The difference in patterns between proteome 2D-gels has little or no relation to phylogeny.
In a broad study of >100 eukaryotic and prokaryotic species in this issue of PNAS, Knight et al. (1) characterize eukaryotic and prokaryotic proteomes by “theoretical 2D-gels,” two-dimensional plots where each protein is represented by its molecular weight and isoelectric point. The difference in patterns between proteome 2D-gels turns out to have little or no relation to phylogeny. Instead, a variety of comparisons seem to indicate that theoretical 2D-gels are shaped by the cellular environment and the ecological niche of the corresponding species. Knight et al. conclude that environmental variability causes a greater variability in the general properties of membrane proteins (at the cell–environment interface) and that metabolic specialization (evidenced by differential usage of carbon substrates) correlates better with the general properties of nonmembrane proteins. The notion that environmental factors influence the properties of a proteome is not new. It was developed in relation to protein characters related to the dimensions represented in the theoretical 2D-gels of Knight et al., i.e., amino acid usages (more specifically the usage of the “charged” residues Asp, Glu, Lys, Arg, and His and of the other ionizable residues Cys and Tyr) and protein length.
Amino Acid Composition
From the seminal studies of Sueoka (2) to the most recent analyses (3, 4), it has been verified that amino acid usages are influenced by the G + C content of a genome. It also has been verified, however, that they reflect adaptations to specific environmental conditions. Thus, acidic residues predominate over basic residues in halophilic prokaryotes (5–7). A variety of amino acid compositional properties further distinguish thermophilic vs. mesophilic species. These properties include more hydrophobic interactions and salt bridges corresponding to higher frequency of long-range interactions, more hydrogen bonds and exposed polar residues corresponding to higher solubility, and higher frequency of branched residues (see ref. 8 for review and references). Many comparisons of thermophilic vs. mesophilic proteins have established preferential usage of specific amino acids in thermophilic vs. mesophilic proteins (e.g., refs. 3, 7, 9, and 10). A recent analysis of amino acid usages in broad collections of complete proteomes (4) suggests that genome C + G content and living temperature explain most of the variability in amino acid usage observed among species and identifies thermophiles for their preferential use of Glu, Ile, Val, Gly, and Arg at the expense of Asn, His, Ser, and especially Gln and Cys. This type of amino acid compositional analyses has been taken one step further by Pe'er et al. (11), who, also investigating the distribution of di- and tripeptide overrepresentations, define species-specific “proteome signatures” in analogy to genome signatures (12).
Protein Length
In relation to the size of proteins in eukaryotic, bacterial, and archaeal proteomes, various studies (4, 7, 13–15) have reported that proteins in eukaryotes are significantly longer than proteins in prokaryotes and are moderately longer in bacteria than in archaea. We find (L.B. and S. Karlin, unpublished data) that these length relations are pervasive in the vast majority of functional categories of proteins. They also are present within homologous protein families common to the three domains as well as among proteins unique to each domain. What can account for these differences in length? A greater length of eukaryote proteins may reflect on the greater complexity of the eukaryote cell compared to the prokaryote cell. Eukaryote proteins are expanded by the addition of sequence motifs or structural domains that act as functional regulators (15). Compared with prokaryote proteins, it also may be more difficult for eukaryote proteins to associate. Fusion of single-function proteins into multidomain units also may facilitate the interaction between functional units in a crowded cytoplasmic space partitioned by a complex array of compartments and may diminish the need to produce proteins in greater amounts to achieve proper concentrations of their complexes.
The overall modest reduction in the median length of archaeal (thermophilic) vs. bacterial (mesophilic) orthologs (≈15–20 aa) is compatible with a length reduction of disordered loops that has been suggested to confer extra stability to thermotolerant proteins (8, 16). However, bacterial proteins tend to be longer than archaeal proteins, also comparing only thermophilic or mesophilic species (L.B. and S. Karlin, unpublished data), suggesting that factors other than temperature distinguish bacterial from archaeal protein size. In this respect it is interesting to observe that, of all sequenced prokaryotes, archaeal species are all free-living, mostly in extreme environments, whereas bacterial species are often obligate or facultative parasites of animals and plants or endocellular parasites whose reduced proteomes include longer proteins than in most other species. It is likely that, besides temperature, other stresses and environmental fluctuations favor the evolution of less complex and more stable proteins among free-living species. Free-living species also are more likely to be subject to starving conditions than parasitic ones are. Akashi and Gojobori (17) and Seligmann (18) provide evidence that protein amino acid composition reflects selection for less expensive amino acids. This effect is more pronounced in highly expressed proteins (17) and is less pronounced in the smaller proteomes of endocellular parasites subject to less intense selection (18). In this perspective, minimizing the length of a protein would effectively reduce its cost. Consistent with this interpretation, the length of proteins from obligate and/or endocellular parasites tends to be greater than that from species subject to starving conditions.
Measures of Size and Charge
The study by Knight et al. (1) differs from these analyses in several respects. First, protein size and charge are represented in this article by their distribution among all proteins in the proteome rather than by an average value. Second, correlations between size and charge are accounted for in their two-dimensional representation. Third, molecular weight and especially isoelectric point are related but different characters than protein length and amino acid composition, respectively. Molecular weight is obviously related to protein length. Indeed, although amino acid composition can influence the nexus of length with molecular weight, their correlation is almost perfect (e.g., among proteins in Escherichia coli, r = 0.9988). Less obvious is the relation of isoelectric point (pI) and amino acid composition. In this respect it is interesting to examine the relations that isoelectric point and frequency of charged residues (Asp, Glu, Lys, and Arg) have with the net charge of a protein, the character that is most likely to be directly relevant to the functionality of the protein.
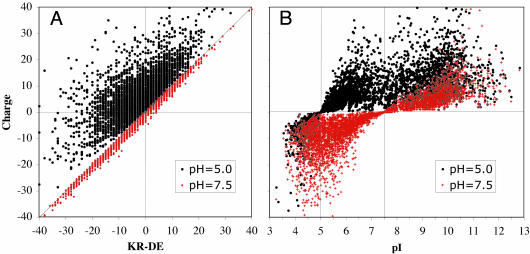
The theoretical net charge of a protein at a given pH is estimated on the basis of the Henderson–Hasselbach equation [pK = pH + log([A]/[B])], [A] being the concentration of an acid and [B] the concentration of its conjugated base] to all acidic and basic groups of the protein, and its theoretical isoelectric point is defined as the pH at which the protein has no charge. The relations of protein charge with isoelectric point or counts of Lys + Arg - Glu - Asp are exemplified in Fig. 1 for the E. coli K12 proteome, where protein charge has been estimated at physiological pH = 7.5 and also at pH = 5.0. The pH value 5.0 was chosen for its biological relevance, because it corresponds to the cytoplasmic pH maintained in E. coli when it crosses the highly acidic environment of the stomach. It is likely to be biologically relevant that although at physiological pH most of the proteins in E. coli, and in the majority of prokaryotic species (1, 7, 19), are negatively charged, most of them become positively charged at acidic pH. As seen in Fig. 1 A, at near neutral pH the difference in counts of basic vs. acidic residues correlates very well to the theoretical charge of a protein, and it estimates the correct proportion of proteins that have a positive net charge (36.6%). However, at pH = 5.0 it grossly underestimates the percentage of proteins carrying a positive charge (81.2%). Fig. 1B shows that isoelectric point values are always poor indicators of the quantitative charge of a protein. However, contrary to counts of charged residues, the isoelectric point value is a much better predictor of whether a protein is negatively or positively charged at any pH value.
Fig. 1.
The protein charge of all proteins of the E. coli K12 proteome is calculated at pH 7.5 and at pH 5.0 and is compared with the usage of basic residues Arg and Lys minus acidic residues Asp and Glu (KR - DE) (A) and the theoretical isoelectric point (pI) of the protein (B).
The conditions that affect the distribution of protein characters such as size, charge, isoelectric point, and amino acid composition within each proteome are likely to result from species-specific adaptations to complex and multivariate environmental conditions and lifestyles. The article by Knight et al. (1) utilizes a novel representation of proteomic features and points to niche qualifiers more complex than temperature and salt concentration as possible determinants of its general shape. Among these qualifiers, those related to free-living vs. parasitic lifestyles appear to be promising candidates, but there is a large space of possibilities that still needs to be explored.
See companion article on page 8390.
References
- 1.Knight, C. G., Kassen, R., Hebestreit, H. & Rainey, P. B. (2004) Proc. Natl. Acad. Sci. USA 101, 8390-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sueoka, N. (1961) Cold Spring Harb. Symp. Quant. Biol. 26, 35-43. [DOI] [PubMed] [Google Scholar]
- 3.Kreil, D. P. & Ouzounis, C. A. (2001) Nucleic Acids Res. 29, 1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tekaia, F., Yeramian, E. & Dujon, B. (2002) Gene 297, 51-60. [DOI] [PubMed] [Google Scholar]
- 5.Gandbhir, M., Rasched, I., Marliere, P. & Mutzel, R. (1995) Res. Microbiol. 146, 113-120. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy, S. P., Ng, W. V., Salzberg, S. L., Hood, L. & DasSarma, S. (2001) Genome Res. 11, 1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlin, S., Brocchieri, L., Trent, J., Blaisdell, B. E. & Mrazek, J. (2002) Theor. Popul. Biol. 61, 367-390. [DOI] [PubMed] [Google Scholar]
- 8.Kumar, S. & Nussinov, R. (2001) Cell. Mol. Life Sci. 58, 1216-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar, S., Ma, B., Tsai, C. J., Sinha, N. & Nussinov, R. (2000) Protein Sci. 9, 10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das, R. & Gerstein, M. (2000) Funct. Integr. Genomics 1, 76-88. [DOI] [PubMed] [Google Scholar]
- 11.Pe'er, I., Felder, C. E., Man, O., Silman, I., Sussman, J. L. & Beckmann, J. S. (2004) Proteins 54, 20-40. [DOI] [PubMed] [Google Scholar]
- 12.Campbell, A., Mrazek, J. & Karlin, S. (1999) Proc. Natl. Acad. Sci. USA 96, 9184-9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin, M. Y., Tatusov, R. L. & Koonin, E. V. (1999) in Organization of the Prokaryotic Genome, ed. Charlebois, R. L. (Am. Soc. Microbiol., Washington, DC).
- 14.Skovgaard, M., Jensen, L. J., Brunak, S., Ussery, D. & Krogh, A. (2001) Trends Genet. 17, 425-428. [DOI] [PubMed] [Google Scholar]
- 15.Zhang, J. (2000) Trends Genet. 16, 107-109. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, M. J. & Eisenberg, D. (1999) J. Mol. Biol. 290, 595-604. [DOI] [PubMed] [Google Scholar]
- 17.Akashi, H. & Gojobori, T. (2002) Proc. Natl. Acad. Sci. USA 99, 3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seligmann, H. (2003) J. Mol. Evol. 56, 151-161. [DOI] [PubMed] [Google Scholar]
- 19.Karlin, S., Blaisdell, B. E. & Bucher, P. (1992) Protein Eng. 5, 729-738. [DOI] [PubMed] [Google Scholar]