram—showing proteins accumulated in the cell that had been visualized by (silver) staining techniques was pseudocolored green. The second image of an autoradiograph showing the proteins synthesized during a 5-min L-[35S]-methionine pulse label was pseudocolored red. As a result, accumulated as well as newly synthesized vegetative proteins took on a yellow color. After imposition of a glucose-starvation stimulus, however, proteins newly synthesized in response to that stimulus are red because they have not yet accumulated in the cell. Proteins whose synthesis has been switched off by the stimulus change their color from yellow to green.
Exponentially Growing Cells
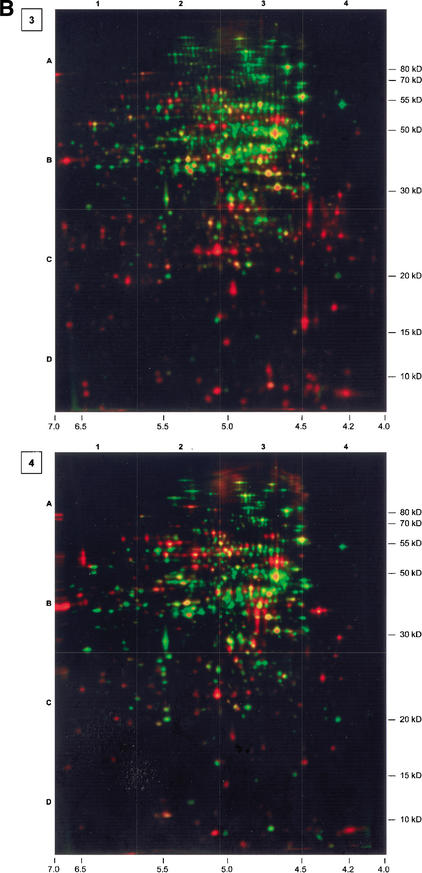
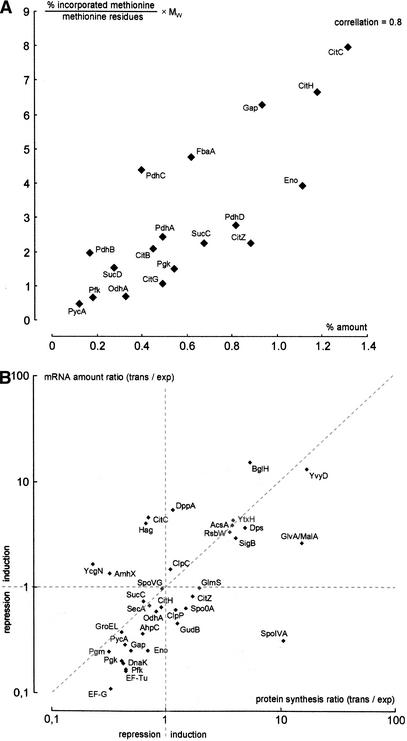
As long as glucose and other nutrients are available, B. subtilis reaches a generation time of ∼1 h at 37°C using the described minimal medium (Fig. 1). The first sample (Fig. 2, Gel 1) illustrates that during this growth phase mainly vegetative proteins are synthesized (autoradiogram, red) and accumulated (densitogram, green; both together, yellow), among them the main catabolic enzymes for glucose utilization, enzymes synthesizing amino acids, proteins, and nucleotides, as well as proteins of the translational machine, and so on (Fig. 2, Gel 1; Büttner et al. 2001). As long as sufficient glucose is available to support exponential growth, the synthesis and amount of protein show a reasonable correlation (Fig 3A).
Figure 1.
Growth of B. subtilis 168 and the isogenic sigB-mutant strain ML6. Numbers indicate the developmental stages from which samples were taken and subsequently labeled with L-[35S]-methionine. Columns show the amount of radioactivity incorporated into protein during 5 min.
Figure 2.
Series of dual channel images (silver-stained protein amount, green; autoradiogram of currently synthesized protein, red). Numbers in boxes associate the images to the corresponding growth phase. For further information, see Figure 1. Gel 3 was complemented by a separate autoradiogram showing protein spots that had already been identified. The colors of the text boxes indicate the relative synthesis compared with exponential growth conditions (black, not detected; red, induced; yellow, unchanged; light green, slightly repressed; green, repressed).
Figure 3.
Scatterplot (A) illustrating the correlation of amount and synthesis of proteins involved in basic carbon metabolism after 2.5 h of exponential growth (Fig. 2, Gel 1). The X-axis shows the protein amount in percentage of the whole detectable protein on the 2D gel; the Y-axis the L-[35S]-methionine incorporation corrected by the methionine content and the protein size. Scatterplot (B) shows a representative sample of mRNAs/proteins changing their expression during transient phase. The Y-axis displays the fold change of the mRNA-amount, the X-axis the fold change of methionine incorporation (protein synthesis).
Because of decreasing glucose concentration (see Fig. 1), generation time increases and L-[35S]-methionine incorporation decreases. However, more or less the same proteins are synthesized until glucose exhaustion (Fig. 2, Gel 2).
Glucose-Starvation Response
The proteomics approach resulted in identification of two main groups of proteins (regulons) synthesized in response to stress and starvation (see Hecker et al. 1996; Antelmann et al. 1997; Hecker and Völker 1998): Proteins induced by only one stimulus (or by a group of strongly related stimuli) may have a specific protective function against this specific stress or starvation factor only. Each stimulus induces a typical set of proteins (heat-stress-specific, glucose-starvation-specific, etc.). The second group encompasses general stress/starvation proteins induced by different environmental stimuli. These general stress/starvation proteins may have a rather nonspecific but nevertheless very important protective function under stress or starvation to ensure cell viability in a nongrowth state regardless of the specific stress/starvation stimulus that induced the nongrowth situation. Proteins induced or repressed in response to growth-restricting factors form the complex adaptational network composed of a large number of tightly connected specific and general stress/starvation regulons.
Accordingly, extensive changes in protein synthesis patterns occurred after entering the transient phase (Fig. 2, Gel 3). Only one-fifth of the methionine was still incorporated into protein compared with exponentially growing cells (see Fig. 1). When cells enter the stationary phase >150 detectable proteins (red) are sequentially induced (Fig. 2, Gels 3–7), and at least 400 proteins produced during exponential growth are repressed (green). The induction/repression ratios of protein synthesis are in good correlation to the changes of the mRNA levels derived from the DNA-array experiments carried out in parallel (Fig. 3B). These proteins can be assigned to quite different glucose-starvation-specific and nonspecific regulons forming a strong adaptational network to survive glucose starvation.
Nonspecific Glucose-Starvation Responses
General Stress/Starvation Response
One of the most obvious protein groups belonging to the general stress/starvation regulon is induced at the beginning of glucose starvation. This regulon, controlled by the alternative ς factor ςB, the regulator for the general stress response, is not induced in glucose-starved cells of the sigB-mutant strain B. subtilis ML6 (Fig. 4). This induction is only transient, probably because utilization of alternate carbon sources might increase the ATP level that down-regulates ςB activity (Fig. 2, Gel 3, red spots; Fig. 4, red spots). For this purpose, almost 20% of the available translational capacity is used. These general stress proteins are also induced by a different set of stress and starvation signals including salt, acid, or heat stress, or energy limitation in general (Price 2000). However, a few general stress proteins, such as thioredoxin or the Clp protease/ATPase ClpP/ClpC strongly induced by heat stress, are not significantly induced in glucose-starved cells, probably because still active repressors block the ςB-dependent promoters (Fig. 4, red spots; Table 1). The induced proteins are expected to provide the nongrowing cell with a multiple, nonspecific, and prospective stress resistance to be prepared for and protected against future stress (Hecker and Völker 1998). Some of the proteins, for instance, are necessary for adaptation to oxidative stress, which may be a common environmental situation in glucose-starved B. subtilis cells living in the upper layers of soil. Whereas exponentially growing B. subtilis cells are highly sensitive to hydrogen peroxide, glucose-starved cells became resistant. The development of oxidative stress resistance mainly depends on ςB. The very early induction of these stress proteins starting in the transient phase may be one of the earliest indicators of glucose starvation. Lowering of ATP concentration or alternative energy starvation indicators may be the cellular signals for induction of the regulon (for review, see Price 2000). It is interesting to note that, at least under these conditions, ςB-dependent general stress proteins are not significantly accumulated in a detectable amount during glucose starvation (Fig. 2, Gel 3–7; Fig. 4; Fig. 5, ςB-dependent proteins, no change to yellow color for almost all proteins).
Figure 4.
Dual channel image of autoradiograms (protein synthesis) of B. subtilis 168 (red) and the isogenic sigB-mutant strain ML6 (green), transformed with the Delta2D two-dimensional gel analysis software. Samples were taken from cultures in the transient growth phase. Already identified spots synthesized at a higher rate in the wild type are indicated by text labels, and nonidentified spots by white circles. For spots induced in a ςB-dependent manner, see also Table 1.
Table 1.
List of Protein Spots Influenced by Regulators ςB (A), ςH (B), RelA (C), CcpA (D), and CodY (E)
| Valid gene name | Gel 1–9 | Synonym | B. SubtiList acc no. | Description |
| A. ςB-Dependent Genes (Wild type/ςB-Mutant Comparison) | ||||
| clpC | − | mecB | BG10148 | Class III stress response-related ATPase |
| clpP | − | yvdN | BG19016 | ATP-dependent Clp protease proteolytic subunit (class III heat-shock protein) |
| ctc | + | BG10115 | General stress protein | |
| dps | + | ytkB | BG12584 | Stress- and starvation-induced gene controlled by ςB |
| gsiB | + | BG10826 | General stress protein | |
| gspA | + | ipa-12d ywaG | BG10558 | General stress protein |
| gtaB | + | BG10402 | UTP-glucose-1-phosphate uridylltransferase | |
| katE | + | katB | BG11102 | Catalase 2 |
| katX | + | yxll | BG11945 | Major catalase in spores |
| nadE | + | gsp-81 outB tscBGH | BG10694 | NH3-dependent NAD+ synthetase |
| rsbV | + | BG10733 | Positive regulator of ςB activity (anti-anti-ς factor) | |
| rsbW | + | BG10734 | Negative regulator of ςB activity (switch protein/serine kinase, anti-ς factor) | |
| sigB | + | rpoF | BG10735 | RNA polymerase general stress ς factor |
| sodA | − | yqgD | BG11676 | Superoxide dismutase |
| ycdF | + | BG12761 | Unknown; similar to glucose 1-dehydrogenase | |
| ydaE | + | BG12053 | Unknown | |
| ydbD | + | BG12071 | Unknown; similar to manganese-containing catalase | |
| yfkM | + | BG12929 | Unknown; similar to unknown proteins | |
| yflT | + | BG19020 | Unknown | |
| yhdN | + | BG13020 | Unknown; similar to aldo/keto reductase | |
| yitT | + | yuxA | BG10850 | Unknown; similar to unknown proteins |
| ykzA | + | yzzE | BG19021 | Unknown; similar to organic hydroperoxide resistance protein |
| yocK | + | BG13524 | Unknown; similar to general stress protein | |
| ysnF | + | BG12337 | Unknown; similar to unknown proteins | |
| ytxH | − | csb40 | BG10975 | Unknown; similar to general stress protein |
| yvyD | − | BG10740 | Unknown; similar to ς54 modulating factor of Gram-negative bacteria | |
| B. ςH-Dependent Genes | ||||
| yvyD | □□▪▪▪□▪□□ | BG10740 | General stress protein | |
| spoVG | ▪▪▪▪▪▪▪▪▪ | BG10112 | Sporulation | |
| spo0A | □□▪▪▪▪▪▪□ | sof-1 spo0C spo0G spoIII | BG10765 | Two-component response regulator |
| ytxH | □□▪▪▪▪▪▪▪ | csb40 | BG10975 | Unknown function, similar to general stress protein |
| citG | ▪▪▪▪□□□□□ | BG10384 | Fumarase | |
| C. relA-Dependent Genes Class I: RelA-dependent transcriptional repression (negative regulation) | ||||
| rplA | X | BG10164 | Ribosomal protein L1 | |
| rplJ | ▪▪□□□□□□□ | BG11220 | Ribosomal protein L10 | |
| rpoB | ▪▪□▪□▪▪▪▪ | BG10728 | RNA polymerase (β subunit) | |
| fus | ▪▪□□□□□▪▪ | EF-G | BG11939 | Elongation factor G |
| tufA | ▪▪□□□□□▪▪ | EF-Tu | BG11056 | Elongation factor Tu |
| rplE | X | BG10760 | Ribosomal protein L5 | |
| rplF | X | BG11408 | Ribosomal protein L6 | |
| adk | ▪▪▪□□□□□□ | BG10446 | Adenylate kinase | |
| rpoA | X | BG10732 | RNA polymerase (α subunit) | |
| rpsB | ▪▪□□□□□□□ | BG19004 | Ribosomal protein S2 | |
| tsf | ▪▪□□□□□▪▪ | EF-Ts | BG19025 | Elongation factor Ts |
| frr | weak | BG12587 | Ribosomal recycling factor | |
| rpsF | X | BG10049 | Ribosomal protein S6 | |
| tig | ▪▪□□□□□▪▪ | yzzH | BG19023 | Trigger factor (peptidyl prolyl isomerase) |
| ylaG | ▪▪□□□□□▪▪ | BG13344 | Similar to GTP-binding elongation factor/unknown | |
| yugI | weak | BG12363 | Similar to polyribonucleotide nucleotidyltransferase/unknown | |
| bkdAA | X | bfmb1a bfmbaa bkd | BG10307 | Branched-chain α-keto acid dehydrogenase E1 subunit (2-oxoisovalerate dehydrogenase α subunit) |
| pnpA | ▪▪□□□□□▪▪ | comR | BG11491 | Polynucleotide phosphorylase |
| atpA | ▪▪□▪▪□□▪▪ | BG10819 | ATP synthase (α subunit) | |
| atpD | ▪▪□□□□□□□ | BG10821 | ATP synthase (β subunit) | |
| yjlD | ▪▪□□□□□□□ | BG13203 | Similar to NADH dehydrogenase/unknown | |
| Class II: RelA-dependent transcriptional induction (positive regulation) | ||||
| ilvB | ▪▪□▪▪▪▪▪▪ | BG10670 | Acetolactate synthase (large subunit) | |
| ilvC | ▪▪□□□□□▪▪ | BG10672 | Ketol-acid reductoisomerase | |
| leuA | ▪▪□□□□□▪▪ | BG11948 | 2-Isopropylmalate synthase | |
| leuB | ▪▪□□□□□▪▪ | leuC | BG10675 | 3-Isopropylmalate dehydrogenase |
| leuC | ▪▪□□□□□▪▪ | BG11949 | 3-Isopropylmalate dehydratase (large subunit) | |
| leuD | ▪▪□□□□□▪▪ | BG11950 | 3-Isopropylmalate dehydratase (small subunit) | |
| ywaA | ▪▪□□▪□□▪▪ | ipa-0r | BG10546 | Similar to branched-chain amino acid aminotransferase/unknown |
| guaB | ▪▪▪□□□□□▪ | guaA | BG10073 | Inosine-monophosphate dehydrogenase |
| ald | ▪▪□□□□□▪▪ | spoVN | BG10468 | L-alanine dehydrogenase |
| spoVG | ▪▪▪▪▪▪▪▪▪ | Ipa-12d ywaG | BG10558 | Stage V sporulation protein, required for spore cortex synthesis, inhibitor of sporulation |
| spo0A | □□□□▪▪▪▪▪ | BG14002 | Two-component response regulator | |
| gspA | □□▪▪▪▪▪▪□ | gcaA ybxD | BG10948 | General stress protein |
| yvyD | □□▪▪▪▪▪□□ | BG12558 | Similar to ς54 modulating factor of Gram-negative bacteria, similar to ribosomal proteins | |
| ytxH | □□▪▪▪▪▪▪□ | yfjI | BG12560 | Similar to general stress protein/unknown |
| yurP | ▪▪□□□□□▪▪ | BG12680 | Similar to glutamine-fructose-6-phosphate transaminase/unknown | |
| D. ccpA-Dependent Genes | ||||
| glmS | □□□▪▪▪▪▪□ | gcaA ybxD | BG10948 | L-Glutamine-D-fructose-6-phosphate amidotransferase |
| acoA | □□□▪▪□□□□ | yfjK | BG12558 | Acetoin dehydrogenase E1 component (TPP-dependent α subunit) |
| acoC | □□□▪▪□□□□ | yfjI | BG12560 | Acetoin dehydrogenase E2 component (dihydrolipoamide acetyltransferase) |
| sucC | ▪▪▪▪▪▪▪▪▪ | BG12680 | Succinyl-CoA synthetase (β subunit) | |
| sucD | ▪▪▪□□□□□▪ | BG12681 | Succinyl-CoA synthetase (α subunit) | |
| gapB | □□▪▪▪▪▪□□ | BG12592 | Glyceraldehyde-3-phosphate dehydrogenase | |
| rbsK | n.i. | BG10877 | Ribokinase | |
| rbsA | □□▪▪▪□▪□□ | BG10879 | Ribose ABC transporter (ATP-binding protein) | |
| rocA | □□▪▪□□□□□ | ipa-76d | BG10622 | Pyrroline-5 carboxylate dehydrogenase |
| rocG | n.i. | ipa-75d; yweB | BG10621 | Glutamate dehydrogenase (major) |
| iolH | n.i. | yxdG | BG11123 | Unknown function in myo-inositol catabolism |
| iolA | n.i. | mmsA yxdA | BG11117 | Oxidative decarboxylase in myo-inositol catabolism |
| iolS | n.i. | yxbF | BG11363 | Unknown function in myo-inositol catabolism |
| rocF | □□□▪▪□□□□ | BG10932 | Arginase | |
| rocD | ▪▪▪▪▪□□▪▪ | BG10722 | Ornithine aminotransferase | |
| E. CodY-Dependent Genes | ||||
| dppA | □□▪▪▪▪□□□ | BG10722 | D-alanyl-aminopeptidase | |
| hutI | □□□□□▪□□□ | BG11100 | Imidazolone-5-propionate hydrolase | |
| hutU | □□▪▪▪▪□□□ | BG10668 | Urocanase | |
| hag | ▪▪▪▪▪□□□▪ | BG10655 | Flagellin protein | |
| srfAD | ▪▪▪□▪▪▪▪▪ | BG10171 | Required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation | |
| citB | □□□□□▪▪▪▪ | BG10478 | Aconitate hydratase | |
| spo0A | □□▪▪▪▪▪▪□ | sof-1 spo0C spo0G spoIII | BG10765 | Two-component response regulator |
Plus signs in the column [Gel 1–9] (A) indicate a clear induction in a ςB-dependent manner at the protein level in the wild type compared to the ςB-mutant strain (see also Fig. 3). Minus signs show an overlaying effect of additional regulators masking the ςB dependency. Filled squares at positions 1 to 9 in the column [Gel 1–9] (B–E) point out high synthesis in the corresponding gels (see Figs. 1 and 2). X signs indicate very weak expression levels of the corresponding spots.
Figure 5.
Patterns of amount (green) and synthesis (red) of general stress/starvation and glucose starvation-specific proteins during different growth stages (columns correspond to the numbers in Fig. 1). Diagrams show protein synthesis in percentage normalized to the whole synthesis detectable on a 2D gel as described in Methods.
Negative Stringent Control
The second general starvation response strongly activated in glucose-starved cells is the stringent response, also induced by amino acid starvation or oxygen limitation (Nishino et al. 1979; Hecker et al. 1987; Wendrich and Marahiel 1997). Mediated by guanosine tetraphosphate (ppGpp), the stringent response prevents wastage of nutrients during nutritional deprivation. Many genes typically expressed in growing cells are repressed because continuous synthesis of those proteins already available in sufficient levels is unnecessary in nongrowing cells. This negative stringent control can be observed for many vegetative proteins (change from yellow to green), among them components of the translation apparatus (Ef-Tu, ribosomal proteins, etc.; Fig. 5, translational machinery) or proteins involved in amino acid anabolism. It has to be mentioned that not all proteins changing color from yellow to green with onset of glucose starvation belong to the stringent response regulon. This was demonstrated by analysis of a relA mutant no longer able to synthesize ppGpp in response to starvation (Table 1, relA-dependent transcriptional repression; Eymann and Hecker 2001). Synthesis of glycolytic enzymes (see Fig. 5, glycolysis), for instance, is repressed in the wild type as well as in an relA mutant (Eymann et al. 2002).
Positive Stringent Control, Spo0A, and Other General Stationary-Phase Responses
There are a few proteins whose synthesis is induced by nutrient starvation only in the wild type but not in the relA mutant, indicating positive stringent control. Among these proteins induced RelA-dependently in glucose-starved cells is the response regulator Spo0A. The concentration and phosphorylation state of Spo0A are crucial for many stationary growth phase events (Hoch 1995; Greene and Spiegelmann 1996; Sonenshein 2000). Phosphorylated Spo0A stimulates transcription of spo0H encoding ςH. However, only a very few ςH-dependent genes seem to be activated in glucose-starved cells, among them YvyD and YtxH or Spo0M.
The ytxH operon and yvyD, which are under double control by sigH and sigB, are activated by ςB in glucose-starved cells; however, the extended transcription pattern compared with gsiB indicates the replacement of ςB by ςH (see Figs. 2 and 4). The ςB activity is indicated by the expression kinetics of GsiB solely dependent on ςB. The high YvyD expression after this period of ςB activity is probably caused by ςH. In contrast to GsiB, which is not accumulated, this prolonged expression pattern of YvyD leads to a distinct protein accumulation (see also Drzewiecki et al. 1998).
The induction of DppA indicates that the global regulator CodY is no longer able to repress the gene (probably triggered by a drop in the GTP level; Ratnayake-Lecamwasam et al. 2001). For other general regulons probably activated in stationary-phase cells, marker proteins indicating their activity under our experimental conditions were not found. The application of DNA-array techniques is necessary to make a final decision about their activity. ςH in cooperation with Spo0A∼P also activates the spoIIA operon, thereby triggering sporulation. Few sporulation proteins appeared after a few hours of glucose starvation, for example, SpoIVA (ςE-dependent, spore cortex formation), YrbA (similar to spore coat protein; Fig. 2, Gels 3–7; Fig. 5, sporulation), indicating that at least a portion of the cell population already initiated sporulation. Analysis of sporulation gene expression in single cells should indicate how many cells really triggered sporulation.
Glucose-Starvation-Specific Responses
Besides the more general responses in stationary-phase cells induced by a set of different stress/starvation stimuli such as general stress response (ςB), stringent response (RelA), or sporulation (Spo0A∼P and others), some specific reactions typical only of carbon source starvation have been induced.
Usage of Alternative Carbon Sources
The first essential glucose-starvation-specific response is switching to utilization of alternative carbon sources mediated mainly by the catabolite control protein CcpA (Stülke and Hillen 2000). Cells growing on excess glucose synthesize ATP mainly via substrate phosphorylation, and enzymes of the TCA cycle are expressed at a relatively low level (Tobisch et al. 1999; Ludwig and Stülke 2001). Under these conditions the task of the TCA cycle consists primarily in production of anabolic intermediates necessary for cell growth. Excess glucose intermediates do not enter the repressed TCA cycle but are excreted as acetoine, lactate, acetate, or other overflow metabolites, resulting in an acidification of the extracellular medium (Tobisch et al. 1999). After exhaustion of glucose the glycolytic pathway is repressed because of the need for a high glucose concentration for expression of the gapA operon (Tobisch et al. 1999; Ludwig and Stülke 2001). This is shown in Figure 5 for the glycolytic enzyme glycerine aldehyde 3-phosphate-dehydrogenase GapA (yellow to green). On the other hand, the gluconeogenic pathway seems to be induced, visualized by the strong induction of GapB, the gluconeogenic glycerine aldehyde-3-phosphate-dehydrogenase (Fillinger et al. 2000). Gluconeogenesis seems to be necessary as a result of exhaustion of glucose because starved cells start to use secondary carbon sources, for example, acetoine, although they are no longer able to grow on the excess metabolites. Induction of the acetoine dehydrogenase subunits AcoA, AcoB, and AcoC (Fig. 2, Gels 4 and 5), which are ςL-dependent, indicates the beginning of this reutilization phase (Ali et al. 2001). Monitoring the color of corresponding spots (Fig. 5, overflow metabolite utilization) demonstrates expression kinetics of this enzyme complex, whose subunits are encoded by genes organized into one operon. First, they appear as red spots induced 10 min after glucose exhaustion, followed by red spots with yellow cores indicating protein accumulation. A color change from red to yellow appears 30 min later, showing a balance between synthesis and accumulation. Finally, the spots change color continuously from yellow to green, demonstrating the stepwise switch-off of their synthesis. As mentioned above, besides acetoine, other overflow metabolites like acetate are also available and can be used as secondary carbon sources. This is indicated by the induction of acsA, encoding acetylCoA-synthetase (Fig. 2, Gels 3–7; Fig. 5, overflow metabolite utilization), during entry into the stationary phase.
Interestingly, catabolic genes also seem to be derepressed in glucose-starved cells without any obvious external inducer. Examples are rbsA, the cytosolic component of ribose ABC transporter, bglH, encoding a β-glucoside-degrading enzyme, or MalA (GlvA), encoding a phospho-α-glucosidase (Fig. 2, Gels 3–7; Fig. 5, alternative carbon source utilization). Possible substrates might be cell-wall turnover products containing ribose or other compounds with α- or β-glycosidic linkages that are excreted during exponential growth, storage metabolites, or lysis products of dead cells. Many of those genes whose products allow the utilization of alternative carbon sources are under CcpA control (see Table 1). These genes are active—in most cases dependent on an additional inducer—when glucose is no longer present.
During the process of alternative carbon and energy source utilization, L-[35S]-methionine incorporation increases transiently to ∼35% of the growth level (Fig. 1; Fig. 2, Gels 4 and 5). This transiently increased global protein synthesis is coupled to a decrease of the synthesis of ςB-dependent proteins as observed in the transient phase (Fig. 2, Gel 3). After exhaustion of all carbon sources 35S-methionine incorporation decreases (Fig. 2, Gels 6 and 7) to less than 1/20 of the growth level measured during exponential growth.
Proteomic Signatures for Protein Stress or Oxidative Stress in Glucose-Starved Cells
During glucose starvation there appears to be no protein stress or oxidative stress, indicated by the absence of proteomics signatures for these stressors. In contrast, Escherichia coli suffers from oxidative stress after longer periods of glucose exhaustion, as shown by the induction of members of the oxidative stress stimulon (Ballesteros et al. 2001; Nyström 2001).
Recovery of Growth
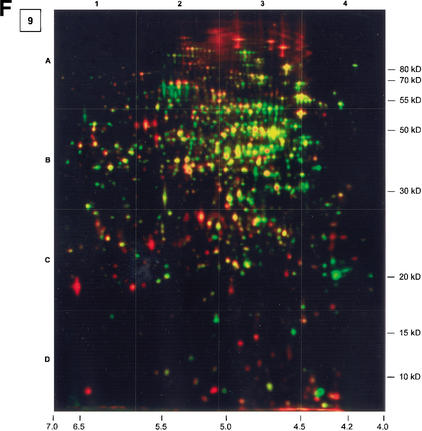
Addition of glucose leads to immediate recovery of cell growth. Synthesis of enzymes for utilization of alternative nutrient sources is instantly turned off, and synthesis of almost all proteins turned off while cells entered stationary phase is reinitialized and colored yellow (Fig. 2, Gels 8 and 9). This is shown for EF-Ts as an example for stringent control, or for the glycolytic enzyme GapA, whereas the synthesis of its gluconeogenesis counterpart, GapB, has been switched off (Fig. 5).
Besides synthesis of the vegetative proteins involved, for example, in glycolysis (Fig. 5, glycolysis) and other basic functions in vegetative cell metabolism, synthesis of PycA (Fig. 5, refill of TCA and recovery of growth), the pyruvate carboxylase-generating oxaloacetate from pyruvate, drastically increases. This may indicate an enormously increased need for the synthesis of intermediates derived from TCA cycle reactions. PycA, whose synthesis has been switched off with entry into the stationary phase, is still present in glucose-starved cells, but its activity is unsolved here. This enzyme is not necessary for growth on substrates such as acetoine or acetate.
DISCUSSION
We found that the transition from growing cells to glucose-starved cells is accompanied by an almost complete reorganization of the gene expression program (Gasch et al. 2000). At least 400 proteins change their color from yellow to green because their synthesis has been switched off in the stationary-growth phase. These expression kinetics are typical of proteins with housekeeping functions necessary for growth and the cell cycle. The cellular level of those almost stable proteins whose synthesis was switched off was arrested in cells entering the stationary phase and no longer able to grow. On the other hand, >150 proteins have been induced in a sequential order as indicated by the appearance of new protein spots colored red. These 150 proteins belong to different general and specific regulation groups, which was shown by the analysis of mutants in genes encoding global regulators. Because the induction of the single regulons occurs in a sequential manner, this reprogramming of gene expression can only be detected by a kinetic study (Figs. 2 and 5). The powerful dual channel imaging technique not only allows the description of stimulons or regulons but also allows us to follow the kinetics of each single protein with the three colors: Red means newly induced but not yet accumulated, yellow means synthesized and accumulated, and green means no longer being synthesized, but still present and probably still active. The expression kinetics of AcoB shown in Figure 5 is typical of most of the glucose-starved inducible proteins considered in this study (see MalA, BglH, RbsK). After a more or less short induction period, the gene expression has been switched off again, but this transient induction seems to be sufficient for the accumulation of the protein needed during glucose starvation. Other glucose-starvation-specific proteins showed more extended expression kinetics, such as GapB or AcsA (Fig. 5).
The total reprogramming of the gene expression network was confirmed by the application of DNA macroarray techniques (data not shown). The data clearly show that about the half of all B. subtilis genes are involved in the process, changing their expression pattern. Almost 1000 (vegetative) genes were switched off in cells entering glucose starvation, and the same number was induced on a different time scale. A similar reprogramming of the gene expression pattern was observed for starved Saccharomyces cerevisiae cells (Gasch et al. 2000). The earliest response to glucose starvation was the induction of the stringent response as well as of the ςB-dependent general stress regulon followed by diverse glucose-starvation-specific responses. The RNA profiling data confirm our findings from proteomics: The interplay between specific and more general reactions to glucose starvation ensures survival in the starvation period. Whereas the specific responses guarantee a direct and specific interaction with the stimulus, the general ςB-dependent response protects the nongrowing cell during a long survival period. This combination of mRNA profiling and proteomics is an extremely useful approach to visualize what is happening in the cell during growth and development, shown here for our model case, glucose starvation. The DNA-array data represent a more complete description of the changes in gene expression, but the proteomic data allow a more convenient (visual) inspection of the gene expression patterns, which is quite sufficient for getting an overview of cell physiology. Furthermore, the proteomic approach, which more closely reflects the final level of gene expression, depicts reactions that can never be visualized by using the DNA-array techniques, such as the large number of proteins still present and probably active in nongrowing cells but no longer being synthesized (the green proteins), or posttranslational reactions such as protein modification, stability, or protein targeting not considered in this study (see Antelmann et al. 2001; Büttner et al. 2001). The dual channel imaging technique (Bernhardt et al. 1999) alone, visualizing synthesis and accumulation of proteins, is an excellent tool for describing not only the fate of single regulons but also the fate of each single protein during the growth and development process, including proteins that are no longer being synthesized in nongrowing cells. The combination of the proteomic as well as transcriptomic approach should be the state of the art for a comprehensive analysis and description of growth and development.
One of the key questions to be addressed in those studies is the relationship among mRNA, protein synthesis, and protein levels in cells. The approach we used offers the opportunity to compare the protein synthesis level with the protein amount at each single protein spot on the 2D gels at each time point on the growth curve. For this purpose we selected some model proteins from basic carbohydrate metabolism. Comparison shows a reasonable correlation of protein synthesis with protein amount during the exponential growth phase (Fig. 3, top). In glucose-starved cells, however, this good correlation no longer exists (Gap, Eno, Ef-Tu, Ef-Ts, EF-G, etc.; see also Fig. 2, Gel 3 and annotated Gel 3). Most of the vegetative proteins synthesized specifically during growth and the cell cycle are still present but are no longer being synthesized. On the other hand, many proteins are strongly induced in response to glucose starvation but not yet accumulated (see Fig. 2, Gel 3). These data allow us to conclude that the amount of the single proteins is a reasonable indicator for their protein synthesis rate only in cells that were grown under constant conditions over a longer time span. In cells entering the stationary phase, the protein level does not necessarily reflect the protein synthesis rate. To analyze this gene expression pattern in nongrowing cells, mRNA profiling or pulse-labeling studies with radioactively labeled amino acids is required. Our data show that these global changes in the gene expression pattern can be visualized by both approaches, both providing not exactly the same but more or less similar relationships (Fig. 3, bottom).
Two groups of proteins synthesized in response to glucose starvation can be distinguished: proteins strongly induced at the beginning of glucose exhaustion but not accumulated in later stages of starvation, and proteins induced as well as accumulated at this period. To our surprise, the ςB-dependent general stress proteins belonging to the first group have not been significantly accumulated during glucose starvation, except YvyD (see Fig. 2, Gel 3 and following). In accordance with this finding is the only transient induction of the proteins followed by a strong reduction of gene expression. We suggest that a relatively low level of the proteins may be sufficient for fulfilling their physiological function under the physiological conditions of the experiment. The synthesis of the other group is maintained for a longer time period in the stationary phase (see, e.g., AcoB in Fig. 2, Gels 4–7), resulting in a significant accumulation of the proteins.
To summarize, simple staining of the 2D gels is a reasonable approach to obtain information on the gene expression pattern in growing cells because more sensitive mRNA or protein synthesis studies will give more or less similar results with the exception of rare cases (e.g., unstable proteins). This is no longer true for cells entering the stationary phase. To obtain an overview on the gene expression pattern of nongrowing cells, the prevailing physiological state of cells in nature, mRNA profiling or pulse-labeling experiments have to be chosen, but both approaches are not sufficient to learn what proteins are available and probably active in those cells. To get this kind of information, these studies have to be complemented with protein staining techniques that will provide information on the actual concentration of proteins in the nongrowing cell.
METHODS
Bacterial Strains and Culture Conditions
Wild-type B. subtilis 168 (trpC2; Anagnostopoulos and Spizizen 1961) and the isogenic sigB-mutant strain B. subtilis ML6 (Igo et al. 1987) were grown under vigorous agitation at 37°C in a synthetic minimal medium as described earlier (Stülke et al. 1993). Citrate and glutamate were excluded to avoid usage of these compounds as additional carbon sources and to exclude diauxic growth phenomena. Glucose was supplemented to a final concentration of 0.05%. These conditions allow growth to an optical density of 1.0 at 500 nm (OD500). After 3 h in the stationary phase, glucose was added to a final concentration of 0.05%, and the recovery of growth was monitored.
2D Electrophoresis, Gel Staining, and Image Generation
Analytical Electrophoresis
Samples were taken along the growth curve as indicated in Figure 1. Pulse-labeling was performed for 5 min by using 10 μCi of L-[35S]-methionine per milliliter. Then, 5 min later, L-[35S]-methionine incorporation was stopped by adding unlabeled methionine (1 mM) and chloramphenicol (100 μg/mL) and subsequently by transferring the sample to ice. After disruption of the harvested cells by sonication, the protein solution was separated from cell debris by centrifugation, and the protein amount (Bradford 1976) and incorporated radioactivity were determined. Crude protein extracts (60 μg of protein) were loaded onto Pharmacia ready-made IPG strips (pI range 4 to 7) for the isoelectric focusing of 2D gel electrophoresis as recommended by the manufacturer. The separation in the second dimension was carried out as described previously (Schmid et al. 1997; Büttner et al. 2001). After fixing and silver staining (Bloom et al. 1987), the wet gels were scanned with a Hewlett-Packard ScanJet 6000 in transmission mode at a resolution of 200 dpi and a color depth of 8 bits–256 gray levels (10 bits–1024 gray levels internally). Silver-stained gels were dried on a chromatography paper backing by using a heated vacuum dryer. For autoradiography of the radiolabeled protein pattern, dried gels were exposed to storage phosphor screens (Molecular Dynamics Storage Phosphor Screen, 20 by 25 cm) for a time span ensuring usage of the whole dynamic range by the strongest spot and corresponding to the amount of radioactivity separated on the gel. Screens were scanned using a PhosphorImager SI (Molecular Dynamics) at a resolution of 200 μm and a color depth of 16 bits (65,536 gray levels). Samples were prepared and analyzed in parallel from three independent cultures.
For quantitation of protein levels, 100 μg of protein was separated per 2D gel. Gels were stained by using SyproRuby 2D gel stain (Molecular Probes) according to the protocol of the manufacturer. Protein levels were detected with an Amersham Typhoon Scanner at a resolution of 200 μm and a color depth of 16 bits. Excitation occurred at 457 nm, and emission was measured with the 610-nm-bp filter with a 500-V photo multiplier tube voltage.
Preparative Electrophoresis
For protein identification by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI TOF mass spectrometry), protein spots were isolated from samples that were taken at the end of the stationary-growth phase prior to growth recovery and at 2.5 h after growth recovery. Samples containing 500 μg of protein were prepared and separated as described above and stained with Coomassie Brilliant Blue R-250.
2D Gel Analysis
Dual Channel Imaging and Gel Warping
For better visualization of changes in the protein pattern, Dual Channel Imaging was used. This technique involves overlaying the protein level image (silver stain, pseudocolored green) and the protein synthesis image (autoradiogram, pseudocolored red) onto each other. To ensure that results were not influenced by spot mismatches, distorted gels were adjusted by using the program DECODON Delta2D 3.0 β. Using new algorithms for image analysis, the program removes experimentally generated distortions from 2D gels to ensure total congruency. After pseudocolor overlaying by DECODON Delta2D, in addition the histograms of the silver-stained densitogram and the autoradiogram were normalized by gray-level integration. This generates a dual channel image with an equal representation of detectable quantities of each subimage. It has to be considered, however, that the detectable quantities of proteins by silver staining do not exactly correspond to the real amount of separated proteins because of the nonlinear characteristics and saturation effects in silver-stained gels (Dutt and Lee 2001). For quantitative evaluation of the protein amount during exponential growth, the Sypro Ruby-stained 2D gels were used.
Protein Identification by Using Peptide Mass Fingerprinting
For MALDI TOF mass spectrometry, spots from the preparative 2D gels were cut out manually and digested using a peptide-collecting device (Otto et al. 1996). Peptide solution (0.5 μL) was prepared using an identical volume of cyano-4-hydroxy-cinnamic acid in 50% acetonitril–0.1% trifluoroacetic acid on a sample plate of the Voyager DE-STR MALDI TOF mass spectrometer (Perseptive Biosystems). The obtained peptide mass fingerprints were analyzed with the Protein Prospector Software (available at http://prospector.ucsf.edu). Spots already identified from other gels were reallocated using the master gel of B. subtilis, which is available at Sub2D (available at http://microbio2.biologie.uni-greifswald.de:8080/sub2D.htm; Büttner et al. 2001).
Transcriptome Analysis by DNA Macroarray Hybridization
B. subtilis strain 168 was grown in the described minimal medium. Samples were harvested during the exponential growth phase (texp = 150 min) and during the transient phase (ttrans = 240 min). Preparation of RNA, synthesis of radioactively labeled cDNA, and hybridization of B. subtilis whole genome macroarrays (Sigma-Genosys) were performed as described by Eymann et al. (2002).
Exposed MD storage phosphor screens were scanned using a Storm 840/860 PhosphorImager (Molecular Dynamics) at a resolution of 50 μm and a color depth of 16 bits. Quantitation of the hybridization signals was carried out with the ArrayVision software (Imaging Research, Inc.) after direct import of the phosphorimager files. After background subtraction the overall-normalization function of ArrayVision was used to calculate the normalized intensity values of individual spots in order to compare results from different hybridizations and filters.
To avoid extreme intensity ratios for genes close to or below the detection limit (signal to noise S/N ratio < 1.0), the data sets were corrected as follows: The normalized intensity values (nARVOL) were scaled up to a value corresponding to an S/N ratio = 1.0.
Further analyses were carried out using the GeneSpring 4.0 software (Silicon Genetics). Genes exhibiting S/N ratios > 3ς under at least one growth condition were considered to be significantly expressed.
Availability of Data
The dual channel images from this work and the master gel of B. subtilis are available in Sub2D, the 2D-protein index of Bacillus subtilis (at http://microbio2.biologie.uni-greifswald.de:8080/sub2D.htm). An animation of the proteome changes during growth and in glucose-starved cells in stationary phase is available at http://microbio1.biologie.uni-greifswald.de/starv/movie.htm.
WEB SITE REFERENCES
http://microbio1.biologie.uni-greifswald.de/starv/movie.htm; movie of Bacillus subtilis growth/glucose-starvation response.
http://microbio2.biologie.uni-greifswald.de:8880/sub2d.htm; Bacillus subtilis master gel in Sub2D 2D protein database.
http://prospector.ucsf.edu; Protein Prospector.
Acknowledgments
We thank Karin Binder for excellent technical assistance and Christine Eymann and Michael Berlin for providing information on new analysis of RelA- and CodY-dependent genes, respectively. Furthermore we are thankful to DECODON GmbH Greifswald for making a prerelease of the Delta2D 3.0 gel analysis software available to us. Additionally, we thank Lindsay Winkler and Volker Brözel for stylistic corrections of the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (HE 1887/6-1) and the European Commission (QLG2-CT-1999-01455) to M.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL hecker@uni-greifswald.de; FAX 49 3834 864202.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.905003.
REFERENCES
- 1.Ali N.O., Bignon, J., Rapoport, G., and Debarbouille, M. 2001. Regulation of the acetoin catabolic pathway is controlled by ςL in Bacillus subtilis. J. Bacteriol. 183: 2497-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos C. and Spizizen, J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81: 741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann H., Bernhardt, J., Schmid, R., Mach, H., Völker, U., and Hecker, M. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18: 1451-1463. [DOI] [PubMed] [Google Scholar]
- 4.Antelmann H., Tjalsma, H., Voigt, B., Ohlmeier, S., Bron, S., van Dijl, J.M., and Hecker, M. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11: 1484-1502. [DOI] [PubMed] [Google Scholar]
- 5.Ballesteros M., Fredriksson, A., Henriksson, J., and Nyström, T. 2001. Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 20: 5280-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhardt J., Büttner, K., Scharf, C., and Hecker, M. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20: 2225-2240. [DOI] [PubMed] [Google Scholar]
- 7.Bloom H., Beier, H., and Gross, H.S. 1987. Improved silver staining of plant protein, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93-99. [Google Scholar]
- 8.Bradford M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72: 248-254. [DOI] [PubMed] [Google Scholar]
- 9.Büttner K., Scharf, C., Bernhardt, J., Völker, U., and Hecker, M. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22: 2908-2935. [DOI] [PubMed] [Google Scholar]
- 10.Drzewiecki K., Eymann, C., Mittenhuber, G., and Hecker, M. 1998. The yvyD gene of Bacillus subtilis is under dual control of ςB and ςH. J. Bacteriol. 180: 6674-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutt M.J. and Lee, K.H. 2001. The scaled volume as an image analysis variable for detecting changes in protein expression levels by silver stain. Electrophoresis 22: 1627-1632. [DOI] [PubMed] [Google Scholar]
- 12.Eymann C. and Hecker, M. 2001. Induction of sigB-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis. FEMS Microbiol. Lett. 199: 221-227. [DOI] [PubMed] [Google Scholar]
- 13.Eymann C., Homuth, G., Scharf, C., and Hecker, M. 2002. Bacillus subtilis functional genomics: Global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184: 2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fillinger S., Boschi-Muller, S., Azza, S., Dervyn, E., Branlant, G., and Aymerich, S. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 275: 14031-14037. [DOI] [PubMed] [Google Scholar]
- 15.Gasch A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene E.A. and Spiegelman, G.B. 1996. The Spo0A protein of Bacillus subtilis inhibits transcription of the abrB gene without preventing binding of the polymerase to the promoter. J. Biol. Chem. 271: 11455-11461. [DOI] [PubMed] [Google Scholar]
- 17.Hecker M. and Völker, U. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the ςB regulon. Mol. Microbiol. 29: 1129-1136. [DOI] [PubMed] [Google Scholar]
- 18.___, 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44: 35-91. [DOI] [PubMed] [Google Scholar]
- 19.Hecker M., Richter, A., Schroeter, A., Wölfel, L., and Mach, F. 1987. Synthesis of heat shock proteins following amino acid or oxygen limitation in Bacillus subtilis relA+ and relA strains. Z. Naturforsch. 42: 941-947. [PubMed] [Google Scholar]
- 20.Hecker M., Schumann, W., and Völker, U. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19: 417-428. [DOI] [PubMed] [Google Scholar]
- 21.Herendeen S.L., vanBogelen, R.A., and Neidhardt, F.C. 1979. Levels of major proteins of Escherichia coli during growth at different temperatures. J. Bacteriol. 139: 185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoch J.A. 1995. Control of cellular development in sporulating bacteria by the phosphorelay two-component signal transduction pathway. In Two-component signal transduction (eds. J.A. Hoch & T.J. Silhavy), pp. 129–144. ASM Press, Washington, DC.
- 23.Igo M., Lampe, M., Ray, C., Schafer, W., Moran, C.P., Jr., and Losick, R. 1987. Genetic studies of a secondary RNA polymerase ς factor in Bacillus subtilis. J. Bacteriol. 169: 3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klose J. 1975. Protein mapping by combined isoelectric focusing and electrophoresis in mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik 26: 231-243. [DOI] [PubMed] [Google Scholar]
- 25.Klose J. and Kobalz, U. 1995. Two-dimensional electrophoresis of proteins: An updated protocol and implications for a functional analysis of the genome. Electrophoresis 16: 1034-1059. [DOI] [PubMed] [Google Scholar]
- 26.Kunst F., Ogasawara, N., Moszer, I., Albertini, A.M., Alloni, G., Azevedo, V., Bertero, M.G., Bessieres, P., Bolotin, A., Borchert, S., et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390: 249-256. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig H. and Stülke, J. 2001. The Bacillus subtilis catabolite control protein CcpA exerts all its regulatory functions by DNA-binding. FEMS Microbiol. Lett. 203: 125-129. [DOI] [PubMed] [Google Scholar]
- 28.Msadek T. 1999. When the going gets tough: Survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 7: 201-207. [DOI] [PubMed] [Google Scholar]
- 29.Nishino T., Gallant, J., Shalit, P., Palmer, L., and Wehr, T. 1979. Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J. Bacteriol. 140: 671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyström T. 2001. Not quite dead enough: On bacterial life, culturability, senescence, and death. Arch. Microbiol. 176: 159-164. [DOI] [PubMed] [Google Scholar]
- 31.O'Farrell P.H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250: 4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 32.Otto A., Thiede, B., Müller, E.C., Scheler, C., Wittmann-Liebold, B., and Jungblut, P. 1996. Identification of human myocardial proteins separated by two-dimensional electrophoresis using an effective sample preparation for mass spectrometry. Electrophoresis 17: 1643-1650. [DOI] [PubMed] [Google Scholar]
- 33.Price C.W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related Gram-positive bacteria. In Bacterial stress responses (eds. G. Storz & R. Hengge-Aronis), pp. 179–197. ASM Press, Washington, DC.
- 34.Ratnayake-Lecamwasam M., Serror, P., Wong, K.W., and Sonenshein, A.L. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes & Dev. 15: 1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid R., Bernhardt, J., Antelmann, H., Völker, A., Mach, H., Völker, U., and Hecker, M. 1997. Protein identification of vegetative proteins for a two-dimensional protein index of Bacillus subtilis. Microbiology 143: 991-998. [DOI] [PubMed] [Google Scholar]
- 36.Sonenshein A.L. 2000. Bacterial sporulation: A response to environmental signals. In Bacterial stress responses (eds. G. Storz & R. Hengge-Aronis), pp. 199–221. ASM Press, Washington, DC.
- 37.Stülke J. and Hillen, W. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54: 849-880. [DOI] [PubMed] [Google Scholar]
- 38.Stülke J., Hanschke, R., and Hecker, M. 1993. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139: 2041-2045. [DOI] [PubMed] [Google Scholar]
- 39.Tobisch S., Zühlke, D., Bernhardt, J., Stülke, J., and Hecker, M. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181: 6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.vanBogelen R.A. and Neidhardt, F.C. 1990. Global systems approach to bacterial physiology: Protein responders to stress and starvation. FEMS Microbiol. Ecol. 74: 121-127. [Google Scholar]
- 41.Wendrich T.M. and Marahiel, M.A. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26: 65-79. [DOI] [PubMed] [Google Scholar]
- 42.Werner H. and Bernhardt, J. 1998. WWW-basierte Proteindatenbank des Bacillus subtilis. In Datenbanken in Rechnernetzen (ed. A. Fröhlich), pp. 253–282. GingkoPark Mediengesellschaft, Berlin, Germany.
- 43.Wilkins M.R., Sanchez, J.C., Gooley, A.A., Appel, R.D., Humphery-Smith, I., Hochstrasser, D.F., and Williams, K.L. 1996. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 13: 19-50. [DOI] [PubMed] [Google Scholar]