Abstract
Genomic and full-length cDNA sequences provide opportunities for understanding human gene structure and transcriptional regulatory elements. The simplest regulatory elements to identify are promoters, as their positions are dictated by the location of transcription start sites. We aligned full-length cDNA clones from the Mammalian Gene Collection to the human genome rough draft sequence to estimate the start sites of more than 10,000 human transcripts. We selected genomic sequence just upstream from the 5′ end of these cDNA sequences and designated these as putative promoters. We assayed the functions of 152 of these DNA fragments, chosen at random from the entire set, in a luciferase-based transfection assay in four human cultured cell types. Ninety-one percent of these DNA fragments showed significant transcriptional activity in at least one of the cell lines, whereas 89% showed activity in at least two of the lines. We analyzed the distributions of strengths of these promoter fragments in the different cell types and identified likely alternative promoters in a large fraction of the genes. These data indicate that this approach is an effective method for predicting human promoters and provide the first set of functional data collected in parallel for a large set of human promoters.
[Supplemental material is available online at www.genome.org and http://www-shgc.stanford.edu/myerslab/.]
Gene expression in eukaryotes is a highly coordinated process involving regulation at many different levels. The regulation of transcription initiation is an important, and often the rate-limiting step in this process. Although several types of cis-acting DNA sequence elements contribute to this regulation, the simplest elements to locate may be promoters, as they are located just upstream from transcription start sites. Until recently, most functional studies of promoters were conducted on a gene-by-gene basis, yielding such data sets as the Eukaryotic Promoter Database (http://www.epd.isb-sib.ch/), which contains ∼300 human promoters. There have also been recent attempts to identify promoters on a large-scale with strictly computational methods (Davuluri et al. 2001; Ohler and Niemann 2001; Down and Hubbard 2002).
Due to the availability of the draft sequence of the human genome and full-length cDNA libraries, an alternative strategy to identify promoters would be to align full-length cDNA sequences to the human genome sequence, predict transcription start sites, and identify the sequences immediately upstream from these start sites. In one such application of this approach, Suzuki and colleagues used an oligo-capping approach to enrich for full-length cDNAs (Suzuki and Sugano 2001). They mapped these cDNAs onto the genomic sequence to predict transcription start sites (TSS) and used these TSSs to infer the location of potential promoter regions of 1031 human genes (Suzuki et. al. 2001). They compiled this information in the DataBase of Human Transcriptional Start Sites (DBTSS; http://elmo.ims.u-tokyo.ac.jp/dbtss/home.html), which currently contains start sites for 8996 genes (Suzuki et al. 2002).
In this study, we used a similar approach to identify putative human promoters, but also combined it with an experimental reporter gene transfection assay to test whether the predicted sequences contain promoter activity. We chose to use the Mammalian Gene Collection (MGC; Strausberg et al. 1999) data set, which is another resource for putative full-length human cDNA clones. Our data indicate that a very high fraction of the full-length cDNAs in the MGC data set are directly adjacent to or very near transcription promoters and that this assay system, combined with simple computational analysis of genomic and cDNA sequence information, provides a useful tool for analyzing and annotating the human genome.
RESULTS AND DISCUSSION
The success of an approach that identifies putative transcriptional promoters on the basis of their immediate proximity to the 5′ ends of cDNA clones clearly depends on high-quality cDNA sources that contain full or almost full-length transcripts. Because we wanted to use the new data set of full-length cDNAs generated by the Mammalian Gene Collection (http://mgc.nci.nih.gov/index_html), we analyzed the MGC sequences and compared them with the DBTSS data set. First, we compared the lengths of every MGC clone with the corresponding clone of the same gene from the DBTSS data set to see how much overlap and similarity in length there were between them. We found that only 37.5% of the MGC clones have a corresponding DBTSS clone. For the MGC clones that have a corresponding DBTSS clone, 52% have a large transcription start site discrepancy (>500 bp), and the MGC clone is longer on average by ∼1000 bp in this category. However, in the remaining group, in which the clones have no discrepancy or a small TSS discrepancy (<500 bp), the DBTSS clone is longer on average by only 18.8 bp (see Supplementary Material). This analysis indicates that, for the 37.5% of cDNA clones present in both data sets, DBTSS clones are nominally longer when the discrepancy in TSS is small, but MGC clones are significantly longer when the discrepancy in TSS is large. Furthermore, the large fraction of genes represented by MGC clones that are not present in the DBTSS indicate that the MGC is a rich source of full-length sequences for new genes. This analysis suggests that these two sources are equally effective for predicting TSSs.
To identify putative human transcriptional promoters, we aligned the sequences of the 10,276 full-length MGC cDNA clones to the December, 2001 human draft sequence by using the BLAT algorithm (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start). We then selected 600 bp of genomic sequence located −550 to +50 relative to the 5′ end of each cDNA clone. If a cDNA is truly full length, we expect this sequence to include the basal promoter and nearby upstream regulatory elements. (These sequences and other supplementary material are available at http://www-shgc.stanford.edu/myerslab/ and www.genome.org.)
To determine whether these DNA segments contain transcriptional promoters, we selected 152 of these fragments at random from the list of 10,276 and tested them for their ability to drive transcription of a plasmid-based luciferase reporter gene in cultured cell transfection assays. We amplified each promoter with PCR from the genome and directionally cloned the amplified fragments into a plasmid vector upstream from the luciferase gene. We transfected each promoter/luciferase construct individually, along with a control plasmid expressing the renilla gene, into four human cell lines, an embryonic kidney cell line (293), a cervical carcinoma line (HeLa), a hepatocyte line (HuH7), and a fibrosarcoma line (HT1080). We determined the strengths of these putative promoters by measuring the luciferase to renilla ratio, allowing us to control for transfection efficiency. Analysis of duplicate experiments indicates that these data are highly reproducible (see Methods). We normalized the values within each cell type by the mean of the negative controls, so that the measured strength of each promoter indicates its fold increase over negative control activity. We defined a fragment as a promoter when its strength value was greater than three standard deviation units above the mean of the negative controls for each cell type independently.
With this conservative approach, we found that 91% of the 152 DNA fragments drove the expression of the luciferase reporter gene in at least one of the four cell types, and 89% drove luciferase expression in at least two of the four cell types (Table 1). A total of 14 of the 152 upstream DNA segments that we tested did not produce luciferase signals above the threshold set by our negative controls in any of the four cultured cell lines. These results could be due to a variety of reasons, including the possibility that some MGC cDNA sequences are not full length (and therefore the adjacent segments we chose from the genomic sequence are within the transcribed parts of the genes). Negative results could also come from true promoters if they are very weak, or promoters that are inactive in the cell types we assayed. These results suggest strongly that the vast majority of the sequenced full-length cDNA clones in the MGC set have a 5′ end very near or at a natural start site of transcription for their corresponding gene, and that the sequences just upstream from these 5′ ends have promoter activity.
Table 1.
Percentage of Active Promoters
| Percentage | |
| Positive in ≥ 1 out of 4 cell types | 91 |
| Positive in ≥ 2 out of 4 cell types | 89 |
| Positive in ≥ 3 out of 4 cell types | 82 |
| Positive in ≥ 4 out of 4 cell types | 76 |
Percentage of putative promoters giving positive luciferase signals in cultured cell transfection assays.
Because there are differences in TSS predictions between the MGC data set and the data sets in RefSeq and DBTSS, we stratified the 152 experimental promoters on the basis of the TSS discrepancies (Table 2). In cases in which the MGC clone is the same length as or longer than sequences in RefSeq or DBTSS, we designate the TSS difference as zero. Of the 152 MGC clones, 94 were shorter than the corresponding cDNA sequences in RefSeq or DBTSS. Of those 94, 81 had predicted TSSs in the 5′ UTR of the longest transcript, and only 2 of these 81 failed to show promoter activity in our assay. The remaining 13 of the 94 clones corresponded to a putative TSS that interrupts the ORF predicted by the longest transcript. Surprisingly, 6 of these 13 had significant promoter activity in our assay. If these are true biological promoters, they would produce a truncated or entirely different protein than that encoded by the longest transcript.
Table 2.
Ranges of TSS Discrepancies and Promoter Activities of the 152 Experimental DNA Fragments
| TSS difference (bp) | Experimental promoters | Avg. strength of positives | ||||
| number | percent | negative | positive | % positive | ||
| 0 | 58 | 38.2 | 5 | 53 | 91.4 | 305 |
| 1–100 | 47 | 30.9 | 1 | 46 | 97.6 | 386 |
| 100–300 | 12 | 7.9 | 1 | 11 | 91.7 | 297 |
| 300–500 | 7 | 4.6 | 0 | 7 | 100.0 | 174 |
| 500–1000 | 7 | 4.6 | 0 | 7 | 100.0 | 131 |
| >1000 | 21 | 13.8 | 7 | 14 | 66.7 | 30 |
| Total | 152 | 100.0 | 14 | 138 | 90.8 | 289 |
TSS difference is the discrepancy in length (base pairs) between the putative TSS predicted by MGC clones and the TSS predicted by the longest transcript available from RefSeq or DBTSS. When the TSS difference is 0, the MGC clone is the longest available. Due to the size of the fragments we are testing (∼500 bp), those TSS differences that are less than 500 bp contain sequences just upstream of the TSS of the longest transcript described above.
Eighty-two percent of our 152 experimental promoter fragments are based on TSSs that lie within 500 bp of the TSS predicted by the longest cDNA available from all sources. Therefore, these fragments are likely to contain some or all of the basal promoter elements just upstream of the longest transcript. However, those sequences with TSS differences of 500 bp or more are important to study, as 20/28 of these (14% of all the positives) have promoter activity in our assay. These fragments are likely to function as alternative promoters (Ayoubi and Van de Ven 1996). For some of the cDNA clones in this group, including dystrobrevin α (DTNA) and ribosomal proteins S10 (RPS10) and L3 (RPL3), shorter alternative transcripts corresponding to the TSS we used have been identified experimentally in reports by other groups or are supported by the presence of many independently obtained transcripts in GenBank (Sadoulet-Puccio et al. 1997; Kenmochi et al. 1998; Holzfeind et al. 1999). Interestingly, these alternative promoters are weaker, on average, in our assay system.
Those fragments with less than a 500-bp TSS discrepancy may also correspond to alternative promoters that regulate different transcription start sites. For example, the human C4b-binding protein β-chain gene (C4BPB) has two distinct mRNA species with TSSs that differ by only 376 bp (Hillarp et al. 1993), one of which corresponds to the TSS used in this study. In another example, the same shorter transcripts that we used for ribosomal protein large P0 (RPLP0) are also annotated in DBTSS and RefSeq. Thus, although it might seem prudent to disregard sequences from full-length cDNA libraries that have longer clones in GenBank, our results indicate that doing this would incorrectly remove a substantial number of alternative promoters. These findings suggest that a significant number of human genes use one or more alternative promoters. The use of alternative splicing, along with alternative promoters, provides two mechanisms for greatly increasing the diversity of transcripts.
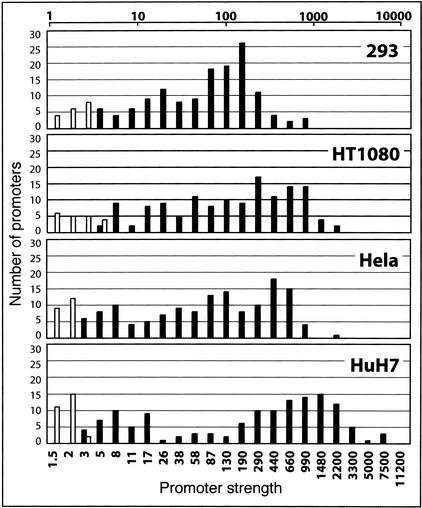
We examined the strengths of all 138 positive promoters in each of the 4 cell types. Not surprisingly, these results indicate that there is a wide range of apparent strengths among these promoters. Furthermore, the distributions of the promoter strengths appear to be non-normal for each cell type, and each cell type has a different distribution (Fig. 1). These results suggest that there are multiple classes of promoters within this set. The multi-modality of promoter strengths seen within a single cell type may be biologically relevant, but we cannot exclude the possibility that this is an artifact of our functional assay. The different classes may represent promoters for which we captured only the basal promoter versus groups for which we isolated the basal promoter and some other upstream regulatory elements. However, we observed no obvious correlation between promoter strengths in our functional assay and the discrepancies in TSS predictions.
Figure 1.
Distribution of promoter strengths. The distribution of the promoter strengths for the 138 positive clones of the 152 we tested is shown at top on a log scale in the 4 cell types tested. The number of promoters that fall within each bin is shown on the Y axis, and bin boundaries are denoted on the X-axis. We calculated promoter strength as a fold increase of luciferase activity over the negative controls in a given cell type. The black bars indicate promoters that fall above our threshold value for a functional promoter, and the white bars indicate those below that threshold (see Methods). Bins that contain both positive and negative promoters have boundaries that span the threshold value.
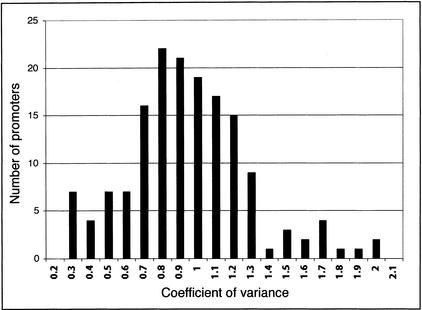
The differences we observe in each individual promoter's strength in the four cell types are less likely to be an experimental artifact, considering that we used the same reporter construct in each cell line and used an internal transfection control. To measure the variability of the activity of a promoter independent of its strength, we calculated the coefficient of variance (CV, the standard deviation divided by the mean) for each promoter in each of the four cell lines. The distribution of the CVs for the 138 fragments that showed promoter activity demonstrated a possible minor mode at the tail containing the highest coefficient of variance (Fig. 2). Of the genes with promoters with the lowest CV, several are known to exhibit widespread expression in many tissue types. Conversely, of those promoters with the highest CV, several are known to be more restricted in their distribution of expression. For example, the ferritin (FTH1) transcript has been detected in 211 distinct cell types (Unigene; http://www.ncbi.nlm.nih.gov/UniGene/), and its promoter had a CV of 0.29 in our analysis. Additionally, mannosidase (MAN2B1) expression has been measured in 84 distinct cell types, and its promoter had a CV of 0.24 (Unigene). In contrast, the kallikrein 5 (KLK5) transcript, which had a high CV (1.65), is expressed primarily in breast, brain, and testes (Yousef and Diamandis 1999). Likewise, the synovial sarcoma (SSX4) transcript has been detected only in the bone, foreskin, and a carcinoma cell line, and its CV was 1.59 (Unigene). Therefore, although this reporter gene transfection system is artificial, it appears not only to verify a large fraction of human promoters, but also to maintain some aspects of cell type-specific regulation.
Figure 2.
Distribution of the variance of promoter strength. We calculated the coefficient of variance for each individual promoter in the four cell types to estimate the variance of promoter strength. By measuring the standard deviation relative to the mean, we compared the variation between both strong and weak promoters. The promoters in the left tail (low variance) and right tail (high variance) of the distribution may be more likely to have constitutive and cell type-specific activities, respectively.
Because this sampling indicates that at least 90% of the DNA fragments are likely to be functional promoters, we analyzed the 600-bp sequence of all 10,276 putative promoters that we derived from the MGC dataset. The overall GC content of this large set of DNA fragments is 57%. We found that 27% of these fragments contain a strict TATA-box sequence (TATA[T/A][T/A]) and 65% include a less-strict TATA-box sequence (TA[T/A][T/A] [T/A][T/A]) (Table 3). Although the TATA-box is often located 25 to 30 bases upstream of the transcription start site, we chose to look for the element in the entire 600 bp of each fragment because of the uncertainty of the exact location of the transcription start site. The percentage of sequences with a TATA-box in our dataset is smaller than the percentage of human promoters containing a TATA-box in the Eukaryotic Promoter Database (51% strict and 76% less strict), indicating that TATA elements may not be as prevalent in human promoters as suggested by the EPD. Interestingly, when we stratified the promoters into three groups based on GC content, there were fewer strict TATA elements than expected by chance in the 25%–45% GC group, and there were more TATA elements than expected by chance in the groups that are 45%–65% GC and 65%–85% GC (Table 3). We obtained similar results when we analyzed the 152 experimental fragments, suggesting that this smaller set is representative of the entire set. The slight increase of promoters in the range of 45%–65% GC in our experimental set is likely due to the bias of PCR primer design and amplification efficiency, which is generally more successful on sequences that are ∼50% GC (Table 3). We observe no correlation between GC content and measured promoter strength, or between the presence of the TATA element and promoter strength.
Table 3.
Sequence Analysis of Putative Promoters
| All | Total | % Total | Strict TATA | Less strict TATA | ||
| obs | exp | obs | exp | |||
| 25%–45% GC | 1462 | 14.2 | 1023 | 1218.80 | 1450 | 1460.90 |
| 45%–65% GC | 6318 | 61.5 | 1577 | 1442.20 | 4535 | 4078.50 |
| 65%–85% GC | 2496 | 24.3 | 219 | 63.9 | 640 | 246 |
| 10276 | 100 | 2819 | 2725 | 6625 | 5785 | |
| Experimental | Total | % Total | Strict TATA | Less strict TATA | ||
| obs | exp | obs | exp | |||
| 25%–45% GC | 13 | 8.6 | 9 | 10.7 | 12 | 13 |
| 45%–65% GC | 119 | 78.3 | 26 | 29.5 | 75 | 81 |
| 65%–85% GC | 20 | 13.2 | 2 | 0.75 | 5 | 2.9 |
| 152 | 100 | 37 | 40.95 | 92 | 96.9 | |
We grouped the putative promoters in the entire dataset of 10,276 and, separately, the 152 experimental promoters into three classes: 25%–45%, 45%–65%, and 65%–85% GC. The number of promoters in each class are shown. Based on the nucleotide frequency within each group, we also calculated the expected number of promoter fragments in which a strict TATA-box (TATA[T/A][T/A]) or a less-strict TATA-box (TA[T/A][T/A][T/A][T/A]) would appear at least once by chance. Then we calculated the number of promoter fragments in both our experimental and total dataset in which these elements appeared at least once. The shaded boxes indicate those cases in which the observed and expected frequencies of TATA elements are significantly different from one another (P < 0.05).
The work described here provides a large collection of DNA fragments that contain a significant fraction of the transcriptional promoters of human genes. These data are a starting point for studying transcription initiation of human genes on a global scale and provide information that will be helpful in annotating the functional elements of the human genome.
METHODS
Promoter Prediction and Negative Controls
We aligned full-length cDNA clones from the Mammalian Gene Collection (MGC) to the human rough draft sequence by using the BLAT algorithm (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start). We collected the predicted promoter sequence −550 to +50 from the 5′ end of the cDNA. We also collected four sequences from the last exons of four random brain-specific genes, and seven random nonrepetitive intergenic sequences to serve as negative controls. The complete predicted promoter dataset, the sequences of promoters that we tested experimentally, and the sequences of negative controls are available as supplementary material at http://www.shgc.stanford.edu/myerslab/ and http://www.genome.org.
PCR Amplification and Digestion
We designed primers to amplify a 500-bp product from a set of 152 of the DNA fragments that we selected randomly from the set of 10,276 putative promoters. BglII and MluI restriction sites were included at the 5′ end of the forward and reverse primers, respectively, to maintain the promoter orientation while cloning into the luciferase test vector. We cleaned up the PCR reactions by using the Qiaquick 96-well cleanup kit (QIAGEN), and digested the products to generate sticky ends for cloning.
Cloning and Vector Preparation
We ligated each digested product upstream to the luciferase gene in the pGL3-basic vector (Promega) and transformed each ligation into Top10 chemically competent bacteria (Invitrogen). To make this as high-throughput as possible, we performed the PCR reactions, digestions, ligations, and transformations in 96-well format. After plating each transformation reaction, we picked colonies, tested for the proper insert, and then grew each clone as an overnight culture. We then made minipreps (QIAGEN) of each culture with an individual column, measured the concentration of DNA in each sample by UV absorption, and diluted each to a concentration of 100 ng/microliter.
Transfection and Luciferase Assay
To control for transfection efficiency, we cotransfected 100 ng of each experimental luciferase plasmid with 8 ng of the renilla-containing pRL-TK control plasmid (Promega) into HeLa, 293, HuH7, and HT1080 human cells (ATCC) by using the FuGene6 Lipofectamine Reagent (Roche). HeLa and HuH7 cells were at 80% confluence, 293 and HT1080 were at 50% confluence at the time of transfection. After 24 h, we prepared lysates from each transfection, and assayed luciferase and renilla activity in a 96-well plate luminometer (Wallace) according to the protocol in the Dual Luciferase Kit (Promega). Cells were grown in 96-well plates and the transfections and luciferase assays were done in 96-well format.
Data Analysis
Luciferase to renilla ratios are available on the Supplementary Information page of Genome Research online. We determined the promoter strength of each DNA fragment by calculating the ratio of luciferase signal to renilla signal from each transfection to control for well-to-well variation in transfection efficiency. We then divided each promoter strength value by the mean of the negative controls in a given cell type so that a normalized promoter strength is a measure of its fold increase in activity over background. Finally, for each cell type, we calculated the standard deviation of the negative control values, and set our threshold for a positive signal as three standard deviation units above the mean of the negatives within each cell type. Therefore, we can be 99.7% confident that any values beyond this threshold are positive promoter signals in that cell type.
Twelve representative fragments were tested in duplicate in each of four cell lines to determine the degree of experimental variation in the data. The average coefficient of variance was 0.105, indicating that our results are highly reproducible. This suggests that most of the differences observed in promoter strengths between cell lines are not due to experimental variation.
Sequence Analysis
We calculated the GC content of each promoter, and then grouped them into three classes: 25%–45%, 45%–65%, and 65%–85% GC. We then determined the nucleotide frequency within each group and calculated the probability of finding at least one randomly occurring TATA element per promoter. Next, we searched our experimental and total datasets to find the number of promoter fragments with at least one of the TATA elements. We then used the χ2 test to determine whether the observed frequencies differed from the expected at a significance cutoff of P <0.05.
WEB SITE REFERENCES
http://elmo.ims.u-tokyo.ac.jp/dbtss/home.html; DBTSS.
http://genome.ucsc.edu/cgi-bin/hgBlat?command=start; UCSC Genome Bioinformatics Site, BLAT.
http://mgc.nci.nih.gov/index_html; Mammalian Gene Collection.
http://www.epd.isb-sib.ch/; Eukaryotic Promoter Database.
http://www.ncbi.nlm.nih.gov/UniGene/; Unigene.
http://www-shgc.stanford.edu/myerslab/; Supplementary Material.
Acknowledgments
We thank members of the Myers Laboratory for discussions and support, and Jeremy Schmutz at the Stanford Human Genome Center for helpful advice. This work was supported by the Stanford Genome Training Program (Training Grant NIH 5 T32 HG00044 to N.D.T.), a Geraldine Jackson Fuhrman Stanford Graduate Fellowship (to S.F.A.), and a National Defense Science and Engineering Graduate Fellowship (to A.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL myers@shgc.stanford.edu; FAX (650) 725-9689.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.794803.
REFERENCES
- 1.Ayoubi T.A.Y. and Van de Ven, W.J.M. 1996. Regulaton of gene expression by alternative promoters. FASEB J. 10: 453-460. [PubMed] [Google Scholar]
- 2.Davuluri R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29: 412-417. [DOI] [PubMed] [Google Scholar]
- 3.Down T.A. and Hubbard, T.J. 2002. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 12: 458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hillarp A., Pardo-Manuel, F., Ruiz, R.R., Rodriguez de Cordoba, S., and Dahlback, B. 1993. The human C4b-binding protein β-chain gene. J. Biol. Chem. 268: 15017-15023. [PubMed] [Google Scholar]
- 5.Holzfeind P.J., Ambrose, H.J., Newey, S.E., Nawrotzki, R.A., Blake, D.J., and Davies, K.E. 1999. Tissue-selective expression of α-dystrobrevin is determined by multiple promoters. J. Biol. Chem. 274: 6250-6258. [DOI] [PubMed] [Google Scholar]
- 6.Kenmochi N., Kawaguchi, T., Rozen, S., Davis, E., Goodman, N., Hudson, T.J., Tanaka, T., and Page, D.C. 1998. A map of 75 human ribosomal protein genes. Genome Res. 8: 509-523. [DOI] [PubMed] [Google Scholar]
- 7.Ohler U. and Niemann, H. 2001. Identification and analysis of eukaryotic promoters: Recent computational approaches. Trends Genet. 17: 56-60. [DOI] [PubMed] [Google Scholar]
- 8.Sadoulet-Puccio H.M., Feener, C.A., Schaid, D.J., Thibodeau, S.N., Michels, V.V., and Kunkel, L.M. 1997. The genomic organization of human dystrobrevin. Neurogenetics 1: 37-42. [DOI] [PubMed] [Google Scholar]
- 9.Strausberg R.L., Feingold, E.A., Klausner, R.D., and Collins, F.S. 1999. The mammalian gene collection. Science 286: 455-457. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki Y. and Sugano, S. 2001. Construction of full-length-enriched cDNA libraries. The oligo-capping method. Methods Mol. Biol. 175: 143-153. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki Y., Tsunoda, T., Sese, J., Taira, H., Mizushima-Sugano, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Nakamura, Y., et al. 2001. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11: 677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki Y., Yamashita, R., Nakai, K., and Sugano, S. 2002. DBTSS: DataBase of human transcriptional start sites and full-length cDNAs. Nucleic Acids Res. 30: 328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yousef G.M. and Diamandis, E.P. 1999. The new kallikrein-like gene, KLK-L2. Molecular characterization, mapping, tissue expression, and hormonal regulation. J. Biol. Chem. 274: 37511-37516. [DOI] [PubMed] [Google Scholar]