Abstract
Many fungi form complex three-dimensional fruiting bodies, within which the meiotic machinery for sexual spore production has been considered to be largely conserved over evolutionary time. Indeed, much of what we know about meiosis in plant and animal taxa has been deeply informed by studies of meiosis in Saccharomyces and Neurospora. Nevertheless, the genetic basis of fruiting body development and its regulation in relation to meiosis in fungi is barely known, even within the best studied multicellular fungal model Neurospora crassa. We characterized morphological development and genome-wide transcriptomics in the closely related species Neurospora crassa, Neurospora tetrasperma, and Neurospora discreta, across eight stages of sexual development. Despite diverse life histories within the genus, all three species produce vase-shaped perithecia. Transcriptome sequencing provided gene expression levels of orthologous genes among all three species. Expression of key meiosis genes and sporulation genes corresponded to known phenotypic and developmental differences among these Neurospora species during sexual development. We assembled a list of genes putatively relevant to the recent evolution of fruiting body development by sorting genes whose relative expression across developmental stages increased more in N. crassa relative to the other species. Then, in N. crassa, we characterized the phenotypes of fruiting bodies arising from crosses of homozygous knockout strains of the top genes. Eight N. crassa genes were found to be critical for the successful formation of perithecia. The absence of these genes in these crosses resulted in either no perithecium formation or in arrested development at an early stage. Our results provide insight into the genetic basis of Neurospora sexual reproduction, which is also of great importance with regard to other multicellular ascomycetes, including perithecium-forming pathogens, such as Claviceps purpurea, Ophiostoma ulmi, and Glomerella graminicola.
Introduction
Many ascomycete fungi sexually reproduce by forming three-dimensional fruiting bodies that produce their sexual spores (ascospores) in sacs called asci. The genetics of fruit-body development in the filamentous fungi has been studied extensively in several fungi including Aspergillus nidulans, Neurospora crassa, Sordaria macrospora, Fusarium graminearum, and Podospora anserina [1]–[9]. Comparative genomics analyses suggest that meiosis machinery and ascospore production are generally conserved within ascomycetes [10]–[17]. Indeed, much of what we know about meiosis in plant and animal taxa has been deeply informed by studies of meiosis in Saccharomyces and, historically, Neurospora [18], [19]. Although some genes involved in fungal fruiting body development have been identified by mutagenesis screens and characterized, the comparative genetic basis for the sexual cycle of these organisms in terms of interactions among gene networks has not been little explored [7], [20]. Expression and comparative transcriptomics studies between N. crassa and N. tetrasperma have revealed key genes involved in asexual development and mating behaviors in these fungi [21], [22]. However, inference and comparison of regulatory pathways for sexual development across these species has been difficult to achieve, partly due to the complex environmental stimuli relevant to sexual development and partly due to a lack of molecular detail regarding relevant gene interactions across sexual development. Neurospora species represent attractive models for elucidating the regulation of fruiting body development by transcriptional profiling and functional analysis due to their simple nutritional requirements, fast vegetative growth, and clearly recognizable stages during sexual development [23], [24]. The most studied species is N. crassa [25], particularly well-known for its use in the original “one gene-one enzyme” experiments by Beadle and Tatum, for its eight-spored ordered asci which enabled centromere mapping of mutants, and for its experimentally tractable and intensively-studied genetic network for circadian rhythm [26], [27].
The life histories of Neurospora spp. span the most common sexual strategies in the fungal kingdom, i.e. heterothallism (self-incompatibility with distinct mating types), pseudohomothallism (self-compatibility in which paired mating types coexist in one mycelium), and homothallism (self-compatibility regardless of mating type). Initiation of sexual reproduction is regulated by mating type genes and leads to cell fusion, nuclear pairing, nuclear fusion, meiosis, and the production of haploid ascospores. The determinant sequences for mating type, mat A and mat a, are at the same genetic locus in a given species but exhibit little to no similarity in nucleotide sequence; that is, they represent idiomorphs rather than alleles [28]. Heterothallic Neurospora species such as N. crassa and N. discreta [23] possess a bipolar mating system, and two strains with opposite mating types, mat A and mat a, must cross to initiate sexual development. Either mating type can produce female structures (protoperithecia with a trichogyne) as well as male reproductive structures (conidia) [29], [30]. Pseudohomothallic species such as N. tetrasperma are generally self-compatible, even though they also require both mating types (A and a) to reproduce. In N. tetrasperma, both mating types are generally found within a single individual, although strains containing a single mating type exist in nature and can be isolated in lab. For N. tetrasperma, mat A and mat a nuclei associated in pairs in four heterokaryotic spores, packaged within an ascus for discharge [31]. Despite mating behavior differences that distinguish N. tetrasperma from N. crassa and N. discreta, phylogenetically N. tetrasperma and N. crassa are closely related and share the most recent common ancestor with N. discreta [32].
The genetic basis of sexual development has been the subject of many investigations, with unresolved controversy arising about the ancestral state of the life style of fungal sexual reproduction, heterothallism or homothallism, and its underlying genetic mechanism(s) [33]–[40]. For example, the heterothallic life style has been suggested to be ancestral within Neurospora [31], because the pseudohomothallic N. tetrasperma does not require a mating partner, but does require both mat idiomorphs to complete the sexual cycle. However, function of mat genes in Neurospora sexual development is not well understood, except their roles in heterothallic species such as N. discreta and N. crassa as regulators of pheromone expression to direct hyphal growth and fusion [4], [41]. For heterothallic species as in N. crassa, the trichogynes, which originate from protoperithecia, change their direction of growth to approach conidia of the opposite mating type and fuse with them. Plasmogamy is followed by development of the perithecium, and later ascal development, karyogamy, and then formation of ascospores [42]–[44]. Two haploid nuclei of opposite mating type fuse in a young ascus resulting in a diploid zygote nucleus that immediately undergoes meioses, followed by a postmeiotic mitosis. The mature ascus delimits eight linearly arranged ascospores, each containing a single nucleus. After mitosis the ascospores become binucleate, gradually grow to their full size, and become pigmented.
Genomes of multiple species of Neurospora have been sequenced, and comparative genomic analyses have been focused on N. crassa and its closely related species [3], [6], [45]. Here we reveal candidate genes involved in fungal development and in the evolution of perithecia by comparisons of the gene expression levels within and across three Neurospora species. We assayed for large-scale differences in morphology and the transcriptomic landscape over the time course of sexual development, identifying putative genes involved in sexual development by comparative gene expression profiling. We also tested for knockout phenotypes of selected candidate genes by assaying knockout strains across sexual development for their ability to produce wild type perithecia. Our results provide insights into the links between gene expression and sexual development of N. crassa and related species, as well as contributing to our understanding of how fungi reproduce sexually.
Materials and Methods
2.1. Strains and culture conditions
Strains of complementary mating types mat a and mat A for N. crassa (FGSC4200, FGSC2489), N. tetrasperma (FGSC2509, FGSC2508) and N. discreta (FGSC8578, FGSC8579) were obtained from the Fungal Genetics Stock Center (FGSC) [46]. The strains were grown on Carrot Agar (CA), made as previously described [47]. The CA petri dish was covered with a cellophane membrane (Fisher Scientific Company) and plugs of agar with strains were deposited on the membrane and incubated at 26°C under constant artificial light from several Ecolux bulbs (F17T8.SP41-ECO, General Electric Company), which provided a net intensity of 14 µMol/m2 S at the media surface. Conidia from the mat a strain on CA were collected and suspended in 2.5% Tween 60 (105–106 conidia/ml). Cultures of the mat A strain on CA were examined using a stereomicroscope for the formation of protoperithecia in 5–7 days, and areas with evenly distributed protoperithecia of a common size were delineated with a marker on the bottom of the plate to be harvested for stage-specific transcriptomics.
Crosses were performed by applying 2 ml of the suspension of mat a conidia in 2.5% Tween 60 (105–106 conidia/ml) to the surface of the mat A protoperithecia plates, at which point considerable disturbance to surface hyphae and other fungal tissues was unavoidable. Sexual development was monitored with a stereomicroscope until fully developed perithecia appeared [48]. Fungal material was harvested by scraping the surface with a razor blade in the areas, where protoperithecia or young perithecia similar in size were densely aggregated, right before the crossing and at 2, 24, 48 h after crossing. Sets of individual perithecia of similar morphological development were picked at 72, 96, and 144 h after crossing. For transcriptomic analysis, all tissues and perithecia were immediately and rapidly frozen in liquid nitrogen as they were sampled, then stored at −80°C.
2.2. Fixation and microscopy
Perithecium development was monitored for all three Neurospora species with a stereomicroscope over the time course of the sexual development. Cultures and crossings were performed as described in 2.1. Pieces of cellophane membrane (about 4 mm×2 mm) carrying 5–20 perithecia of similar size were cut from cultures and fixed in 1.5% formaldehyde and 0.025 M phosphate buffer for at least 48 h. The samples were embedded in resin and prepared for light microscopy as previously described [48]. Briefly, resin blocks were sectioned to a thickness of 1 to 2 µm using a glass knife and stained with 1% toluidine blue. A Leica DM LB microscope (Leica Microsystem Gmbh, Wetzlar, Germany) was used to capture images using a Zeiss AxioCam MRc color camera and AxioVision 4.8.2 (Göttingen, Germany). Image processing and annotation were performed using Adobe Photoshop CS3 (San Jose, CA).
To compare mature perithecia, scanning electron microscopy (SEM) was performed. Perithecia were collected, including the cellophane membrane they were growing on, and fixed immediately in 0.1 M sodium cacodylate buffer (pH 7.2) containing 2% glutaraldehyde over night. The samples were then washed with 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide for 1 h, washed with distilled water, and dehydrated in an increasing series of ethanol. Then, all samples were critical point dried, mounted on an SEM stub, and sputter coated with gold. The samples were examined with a Scanning Electron Microscope (ISI SS-40).
2.3. RNA extraction, cDNA preparation, and transcriptomic sequencing
RNA was isolated from homogenized mycelia with TRI REAGENT (Invitrogen) and RNAeasy Kit (Qiagen) following protocol described by Clark et al. [49] and mRNA was purified using Dynabeads oligo(dT) magnetic separation (Invitrogen). The cDNA libraries for RNA sequencing were prepared according to the Illumina mRNA Sequencing Sample Preparation Guide. In brief, triplicates were prepared for each time point and pooled. Then, mRNA was purified from total RNA and 100 ng (9 µl) were fragmented with 10X fragmentation buffer (Ambion AM8740) and incubated at 70°C for 5 min prior to adding 1 ul stop buffer (Ambion). Fragmented mRNA was precipitated using 100% ethanol with glycogen (Ambion) at −80°C. Random hexamers (N6, Invitrogen) were added to prime reverse transcription of the first strand cDNA separately for each sample, and to recover the second strand cDNA for all samples. After the ligation of standard adapters for Illumina sequencing, all samples were separated on a 2% low melting point agarose gel and processed cDNA fragments of lengths between 200 and 400 bp were selected by gel extraction and purified with Qiaquick gel extraction kit (Qiagen). The quantity of the samples was increased by a PCR using Pfx DNA polymerase (Invitrogen), and 15 cycles of PCR, each cycle comprising 98°C for 10 s, 65°C for 30 s, and 68°C for 30 s. The quantity and quality of the purified PCR products were checked at the Yale Center for Genome Analysis prior to sequencing. Single-end 35 bp reads of N6-primed preparations were separately sequenced, each on eight lanes of an Illumina Genome Analyzer (Yale Center for Genome Analysis).
2.4. Data acquisition and analysis
The libraries were run on eight lanes of an Illumina Genome Analyzer, generating an average of 28 million single-end reads of 36 nucleotides each. Since transcriptomic tags also contain sequences that span exon junctions, the program Tophat v1.1.14 [50] was used to perform spliced alignments of the tags against the N. crassa OR74A genome (NC10; [51]), and those of N. tetrasperma FGSC 2508 (v2) and N. discreta FGSC 8579 (v1) obtained from JGI Genome Portal [52]. We scored results only for tags that mapped to a single unique location in the genome (–max-multihits option was set to 1) with less than three mismatches (–splice-mismatches option was set to 2). We used the default settings for all other Tophat options. We tallied tags aligning to exons of genes with the program HTSeq v0.4.5p6 (Unpublished; http://www-huber.embl.de/users/anders/HTSeq/doc/) and the gene structure annotation file for the reference genome. LOX v1.4 [53] was applied to the tallies for each sample for each gene to estimate gene expression levels and credible intervals across developmental stages. LOX provided relative gene expression levels standardized by the lowest sample, with credible intervals, for all three species (Table S1).
Instead of using simple sequence similarity for homolog identification, we applied a phylogenetic approach that is reliable in calling homologs among closely related genomes, with a cost of power to identify potential homologs for recently evolved gene families and genes experiencing multiple duplication events in their evolutionary history. A total of 2352 orthologous genes were selected by the BranchClust method [54] for N. crassa, N. tetrasperma and N. discreta. BranchClust uses the Reciprocal Best Blast hit method and phylogenetic trees to select putatively orthologous genes using a default threshold e-value of 10−4; we only choose the complete families in those three species. Transcriptomic sequencing revealed expression levels for all orthologous gene triads across sexual development (Table S2). Data is also deposited as accession 239 at the Filamentous Fungal Gene Expression Database (FFGED; [55]) and as accession GSE41484 for N. crassa [56], GSE60256 for N. tetrasperma, and GSE60255 for N. discreta at the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo). More orthologs can be called among these fungi using BLAST approaches, and we also identified more single copy ortholog for these species using a BLAST-based method. However, from our other evolutionary study targeting several gene families with these species, we found that phylogeny-based ortholog calls by BranchClust were more robust and reliable.
A comparative heat map was constructed based on the level of expression of each gene for all three species over the entire time course of sexual development. Gene expression levels for each ortholog were normalized row-by-row by subtraction of the mean and division by the standard deviation. To cluster observed gene expression hierarchically, rows only were clustered by iteratively agglomerating the most similar two gene expression profiles and by averaging the agglomeration for the next iteration (Unweighted Pair Group Method with Arithmetic mean, UPGMA) within which similarity was assessed by the Pearson correlation coefficient of expression between genes. The Functional Catalogue (FunCat: [57]; http://mips.helmholtz-muenchen.de/proj/funcatDB/) annotation scheme was used to group genes according to their functions. The statistically significant overrepresentation of gene groups in functional categories relative to the whole genome was determined by a hypergeometric distribution P value calculation, facilitated by the MIPS FunCat online web application.
2.5. Comparative gene expression analysis of N. crassa, N. tetrasperma, and N. discreta
The LOX estimates across developmental stages for each orthologous gene for each species were assembled to compare gene expression levels across development of N. crassa, N. tetrasperma, and N. discreta. For each developmental stage, two values were calculated using the upper bound of the 95% confidence interval (CI) or the lower bound of the 95% CI from LOX. If the expression of a gene in species 1 was higher than species 2, we calculated the difference between the lower bound of expression in species 1 and the upper bound of expression in species 2. Conversely, if the expression of a gene in species 2 was higher than species 1, we calculated the difference between the lower bound of species 2 and the upper bound of species 1. These differences constitute those of which we can be highly confident. N. crassa was compared with N. tetrasperma, N. crassa with N. discreta, and lastly N. tetrasperma with N. discreta. The calculation described above was performed for all time points (2, 24, 48, 72, 96 and 144 h) for each gene. Finally, for each comparison, all genes were prioritized according to magnitude of difference in a descending order and the first 130 genes with the biggest changes across all time points and common to all comparisons were subsequently selected for knockout and phenotyping. The data for the comparative gene expression analysis can be found in the Tables S3–S5.
2.6. Assessing phenotypes of knock out mutants
Knockout strains for the top 130 candidate genes were compared to wild-type (WT) strains and screened for defects in fruiting body formation. Knockouts of candidate genes were obtained for both mating types in Neurospora crassa from the Fungal Genetics Stock Center (FGSC). These knockouts had been preliminarily assayed for phenotypes by several high-throughput screens as part of a Neurospora knockout project [58], [59], and in that project had not exhibited a mutant phenotype in asexual growth or development. We performed a detailed, controlled screen crossing the two mating types of each deletion strain, cultured on CA in triplicate. The mat A cultures on CA were fertilized with conidia from the mat a strain. Perithecium formation was monitored with stereo- and light microscopy (Nikon Diaphot 300) for the presence and for the orientation of a single beak, black coloration and the typical vase-shape. Mature perithecia were examined in squash mounts for the presence of asci with normal pores, ascospores with normal shapes and numbers, and normal paraphyses. Normal spore development and ascus firing were examined by checking the lid of the Petri dish for black ascospores.
2.7 Cosegregation experiments of knockout mutants showing a phenotype different from the wild type
Knockouts of candidate genes obtained from FGSC were produced using a high-throughput gene deletion strategy in N. crassa strains with deletion mutations of mus-51 and/or mus-52, mutations required for nonhomologous end-joining DNA repair. [58], [59]. A high rate of spontaneous mutations has been observed in in Δmus-51 and/or Δmus-52 strains, and cosegregation experiments were used to demonstrate that the intended knockout deletion was responsible for the mutant phenotype [60]. A hygromycin resistance cassette at the location of the deletion mutation provides a selectable marker. The KO strains showing phenotypes in sexual development were crossed with wild-type strains (FGSC2489 mat A or FGSC4200 mat a). Individual ascospore progeny were isolated for resistance to hygromycin. Their phenotypes were then examined on SCM medium. Cosegregation of hygromycin resistance and the observed phenotype is necessary evidence that the observed phenotype was result of the deletion of the specified gene.
Results
3.1 Perithecium development in N. crassa, N. tetrasperma, and N. discreta on CA follows a common time course
To monitor the sexual development of all three Neurospora species, we crossed wild-type strains of both mating partners of each Neurospora species. The observed perithecial development of N. crassa, N. tetrasperma, and N. discreta aligned with the common time course known for these species [56]. We observed that during perithecial development, grayish to yellowish gray protoperithecia darkened, then blackened once mature perithecia had formed, indicating the biosynthesis of melanin in the time course of perithecial development (Fig. 1). As expected, we also identified the fading of the orange pigmentation in the colony across the time course of sexual development, indicating a reduction in the asexual phase of the life cycle. Additionally, we found no obvious tissue differentiation between protoperithecia and perithecia within 24 h after crossing, except for a slight increase in size and a slight darkening in colour, indicating the biosynthesis of melanin as part of the successful fruiting body formation. Furthermore, the centrum parenchyma of thin-walled cells expanded with increasing perithecial size and differentiated filamentous structures and croziers formed after 48 h to 72 h. Asci containing developing ascospores were visible after 96 h, along with some narrow paraphyses. From 120 h to 144 h post crossing, a beak formed at the apex of the perithecium, and scanning electron microscopy of all three species revealed fully developed fruiting bodies with a beak on top of the perithecium (Fig. 2, panels A–C). Furthermore, squash mounts of perithecia 144 h after crossing revealed that the inside of the perithecium contained mature asci with ascospores (Fig. 2, panels D–F). A major phenotypic difference among species was the number of spores produced, which, as expected in N. tetrasperma was four, in contrast to eight in N. crassa and N. discreta. Additionally, N. discreta produced abundant conidia, which were visible associated with the perithecia after the sexual cycle has concluded (Fig. 2, panels C and F).
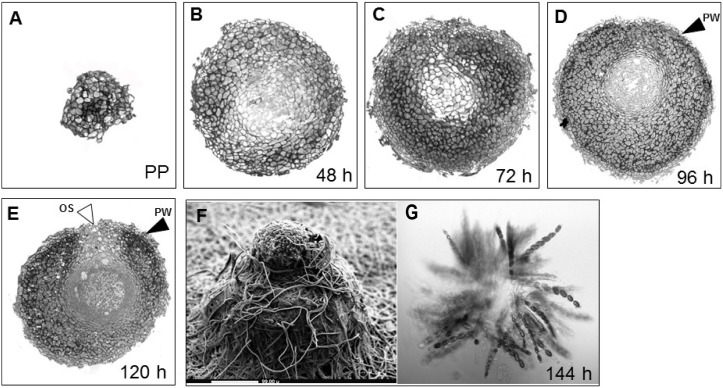
Figure 1. Sexual development of N. crassa.
Cross sections of developing perithecia, from A) the protoperithecium (PP) through a time course of (B) 48 h, (C) 72 h, (D) 96 h, and (E) 120 h after fertilization. These images illustrate the development of several cell layers within the fruiting body such as the perithecium wall (PW, black arrowheads), composed of thick-walled cells, as well as the initial stages of formation of asci and ascospores and the ostiole (OS, white arrowheads) through which spores are released in a later stage. After (F) 144 h, the perithecium including its beak is fully developed, as shown by scanning electron microscopy (SEM). (G) A squash mount of a mature fruiting body, showing asci and ascospores.
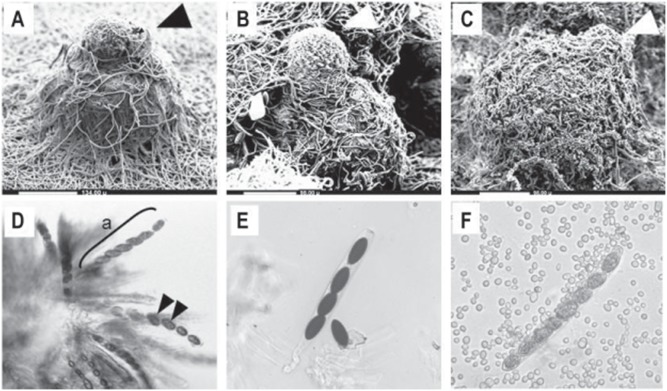
Figure 2. Key morphological characters of N. crassa, N. tetrasperma and N. discreta 144 h after crossing.
Scanning electron micrographs, in which arrowheads indicate the perithecial beaks, of A) N. crassa (bar: 134 µm), B) N. tetrasperma (bar: 98 µm) and C) N. discreta (bar: 98 µm), and light micrographs of squash mounts of (D) N. crassa (a: ascus), (E) N. tetrasperma, and (F) N. discreta (with conidia). At 144 h, some spores are not fully mature.
3.2. Transcriptional profiling of N. crassa, N. tetrasperma, and N. discreta reveals eight functional categories
To determine the gene expression of all three Neurospora species during the time course of sexual development, we performed Illumina next generation transcriptomic sequencing. We prepared samples by isolation of mRNA and performance of reverse transcription of genomic RNA with random hexamers (N6), which were typically consistent in DNA concentration. Single-end 35-base reads of N6-primed preparations were sequenced separately, each developmental stage of each species on one of eight lanes of an Illumina Genome Analyzer, at the Yale Center for Genomic Analysis. A total of 2352 single copy orthologous genes were selected by BranchClust [54] for N. crassa, N. tetrasperma and N. discreta. BranchClust uses the Reciprocal Best Blast hit method and phylogenetic trees to select putatively orthologous genes, with default threshold e-value of 10−4. Our deep transcriptomic sequencing revealed expression in all three species across sexual development for all of these identified orthologous genes. To compare gene expression levels for these genes, we constructed a comparative heatmap with the Hierarchical Clustering Explorer Software [61] based on the level of expression of each gene for all three species over the entire time course of sexual development (Fig. 3) as described in section 2.4. Transcriptional profiling of N. crassa, N. tetrasperma, and N. discreta revealed eight clusters, which we then analyzed separately for functional category representation.
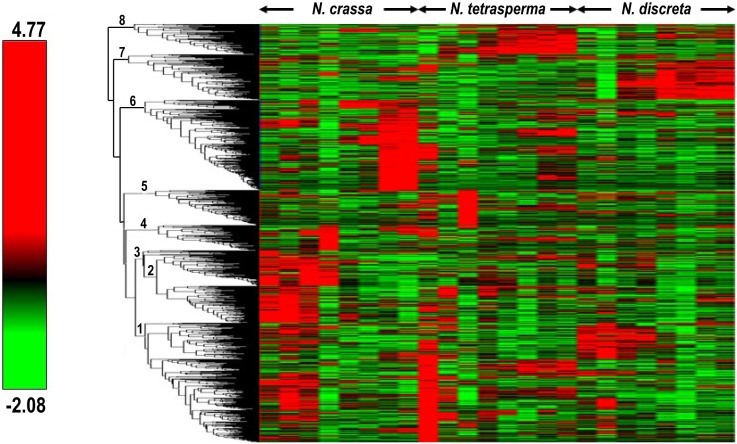
Figure 3. Comparative heat map of N. crassa (N.c), N. tetrasperma (N.t) and N. discreta (N.d) gene expression.
Relative gene expression levels of the orthologous genes in all three species inferred by BranchClust, from Before Crossing (BC) to 144 h after crossing, were normalized row-by-row by subtraction of the mean and division by the standard deviation. Rows were hierarchically clustered by average linkage (UPGMA) applied to the Pearson correlation coefficient. Clusters 1–8 were analyzed for their function with FunCat. The scale of mean-centered relative gene expression ranges from −2.08–4.79, as indicated with the scale bar.
3.2.1 Among genes highly expressed in early stages of perithecium development, the ortholog set was enriched for genes involved in metabolic processes
Where previously identified, genes were assigned cellular or molecular functions by their Functional Catalogue (FunCat) [57] annotation. The statistical significance of overrepresentation of gene groups in functional categories relative to the whole genome was determined using the hypergeometric distribution, facilitated by the Munich Information Center for Protein Sequences (MIPS) FunCat online web application. Eight major clusters were identified based on expression patterns across sexual development for the identified orthologs. In cluster 1 (Fig. 3), 61 of the 671 proteins were unclassified. Of the classified proteins we identified, most were involved in metabolism, cellular transport, cell cycle and DNA processing, protein with binding function, cellular communication and transcription.
Genes in cluster 2 were mainly involved in metabolism, cellular transport, transport facilities and transport routes as well as proteins with binding function, while only 16 of the proteins in cluster 2 were not classified. The metabolic genes were primarily involved in nucleotide/nucleoside/nucleobase metabolism, C-compound and carbohydrate metabolism, lipid, fatty acid and isoprenoid metabolism and metabolism of vitamins, cofactors, and prosthetic groups. The cellular transport-related genes were involved in transported compounds (substrates), protein transport, electron transport, vesicular transport such as the Golgi network and vacuolar/lysosomal transport. Genes encoding proteins with binding functions such as nucleic acid binding, metal binding and complex cofactor/cosubstrate binding. In cluster 3, only 19 out of 199 proteins were not classified, while the remaining proteins were mainly involved in metabolism, especially C-compound and carbohydrate metabolism. Genes in cluster 3 also tended to be involved in cellular transport, including cation and heavy metal ion transport as well as proteins with binding functions, e.g. nucleic acid binding, metal binding and RNA binding. Cluster 4 contained 139 proteins, which were involved mainly in metabolism, especially lipid, fatty acid and isoprenoid metabolism, cellular transport, and cell cycle including DNA restriction or modification. Overall, we observed an enrichment of genes involved in metabolic processes in early stages of perithecium development.
3.2.2 In later stages of the perithecium development, the ortholog set was enriched for genes involved in transcription, cell cycle, protein synthesis and cellular transport
Based on Functional Category (FunCat, [57]) annotations, cluster 5 comprises 198 proteins, of which the majority was involved in nucleotide/nucleoside/nucleobase metabolism, nucleotide/nucleoside/nucleobase metabolism and lipid, fatty acid, and isoprenoid metabolism, but also transcription such as RNA synthesis and RNA processing.
Interestingly, in clusters 6–8 there appears to be a species-specific expression in later stages of the development. Proteins of cluster 6 were mainly involved in transcriptional processes such as RNA synthesis, processing and modification, but also, cell cycle and DNA processing, as well as and metabolism. Cluster 7 contained 259 genes involved in transcription, protein binding, cell type differentiation and cellular transport. In cluster 8 only 13 genes were not identified. The cluster mainly comprised proteins involved in cell differentiation, cellular transport and transcription.
While in the early stages of the perithecium development, the fungi were observed to exhibit an enrichment of expressed genes involved in metabolic processes; in later stages of perithecium development, expressed genes from the ortholog set were enriched for genes involved in transcription, cell cycle, protein synthesis and cellular transport.
3.3 Comparative gene expression analyses revealed genes crucial for the successful development of fruiting bodies
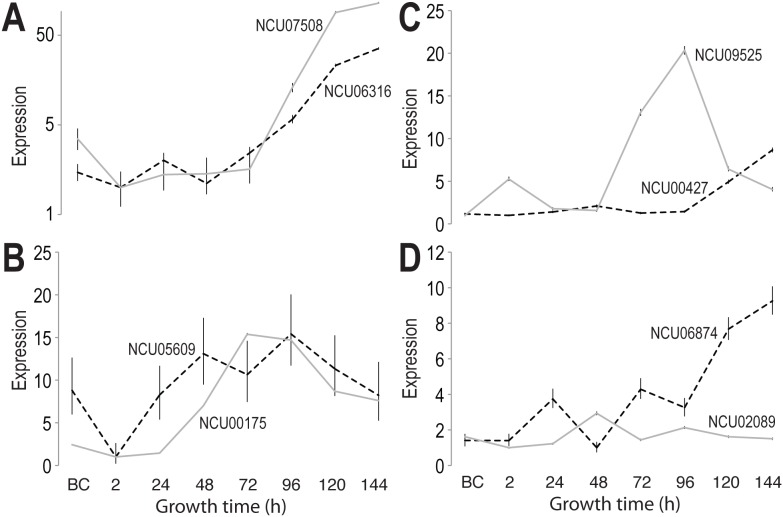
Crossing of the N. crassa WT strains resulted in the successful formation of perithecia with perithecial beak (Fig. 4A). In order to identify genes that are required for the successful development of fruiting bodies, knockouts in N. crassa from the whole genome knockout project [58], [59] of the 130 top candidate genes identified through pairwise comparison of orthologous gene expression in N. crassa, N. tetrasperma, and N. discreta were screened for mutant phenotypes in sexual development. Eight mutant phenotypes were observed that affected perithecium formation. Interestingly, development of NCU06874, encoding a HMG box (high mobility group)-containing protein was arrested after formation of protoperithecia on a fluffy white mycelium. Putative functions of the remaining genes that affect fruiting body formation in N. crassa were determined by a Blast search [62] with the NCBI Blast server of the translated sequence against all known GenBank non-redundant protein sequences (Table 1). However, the deletion of either gene NCU06316, a putative argonaute siRNA chaperone complex subunit which in fission yeast is required for histone H3 Lys9 (H3-K9) methylation, heterochromatin, assembly and siRNA generation [1], or NCU07508, a putative type-2 protein geranylgeranyltransferase subunit, caused the perithecium development to arrest at an early stage between 48 and 72 h (Fig. 4, panels B and C). In the perithecia arising from these KO strains, we observed no perithecial beak and found that the developing perithecia remained spherical, indicating an arrest in development. Furthermore, the CA of these cultures was stained black along the mating zone, suggesting an increase in melanin biosynthesis (Fig. 4D).
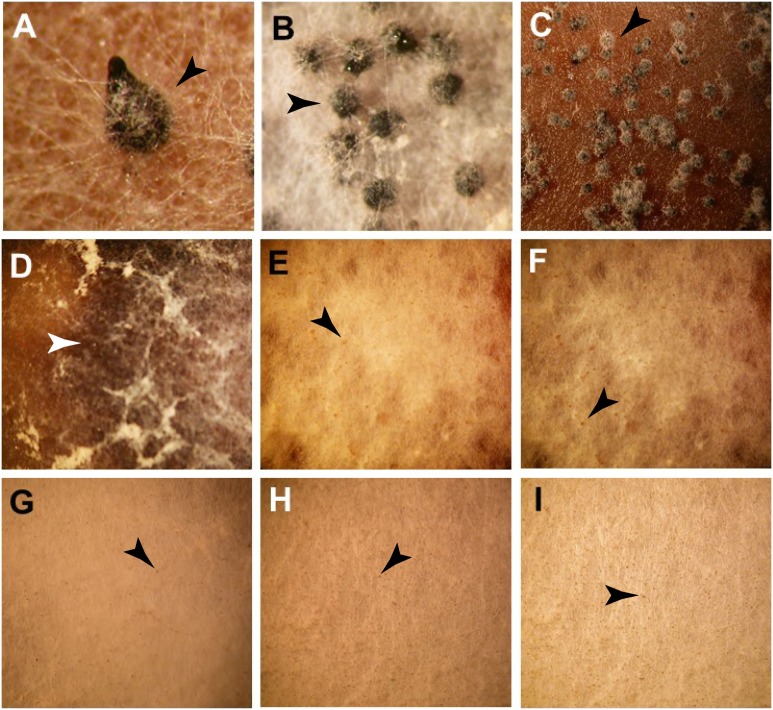
Figure 4. Phenotypes of perithecia from crosses in N. crassa.
(A) WT control, (B) ΔNCU06316: perithecium development was arrested at early stage equally to 48–72 h, (C) ΔNCU07508: perithecium development was arrested at 48–72 h, (D) ΔNCU06874: no perithecia formed, only protoperithecia, though melanin was released into the medium (white arrow heads), (E) ΔNCU05609: no perithecia formed, only protoperithecia, (F) ΔNCU00175: no perithecia formed, only protoperithecia, (G) ΔNCU00427: no perithecia formed, only protoperithecia. (H) ΔNCU02089: protoperithecia failed to develop into perithecia, and (I) ΔNCU09525: protoperithecia only. Perithecia (large, black) and protoperithecia (small, yellowish-gray to gray) are indicated with black arrow heads.
Table 1. Putative functions of genes whose deletion impacts fruiting body formation.
| ID | Annotation by homology | Max | Total | Query | E- | Max |
| score | score | cover | value | iden | ||
| NCU06316 | argonaute siRNA chaperone complex | 246 | 246 | 76% | 8e–72 | 37% |
| subunit Arb1 | ||||||
| [Colletotrichum fioriniae], | ||||||
| XP_007589964.1 | ||||||
| NCU07508 | putative type-2 protein | 145 | 145 | 68% | 1e–34 | 31% |
| geranylgeranyltransferase subunit beta | ||||||
| protein | ||||||
| [Botryotinia fuckeliana], EMR87581.1 | ||||||
| NCU06874 | HMG box-containing protein | 269 | 269 | 42% | 7e–75 | 40% |
| [Magnaporthe oryzae], XP_003718106.1 | ||||||
| NCU05609 | proline-rich protein [Coccidioides | 46.6 | 46.6 | 64% | 0.020 | 29% |
| Posadasii], ACU44647.1 | ||||||
| NCU00175 | repetitive proline-rich cell wall protein | 153 | 205 | 88% | 1e–39 | 44% |
| [Colletotrichum higginsianum], | ||||||
| CCF45612.1 | ||||||
| NCU00427 | YjeF_N domain-containing protein | 734 | 734 | 97% | 0.0 | 53% |
| [Magnaporthe oryzae], | ||||||
| ELQ39075.1 | ||||||
| NCU02089 | hypothetical protein SMAC_04986 | 1276 | 1276 | 99% | 0.0 | 88% |
| [Sordaria macrospora], | ||||||
| XP_003352871.1 | ||||||
| NCU09525 | Bacterial-type extracellular | 284 | 284 | 85% | 2e–93 | 69% |
| deoxyribonuclease [N. haematococca], | ||||||
| XP_003051650.1 |
The expression patterns of the genes whose knockouts showed an impact on fruiting body formation were monitored over the time course of sexual development starting from mature protoperithecia before crossing until 144 h, when wild-type cultures showed mature perithecia. The genes NCU06316 or 07508 exhibited a continuous increase in expression starting at 72 h (Fig. 5A). The increased expression correlated with the arrest of the fruiting body formation in an early stage between 48 and 72 h in deletion mutants.
Figure 5. Expression patterns of eight genes with impact on successful perithecium formation in N. crassa.
A. Expression of NCU06316 (solid line) and NCU07508 (dashed line), knockouts of which lead to early arrest of perithecial development at 48–72 h. Both genes exhibit a continuous increase of expression after 72 h. B. Expression of NCU00175 (solid line) and NCU05609 (dashed line), up-regulated during intermediate stages of perithecial development. C. Expression of NCU09525 (solid line) and NCU00427 (dashed line), both changing significantly in late perithecial development. D. Dynamic changes in expression of NCU02089 (solid line) and NCU06874 (dashed line) across perithecial development. Error bars indicate the inferred 95% credible interval.
Knockouts of NCU05609 (a proline-rich protein 8), 00175 (a repetitive proline-rich cell wall protein), 00427 (a YjeF_N domain-containing protein), 02089 (a hypothetical protein SMAC) and 09525 (a hypothetical protein SMAC) were arrested at the protoperithecium stage, indicating their importance in the formation of fruiting bodies. NCU09525 encodes a putative secreted protein and has been described in Nectria haematococca as bacterial-type extracellular deoxyribonuclease [63]. NCU09525 and 00175 were highly expressed between 48 and 120 h, while expression of NCU05609 peaked at 48 and 96 h after crossing. NCU06874 and 00427 showed an increased expression 96 h after crossing. NCU02089 peaked at 48 h after crossing and remained fairly constant across the remainder of sexual development (Fig. 5C–D). Although only protoperithecia were observed in mutant crosses, the culture medium for the knockout of NCU06874 blackened, indicating the secretion of melanin, and implying that melanin biosynthesis remained functional despite the fact that no perithecia were formed. Genes encoding enzymes in the melanin synthesis pathway showed a highly similar pattern across sexual development within each Neurospora species. While melanin synthesis genes were highly expressed in N. crassa protoperithecia samples, they were expressed at a lower level in N. tetrasperma and N. discreta protoperithecial samples than in late perithecial samples (Table S1). Further research is needed to understand function of melanin synthesis early sexual development in different Neurospora species. The protein encoded by NCU06874 contains a High Mobility Group (HMG) box, a structure often found in proteins involved in the regulation of DNA-dependent processes and DNA repair. Recent studies have demonstrated that deletion of the ortholog of NCU06874 in Podospora anserina and Fusarium graminearum had no effect on perithecium development [64], [65]. However, an effect on the distribution of perithecia in P. anserina was observed [64], [65].
The gene NCU02089 belongs to cluster 5. In N. crassa, this gene is expressed fairly uniformly, showing only a slight peak in expression at 48 h. The corresponding ortholog in N. tetrasperma shows a similar expression pattern. In N. discreta, in contrast, the expression of NCU02089 remains uniform until 96 h, then increases until the end of the sexual development.
The genes NCU06316, 07508, 05609, 00427, 09525, and 06874 belong to cluster 6. In N. crassa, the genes NCU06316 and 07508 are highly expressed after 72 h. In N. tetrasperma, as in N. crassa, the ortholog of NCU07508 also increases in expression after 72 h, while in N. discreta the expression peaks at 48 h, and increases again after 96 h. The expression of the ortholog NCU06316 in N. crassa differs from the one in N. tetrasperma and N. discreta. Over the time course of sexual development, expression of this gene is very low. The gene NCU05609 shows an increasing expression over the time course of the sexual development, peaking at 48 and 96 h. In contrast, the corresponding ortholog in N. tetrasperma does not show any changes in gene expression, while in N. discreta the expression decreases until 24 h, then remains steady until the end of the sexual development. In N. crassa, the expression of the gene NCU00427 remains constant until 96 h after crossing and then increases towards the end of the development. The expression of the corresponding ortholog in N. tetrasperma peaks at 24 and 120 h, while in N. discreta, this gene remains constant over the time course of fruiting body formation. The expression of the gene NCU09525 peaks between 48 and 120 h in N. crassa, while in N. tetrasperma and N. discreta, its expression peaks initially at 2 h, potentially as a consequence of mycelial disruption caused by the spreading of conidia and mating of the strains, and then increases until 96 h after crossing followed by a slight decrease towards the end of the development. The gene NCU06874 shows peaks in gene expression at 24 and 72 h, and an increase of expression from 96 h until the end of perithecium development. In contrast, in both N. tetrasperma and N. discreta, the expression of this gene remains fairly constant over the time course of the sexual development.
The gene NCU00175 belongs to cluster 8 and shows an increased gene expression between 24 and 120 h in N. crassa. A similar pattern was observed for N. tetrasperma, where the expression increased at 48 and 120 h. In N. discreta however, the gene expression increased steadily until 96 h after crossing, remained constant.
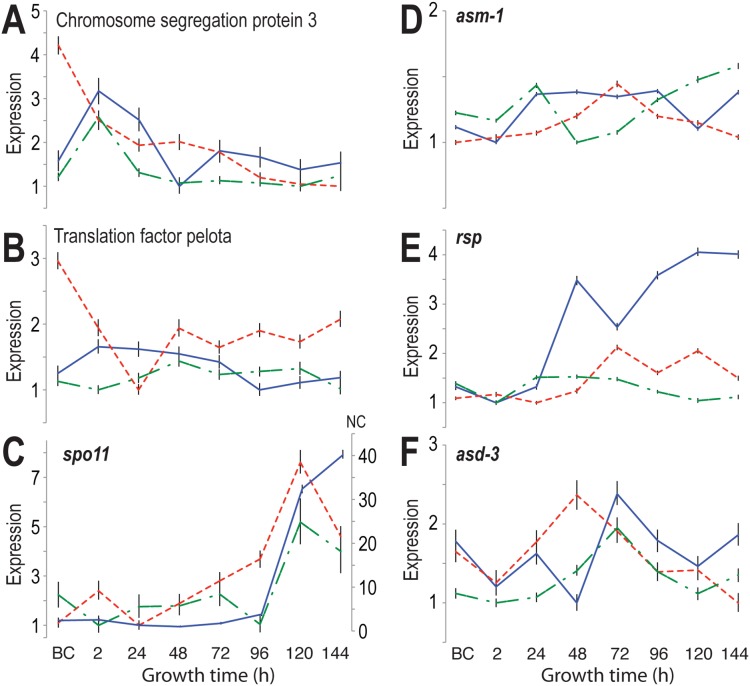
Key differences in gene expression patterns of meiosis-related genes between N. tetrasperma and N. crassa as well as N. discreta were observed (Fig. 6). Many RNA processing genes and meiosis- and mitosis- specific genes have experienced multiple duplications. Only a few single copy meiosis-specific gene homologs have been confirmed with our methods. Among these genes, we observed differences in gene expression between homothallic N. tetrasperma and heterothallic N. crassa and N. discreta. The meiotic chromosome segregation protein 3 homologs, N. tetrasperma Neute1draft_73667 (JGI identifier for the mat A genome), N. crassa NCU01858 (Broad Identifier), and N. discreta Ndisc8579_91937 (JGI identifier) showed a consistent gene expression pattern in N. crassa and N. discreta: two up-regulations at 2 h and 72 h (Fig. 6A). In contrast, the expression of the ortholog of chromosome segregation protein 3 in N. tetrasperma exhibited a steep down-regulation after strains were crossed and only a slight up-regulation 24 h after crossing. Furthermore, the translation factor pelota (Neute1draft_80736, NCU09161, and Ndisc8579_129376), which functions in meiotic cell division, exhibited a generally consistent expression across perithecial development in N. crassa and N. discreta, but a dynamic change in expression in N. tetrasperma during early perithecial development (Fig. 6B). A dramatic up-regulated expression of the meiosis-specific gene spo11 (Neute1draft_98113 m, NCU01120, Ndisc8579_51404) started early in N. tetrasperma 24 h after crossing, but began in N. crassa and N. discreta 96 h after crossing (Fig. 6C). For sporulation-related genes, including asm-1 (ascospore maturation-1), Neute1draft_115825, NCU01414, Ndisc8579_123166), Rsp (round spore, Neute1draft_126958, NCU02764, Ndisc8579_94666), asd-3 (ascus development-3, Neute1draft_103712, NCU05597, Ndisc8579_126512), we also observed up-regulation of expression at the same stages between N. crassa and N. discreta (Fig. 6D–F). Up-regulation of homologs of N. crassa asm-1 and rsp in N. tetrasperma occurred later than in N. crassa and N. discreta, but interestingly the homolog of N. crassa asd-3 exhibited an early up-regulation for N. tetrasperma at 48 h. Similar up-regulation of asd-3 for N. crassa and its homolog in N. discreta occurred at 72 h after crossing. The genes NCU01858 and 09161 belong to cluster 1, the genes NCU01120, 02764 and 05597 to cluster 6 and NCU01414 to cluster 7. Except NCU01858 and 09161, all genes shown in Fig. 6 belong to the candidate list.
Figure 6. Comparative analysis of meiosis-related genes, exhibiting differences in gene expression between N. tetrasperma (red dashed line), N. crassa (blue solid line) and N. discreta (green dash-dotted line).
A) Expression of the N. crassa gene encoding chromosome segregation protein 3 was up-regulated during early sexual development in both N. crassa and in the orthologous gene in N. discreta, but the N. tetrasperma ortholog was down-regulated across sexual development, B) Expression of the gene encoding translation factor pelota in N. crassa and expression of its ortholog in N. discreta was consistent, but the ortholog in N. tetrasperma was dramatically and dynamically differentially expressed. C) Expression of meiosis specific gene spo-11 and its orthologs was up-regulated for all three species from 96 h after crossing, but the up-regulation started early for the N. tetrasperma ortholog. D) Expression of asm-1 (ascus maturation) in N. crassa and its orthologin N. discreta exhibited the same two-peaked pattern, featuring a peak of expression during early development of the perithecium and a second peak at a later stage of perithecial development, but the ortholog in N. tetrasperma exhibited only one peak 72 h after crossing. E) Expression of N. crassa rsp (round spore) and its ortholog in N. discreta exhibited up-regulation preceding 48 h, but upregulation in N. tetrasperma preceded 72 h. F). Expression of N. crassa asd-3 (ascospore development) peaked at 72 h in N. crassa, as did its ortholog in N. discreta, but expression of the ortholog in N. tetrasperma peaked at 48 h. Error bars indicate the inferred 95% credible interval.
3.4 Phenotypes of seven knockout mutants cosegregate with hygromycin resistance
To validate the linkage between the insertional mutation and the phenotype, we followed a strategy developed previously for N. crassa KO strains [56], and backcrossed progeny (ascospores) were selected and germinated. Twelve to twenty single ascospore progeny displayed hygromycin resistance, and complete cosegregation of hygromycin resistance and identified phenotypes was observed for seven out of eight investigated genes (Table S6). The exception was the knockout of NCU09525, for which only a KO strain of mat A was available. No perithecia were produced in this mutant-wild type cross, preventing us from assessing segregation of the cross.
Discussion
Here, we compared the sexual development of three closely related Neurospora species, N. crassa, N. tetrasperma, and N. discreta, focusing on characterizing expression patterns for genes involved in sexual development and identifying new genes or new functions of annotated genes in regulating sexual reproduction, taking advantages of the distinct differences among these otherwise highly similar and closely related species. The comparison of gene expression levels across these species during fruiting body formation revealed eight genes that were shown to be crucial for the successful development of perithecia in N. crassa: their knockouts were unable to produce mature perithecia, and seven of the eight exhibited cosegregation of the phenotype and a hygromycin marker. We identified different expression patterns for meiosis-related genes between N. tetrasperma and N. crassa and N. discreta that correspond with observed differences in meiosis and sexual spore development between pseudohomothallic N. tetrasperma and heterothallic N. crassa and N. discreta. Since meiosis gene sets and sporulation machinery are largely conserved in presence as well as sequence within these genomes, consistent differences among functionally related genes in these species would be strong evidence of dependent associations for the reconstruction of gene networks and thus for understanding the genetic basis of meiosis and sexual sporulation in Neurospora and ascomycetes. Additionally, species-specific expression in later stages of development was observed when clustering the ortholog genes. Future efforts should expand analysis to identify a more complete set of genes involved in meiosis and sporulation based on genetics and reverse genetics on the model N. crassa.
We distinguished eight gene clusters based on similarity of overall expression patterns across sexual development in three Neurospora species. During early sexual development, genes involved in metabolism are enriched, and clusters that appear later during sexual development are enriched with genes involved in metabolism, energy, transcription, cell cycle, and DNA processing. The expression profiles of these clusters correlated with the morphological changes observed during sexual development: at first metabolic genes are upregulated, then transcription and cell cycle related genes are activated, and eventually protein synthesis and transport genes are transcribed to deliver proteins needed to their destination in order to form the complex three dimensional fruiting body.
Our study revealed eight genes that are required for the successful formation of fruiting bodies in N. crassa. Knockouts of two genes, NCU06316 and 07508 resulted in the arrest of perithecium formation between 48 and 72 h after crossing (Fig. 4, panels B and C). In correlation with the gene expression data (Fig. 6A), these genes are of great importance in the later stage of development when structures such as the perithecial beak or ascospores are developed. The developing perithecia for NCU06316 (a putative argonaute siRNA chaperone complex subunit) and 07508 (a putative type-2 protein geranylgeranyltransferase subunit) remain round-shaped without formation of a beak. Six crosses of N. crassa mutant strains showed only protoperithecia and no formation of fruiting bodies. These results correlate with the gene expression observed over the time course of the sexual development in N. crassa. Here, an up-regulation of gene expression during the later stages of the development was observed. Our observations indicate that there are several genes that are required for the successful formation of perithecia.
Conclusions
We have used comparative transcriptomics as a tool to identify genes that are required for the successful formation of fruiting bodies. The impact of the deletion of the candidate genes was demonstrated in phenotypes observed in crosses exhibiting impaired perithecium formation at different stages of the development. Our findings shed light on the developmental process of fruiting body formation, evolution, and spore development in Neurospora species, thus establishing the foundation for future research particularly related to closely related pathogenic fungi. We suggest the potential utility of future research on the eight genes that we have discovered as essential contributors to fruit body development and as candidate genes for targets in the development of fungicides for control of plant pathogens.
Supporting Information
Expression results of all genes of all three Neurospora species.
(XLSX)
Level of Expression (LOX) data of N. crassa, N. tetrasperma and N. discreta orthologues. LOX estimates the Level Of gene eXpression from high-throughput-expressed sequence datasets with multiple treatments or samples. The tables includes the corresponding upper and lower confidence intervals (CI) across developmental stages.
(XLSX)
Comparative gene expression analysis of N. crassa, N. tetrasperma, and N. discreta . The table includes the results from the calculations such as the differences of confidence intervals.
(XLSX)
Comparison of species using the IF statement function. In each comparison the highest value across all time points for each gene was selected (MAX) and the maximum values were then sorted in descending order.
(XLSX)
Prioritization of the maximum value across the time course of the sexual development for each gene in speecies comparisons.
(XLSX)
Cosegregation of hygromycin resistance and identified phenotypes in KO strains of interest.
(DOCX)
Acknowledgments
We thank Dr. Joseph Wolenski and Barry Piekos for providing assistance in the Yale MCDB microscopy facility.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. LOX expression levels of genes in all three species, N. crassa, N. tetrasperma and N. discreta, relative across time points, are available in Table S1. LOX expression levels for the 2352 orthologous gene triads across sexual development are available in the Table S2. Comparative gene expression data are available in Table S3. Comparative gene expression data broken down by time point are available in Table S4. Our ranked, prioritized list based on these calculations is available in Table S5. Data are also deposited at the Filamentous Fungal Gene Expression Database (FFGED accession 239; [55]) and at the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) as accession GSE41484 for N. crassa [56], GSE60256 for N. tetrasperma, and GSE60255 for N. discreta.
Funding Statement
This study was supported by a Gaylord Donnelley Environmental Fellowship of the Yale Institute of Biospheric studies to NL, and National Science Foundation (NSF) Grant MCB 0923797 to FT and JPT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Busch S, Braus GH (2007) How to build a fungal fruit body: from uniform cells to specialized tissue. Mol Microbiol 64: 873–876. [DOI] [PubMed] [Google Scholar]
- 2. Busch S, Schwier EU, Nahlik K, Bayram O, Helmstaedt K, et al. (2007) An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proc Natl Acad Sci U S A 104: 8089–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coppin E, Berteaux-Lecellier V, Bidard F, Brun S, Ruprich-Robert G, et al. (2012) Systematic deletion of homeobox genes in Podospora anserina uncovers their roles in shaping the fruiting body. PLoS One 7: e37488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debuchy R, Berteaux-Lecellier V, Silar P (2010) Mating systems and sexual morphogenesis in Ascomycetes. In: Borkovich KA, Ebbole DJ, editors. Cellular and Molecular Biology of Filamentous Fungi. Washington, DC: ASM Press. pp. 501–535.
- 5. Harting R, Bayram O, Laubinger K, Valerius O, Braus GH (2013) Interplay of the fungal sumoylation network for control of multicellular development. Mol Microbiol 90: 1125–1145. [DOI] [PubMed] [Google Scholar]
- 6. Jamet-Vierny C, Debuchy R, Prigent M, Silar P (2007) IDC1, a pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet Biol 44: 1219–1230. [DOI] [PubMed] [Google Scholar]
- 7.Pöggeler S, Nowrousian M, Kück U (2006) Fruiting-body development in Ascomycetes, in The Mycota I. Growth, Differentiation and Sexuality. In: Kües U, Fischer R, editors. The Mycota I. (2nd edition). Growth, Differentiation and Sexuality. Berlin, Heideberg: Springer-Verlag. pp. 325–355.
- 8.Trail F, Gardiner DM (2014) Applicatioin of genomics to the study of pathogenecity and development in Fusarium. In: Nowrousian M, Esser K. editors. The Mycota: A comprehensive treatise on fungi as experimental systems for basic and applied research Fungal Genomics XIII. Second edition. Berlin, Heidelberg: Springer-Verlag. pp. 267–300.
- 9.Trail F (2013) Sex and fruiting in Fusarium In: Brown D, Proctor R, editors. Fusarium: genomics, molecular and cellular biology. Norwich, UK: Horizon Scientific Press and Caister Academic Press.
- 10. Coppin E, Debuchy R, Arnaise S, Picard M (1997) Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev 61: 411–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeLange AM, Griffiths AJ (1980) Meiosis in Neurospora crassa. I. The isolation of recessive mutants defective in the production of viable ascospores. Genetics 96: 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eckert SE, Kubler E, Hoffmann B, Braus GH (2000) The tryptophan synthase-encoding trpB gene of Aspergillus nidulans is regulated by the cross-pathway control system. Mol Gen Genet 263: 867–876. [DOI] [PubMed] [Google Scholar]
- 13. Eckert SE HB, Wanke C, Braus GH (1999) Sexual development of Aspergillus nidulans in tryptophan auxotrophic strains. Arch Microbiol 172: 157–166. [DOI] [PubMed] [Google Scholar]
- 14.Fischer R, Kües U (2003) Developmental processes in filamentous fungi. Genomics of Plants and Fungi. In: Prade RA, Bohnert HJ, editors. New York: Marcel Dekker. pp. 41–118.
- 15. Johnson TE (1979) A Neurospora mutation that arrests perithecial development as either male or female parent. Genetics 92: 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leslie JF, Raju NB (1985) Recessive mutations from natural populations of Neurospora crassa that are expressed in the sexual diplophase. Genetics 111: 759–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nowrousian M, Ringelberg C, Dunlap JC, Loros JJ, Kuck U (2005) Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora . Mol Genet Genomics 273: 137–149. [DOI] [PubMed] [Google Scholar]
- 18. Dodge BO (1927) Nuclear phenomena associated with heterothallism and homothallism in the Ascomycete Neurospora . Journal of Agricultural Research 35: 289–305. [Google Scholar]
- 19. Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE (1996) Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae . Genetics 144: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lumbsch HT (2000) Phylogeny of filamentous ascomycetes. Naturwissenschaften 87: 335–342. [DOI] [PubMed] [Google Scholar]
- 21. Samils N, Gioti A, Karlsson M, Sun Y, Kasuga T, et al. (2013) Sex-linked transcriptional divergence in the hermaphrodite fungus Neurospora tetrasperma . Proc Royal Soc B: Biol Sci 280: 20130862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z, Kin K, Lopez-Giraldez F, Johannesson H, Townsend JP (2012) Sex-specific gene expression during asexual development of Neurospora crassa . Fungal Genet Biol 49: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perkins D, Raju NB (1986) Neurospora discreta, a new heterothallic species defined by its crossing behavior. Experimental Mycology 10: 323–338. [Google Scholar]
- 24. Turner E, Jacobson DJ, Taylor JW (2010) Reinforced postmating reproductive isolation barriers in Neurospora, an Ascomycete microfungus. J Evol Biol 23: 1642–1656. [DOI] [PubMed] [Google Scholar]
- 25. Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, et al. (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68: 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beadle GW, Tatum EL (1941) Genetic Control of Biochemical Reactions in Neurospora . Proc Natl Acad Sci U S A 27: 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dunlap JC, Loros JJ (2004) The Neurospora circadian system. J Biol Rhythms 19: 414–424. [DOI] [PubMed] [Google Scholar]
- 28. Metzenberg RL, Glass NL (1990) Mating type and mating strategies in Neurospora . Bioessays 12: 53–59. [DOI] [PubMed] [Google Scholar]
- 29. Kronstad JW, Staben C (1997) Mating type in filamentous fungi. Annu Rev Genet 31: 245–276. [DOI] [PubMed] [Google Scholar]
- 30. Raju N (1992) Genetic control of the sexual cycle in Neurospora . Mycol Res 96: 241–262. [Google Scholar]
- 31. Raju NB, Perkins DD (1994) Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora, and Podospora . Dev Genet 15: 104–118. [DOI] [PubMed] [Google Scholar]
- 32. Dettman JR, Jacobson DJ, Taylor JW (2003) A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora . Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- 33. Beatty NP, Smith ML, Glass NL (1994) Molecular characterization of mating-type loci in selected homothallic species of Neurospora, Gelasinospora and Anixiella . Mycol Res 98: 1309–1316. [Google Scholar]
- 34. Casselton LA (2008) Fungal sex genes-searching for the ancestors. Bioessays 30: 711–714. [DOI] [PubMed] [Google Scholar]
- 35. Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, et al. (2010) Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell 9: 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nygren K, Strandberg R, Wallberg A, Nabholz B, Gustafsson T, et al. (2011) A comprehensive phylogeny of Neurospora reveals a link between reproductive mode and molecular evolution in fungi. Mol Phylogenet Evol 59: 649–663. [DOI] [PubMed] [Google Scholar]
- 37. Paoletti M, Seymour FA, Alcocer MJ, Kaur N, Calvo AM, et al. (2007) Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans . Curr Biol 17: 1384–1389. [DOI] [PubMed] [Google Scholar]
- 38. Strandberg R, Nygren K, Menkis A, James TY, Wik L, et al. (2010) Conflict between reproductive gene trees and species phylogeny among heterothallic and pseudohomothallic members of the filamentous ascomycete genus Neurospora . Fungal Genet Biol 47: 869–878. [DOI] [PubMed] [Google Scholar]
- 39. Whittle CA, Nygren K, Johannesson H (2011) Consequences of reproductive mode on genome evolution in fungi. Fungal Genet Biol 48: 661–667. [DOI] [PubMed] [Google Scholar]
- 40. Yun SH, Berbee ML, Yoder OC, Turgeon BG (1999) Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci U S A 96: 5592–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim H, Wright SJ, Park G, Ouyang S, Krystofova S, et al. (2012) Roles for receptors, pheromones, G proteins, and mating type genes during sexual reproduction in Neurospora crassa . Genetics 190: 1389–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bistis GN (1981) Chemotropic Interactions between trichogynes and conidia of opposite mating-Type in Neurospora crassa . Mycologia 73: 959–975. [Google Scholar]
- 43. Harris JL, Howe HB Jr, Roth IL (1975) Scanning electron microscopy of surface and internal features of developing perithecia of Neurospora crassa . J Bacteriol 122: 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raju N (1980) Meiosis and ascospore genesis in Neurospora . Eur J Cell Biol 23: 208–223. [PubMed] [Google Scholar]
- 45. Palma-Guerrero J, Hall CR, Kowbel D, Welch J, Taylor JW, et al. (2013) Genome wide association identifies novel loci involved in fungal communication. PLoS Genet 9: e1003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCluskey K, Wiest A, Plamann M (2010) The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci 35: 119–126. [DOI] [PubMed] [Google Scholar]
- 47. Hallen HE, Huebner M, Shiu SH, Guldener U, Trail F (2007) Gene expression shifts during perithecium development in Gibberella zeae (anamorph Fusarium graminearum), with particular emphasis on ion transport proteins. Fungal Genet Biol 44: 1146–1156. [DOI] [PubMed] [Google Scholar]
- 48. Wang Z, Lehr N, Trail F, Townsend JP (2012) Differential impact of nutrition on developmental and metabolic gene expression during fruiting body development in Neurospora crassa . Fungal Genet Biol 49: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clark TA, Guilmette JM, Renstrom D, Townsend JP (2008) RNA extraction, probe preparation, and competitive hybridization for transcriptional profiling using Neurospora crassa long-oligomer DNA microarrays. Fungal Genet Rep 55: 18–28. [Google Scholar]
- 50. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa . Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- 52. Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, et al. (2012) The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40: D26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Z, Lopez-Giraldez F, Townsend JP (2010) LOX: inferring Level Of eXpression from diverse methods of census sequencing. Bioinformatics 26: 1918–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poptsova MS, Gogarten JP (2007) BranchClust: a phylogenetic algorithm for selecting gene families. BMC Bioinformatics 8: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Z, Townsend JP (2010) The filamentous fungal gene expression database (FFGED). Fungal Genet Biol 47: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Z, Lopez-Giraldez F, Lehr N, Farre M, Common R, et al. (2014) Global gene expression and focused knockout analysis reveals genes associated with fungal fruiting body development in Neurospora crassa . Eukaryot Cell 13: 154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ruepp A, Zollner A, Maier D, Albermann K, Hani J, et al. (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32: 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, et al. (2007) Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet 57: 49–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fu C, Iyer P, Herkal A, Abdullah J, Stout A, et al. (2011) Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa . Eukaryot Cell 10: 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seo J, Shneiderman B (2002) Interactively exploring hierarchical clustering results. Computer 35: 80–86. [Google Scholar]
- 62. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 63. Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, et al. (2009) The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet 5: e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ait Benkhali J, Coppin E, Brun S, Peraza-Reyes L, Martin T, et al. (2013) A network of HMG-box transcription factors regulates sexual cycle in the fungus Podospora anserina . PLoS Genet 9: e1003642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Son H, Seo YS, Min K, Park AR, Lee J, et al. (2011) A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression results of all genes of all three Neurospora species.
(XLSX)
Level of Expression (LOX) data of N. crassa, N. tetrasperma and N. discreta orthologues. LOX estimates the Level Of gene eXpression from high-throughput-expressed sequence datasets with multiple treatments or samples. The tables includes the corresponding upper and lower confidence intervals (CI) across developmental stages.
(XLSX)
Comparative gene expression analysis of N. crassa, N. tetrasperma, and N. discreta . The table includes the results from the calculations such as the differences of confidence intervals.
(XLSX)
Comparison of species using the IF statement function. In each comparison the highest value across all time points for each gene was selected (MAX) and the maximum values were then sorted in descending order.
(XLSX)
Prioritization of the maximum value across the time course of the sexual development for each gene in speecies comparisons.
(XLSX)
Cosegregation of hygromycin resistance and identified phenotypes in KO strains of interest.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. LOX expression levels of genes in all three species, N. crassa, N. tetrasperma and N. discreta, relative across time points, are available in Table S1. LOX expression levels for the 2352 orthologous gene triads across sexual development are available in the Table S2. Comparative gene expression data are available in Table S3. Comparative gene expression data broken down by time point are available in Table S4. Our ranked, prioritized list based on these calculations is available in Table S5. Data are also deposited at the Filamentous Fungal Gene Expression Database (FFGED accession 239; [55]) and at the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) as accession GSE41484 for N. crassa [56], GSE60256 for N. tetrasperma, and GSE60255 for N. discreta.