Abstract
Hepatitis C virus (HCV) infection is a leading cause of liver-related mortality. Chronic hepatitis C (CHC) is frequently associated with disturbances in iron homeostasis, with serum iron and hepatic iron stores being elevated. Accumulating evidence indicates that chronic HCV infection suppresses expression of hepatic hepcidin, a key mediator of iron homeostasis, leading to iron overload conditions. Since hepcidin mediates degradation of ferroportin, a basolateral transporter involved in the release of iron from cells, diminished hepcidin expression probably leads to up-regulation of ferroportin-1 (Fpn1) in patients with CHC. In this study, we determined the protein levels of duodenal Fpn1, and found that its expression was significantly up-regulated in patients with CHC. The expression of duodenal Fpn1 is negatively correlated with mRNA levels of hepcidin, and positively correlated with serum iron parameters. Although iron is a critical factor for growth of a variety of pathogenic bacteria, our results suggest that iron overload in blood does not increase the infection rate of bacteria in patients with CHC.
Introduction
As a co-factor for heme and non-heme iron proteins, iron is an important element for all living organisms; however, excess iron can be toxic for the organism [1]. Therefore, iron metabolism must be tightly regulated. Absorption of dietary iron begins with its transport through the plasma membrane by the divalent metal transporter 1 (DMT1), the major iron transporter in the duodenum [2], [3]. After entering the cytoplasm of enterocytes, iron is stored in ferritin or transferred to the portal blood by the basolateral exporter ferroportin-1 (Fpn1). Fpn-1, a principal iron exporter, plays in transporting iron from the duodenum to the serum, and in iron recycling from reticuloendothelial macrophages into the circulation, respectively [4]. Hepcidin, the key mediator of iron homeostasis produced by liver, negatively controls Fpn1 expression levels by a posttranslational mechanism [5]. Hepcidin mediates internalization and induces degradation of Fpn1 in enterocytes and reticuloendothelial macrophages, thus suppressing duodenal iron absorption and macrophage iron release. The expression of hepcidin is significantly up-regulated by iron overload and inflammation [6], [7], and down-regulated in response to erythropoiesis, iron deficiency, and hypoxia [8].
Iron deposition is a pathophysiologic feature during chronic hepatitis C virus (HCV) infection [9]. Excess iron accumulation can be highly toxic, and contributes to liver injury by inducing oxidative stress [10]. Accumulating evidence indicates that both expression of hepcidin mRNA in liver and serum levels of hepcidin are decreased in patients with chronic hepatitis C (CHC) [11]–[13]. Although the precise mechanism underlying inhibition of hepcidin expression by HCV is not fully understood, production of reactive oxygen species (ROS) is believed to be involved in the process [14], [15]. In vitro experiments demonstrate that ROS induced by HCV proteins suppress the expression of hepcidin by inhibiting binding activity of two positive regulators, C/EBPβ and STAT3, and stabilizing the expression of two negative hepcidin regulators, HIF1α and HIF2α in human hepatoma cell lines [14]. In HCV transgenic mice, ROS induced by HCV increase hepatic expression of CHOP and subsequently reduce DNA binding activity of C/EBPα, which leads to down-regulation of hepcidin mRNA levels. Decreased hepcidin expression enhances ferroportin expression in the duodenum and macrophages, resulting in an increase in duodenal iron transport and a decrease in macrophage iron release [15].
While HCV transgenic mice exhibit high levels of duodenal ferroportin [15], not much is known concerning the expression of ferroportin in patients with CHC. In this report, we evaluated the protein expression of Fpn-1 in duodenum biopsy samples from patients with CHC. Since bacterial pathogens need iron to replicate and survive [16]–[18], we also investigated whether increased iron levels in blood could influence bacterial communities in patients with CHC.
Materials and Methods
Liver and duodenum samples
We recruited 131 patients with CHC and 47 patients with nonalcoholic fatty liver disease (NAFLD) (Control group), who were hospitalized between May 2011 and May 2013 at The First Affiliated Hospital, Kunming Medical University, China. Patients with CHC were eligible for inclusion in the study if they were HCV seropositive, had detectable HCV RNA that liver biopsy was performed. Exclusion criteria were viral hepatitis other than hepatitis C, hepatic cirrhosis, alcoholic or drug-induced liver injury, hepatocellular carcinoma, serious cardiopulmonary and renal disease, acute inflammatory disorders, and upper gastrointestinal bleeding. The diagnosis of chronic HCV infection was based on the measurement of serum anti-HCV antibody (Auto delfia1235, PerkinElmer, Inc, Waltham, MA, USA) and HCV RNA (Freedom Evolyzer-z, Tecan Group Ltd., Männedorf, Switzerland). NAFLD is diagnosed according to the following three criteria: non-alcoholic, detection of steatosis ultrasonography and liver biopsy, and appropriate exclusion of other liver diseases [19]. Of these patients, 43 patients with HCV and 22 with NAFLD suffered from persistent or recurrent pain or discomfort centred in the upper abdomen. After clinical examination and routine biochemistry, these patients underwent upper gastrointestinal endoscopy. Exclusion criteria were the presence of duodenal duodenitis and gastroduodenal ulcer on endoscopy. During the upper gastrointestinal endoscopy, duodenal biopsies were taken from 15 patients with HVC and 12 patients with NAFLD, respectively. Furthermore, liver samples were obtained from these 27 patients by needle biopsy for diagnosis of chronic liver diseases. Thus, 27 liver and duodenum samples were included in this study. Written informed consent was obtained from each patient included in the study. The study was approved by the Kunming Medical University Ethics Committee and carried out according to the guidelines.
Assays for inflammatory factors, transaminases and iron parameters
Blood samples of all these subjects were collected in the morning under fasting conditions, and sera were separated immediately after blood collection and stored at −80°C until use. Serum levels of tumor necrosis factor (TNF-α) and interleukin-6 (IL-6) were determined using the commercially available ELISA kits (R&D Systems, Minneapolis, MN). The assays were performed according to the manufacturer’s protocols. Serum iron status, alanine aminotransferase (ALT), aspartate aminotransferase (AST), and bilirubin were determined using an automated Olympus AU5400 biochemistry analyzer (Olympus Corp., Tokyo, Japan). Hemoglobin levels and red blood cell count in blood were determined by ADVIA 120 hematology analyzer (Siemens, Healthcare Diagnostics Inc., Deerfield, IL, USA).
Immunohistochemistry and prussian blue staining
After liver and duodenal biopsies were fixed in 10% buffered formalin, these tissues were embedded in paraffin, and sectioned at 5 µm in thickness. For immunohistochemical analysis, the sections were blocked by 3% BSA-PBS solution at room temperature for 1 h. The sections were then stained with monoclonal anti-prohepcidin (Santa Cruz Biotechnology, Santa Cruz, CA; 1∶250 dilution) or goat anti-Fpn1 antibodies (Santa Cruz Biotechnology; 1∶250 dilution) at 4°C overnight, respectively. After washed with PBS three times for 10 min at room temperature, the sections were incubated with HRP-conjugated anti-mouse or goat IgG (Boster Biol Tech, Wuhan, China) for 60 min at room temperature. The substrate solution diaminobenzidine (Tiangen, Beijing, China) was added until the desired intensity of color was developed. Prussian blue staining for iron was performed using a Prussian blue staining kit (Polysciences, Inc. Warrington PA, USA) according to the manufacturer’s protocol.
Real-time PCR
Total RNA was extracted from liver tissue using Trizol reagent (Invitrogen, Carlsbad, USA). cDNAs were generated by reverse transcription with SuperScript II (Invitrogen). Real-time PCR was performed with the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, USA) using SYBR Premix-Ex TagTM (TaKaRa, Dalian, China). The primers used were: hepcidin, 5′- CTG CAA CCC CAG GAC AGA G -3′ (forward), 5′- GGA ATA AAT AAG GAA GGG AGG GG -3′ (reverse) [20]; Fpn1, 5′- CTA CTT GGG GAG ATC GGA TGT -3′ (forward), 5′- CTG GGC CAC TTT AAG TCT AGC -3′ (reverse) [21]; and GAPDH, 5′- ACT TTG GTA TCG TGG AAG GAC TCA -3′ (forward); 5′- GTA GAG GCA GGG ATG ATG TTC TG -3′ (reverse).
Western blotting
After duodenal samples were homogenized in liquid nitrogen, the homogenate was lysed on ice for 30 minutes in lysis buffer (BioTeKe, Beijing, China). The lysates (25 µg of total protein) were loaded per well and separated on a 10% SDS-polyacrylamide gel. Proteins were then transferred onto a polyvinylidene difluoride membrane. Primary antibodies were goat anti-Fpn1 antibodies (Santa Cruz Biotechnology, 1∶2500 dilution). The secondary antibody was a peroxidase-coupled anti-goat IgG (Boster Biol Tech). The membrane was exposed to Pierce® ECL Western Blotting Substrate (Thermo Scientific, Rockford, lL), and the film was developed.
Sample preparation, pyrosequencing and bioinformatics
Genomic DNAs of bacteria in serum were extracted, the 16S ribosomal RNA (rRNA) gene V1–V2 region was amplified by PCR, and the amplicons was sequenced on a 454 Genome Sequencer FLX Titanium platform, as described in our recent studies [22], [23]. To eliminate artificial contaminations, potential presence of bacteria in all reagents for DNA extract and PCR were detected by amplifying bacterial 16S rRNA using PCR methods as described above. Sequencing reads were quality filtered, OTU clustered, ChimeraSlayer filtered and further analyzed using the QIIME pipeline and RDP-classifier. All OTUs found in 50% of blood samples were retained for the following further analyses. PLS-DA plotting analysis of samples based on microbiota analysis was performed using METAGENassist, a comprehensive web server for comparative metagenomics.
Statistical analysis
Statistical analysis was performed using SigmaPlot 12.0 (Systat Software Inc., London, UK). General characteristics were expressed as median and mean or percentages. Two samples comparisons were performed using t-test (parametric) or Mann-Whitney rank sum test (non-parametric). Correlation analyses between two samples were performed using Spearman’s rank correlation. Statistical significance was set at P<0.05.
Results
Clinical characteristics of the patients
The characteristics of patients with CHC and without HCV infection (the control group) are shown in Table 1. There was no statistical difference in age and sex distribution in the two groups. In patients with CHC, the mean values of HCV RNA was 990±205 kIU/ml. Serum levels of ALT, AST, IL-6, and TNFα in patients with CHC were significantly higher than those in the control group. In contrast, patients with CHC had similar red blood cell counts, hemoglobin, and hematocrit levels, compared to the control group.
Table 1. Clinical characteristics of patients in this study.
| HCV(+) patients | HCV(−) patients | P value | |
| Age, y | 50.6±14.2 (31∼67) | 56.7±13.1 (42∼71) | >0.05 |
| Gender (M/F) | 9/6 | 8/4 | >0.05 |
| ALT (IU/L) | 66.5±25.6 | 42.2±17.3 | <0.05 |
| AST (IU/L) | 60.6±31.4 | 36.7±15.1 | <0.05 |
| Hematocrit (%) | 40.2±4.8 | 42.1±5.3 | >0.05 |
| RBC (104/mm3) | 427±91 | 465±88 | >0.05 |
| Hemoglobin (g/L) | 12.8±1.7 | 13.5±1.8 | >0.05 |
| Serum ferritin (ng/ml) | 208±144 | 106±74 | <0.01 |
| Transferrin saturation (%) | 56±22.4 | 24.5±18.5 | <0.001 |
| IL-6 (pg/ml) | 5.4±4.1 | 2.5±1.4 | <0.01 |
| TNFα (pg/ml) | 11.4±6.1 | 6.8±4.2 | <0.05 |
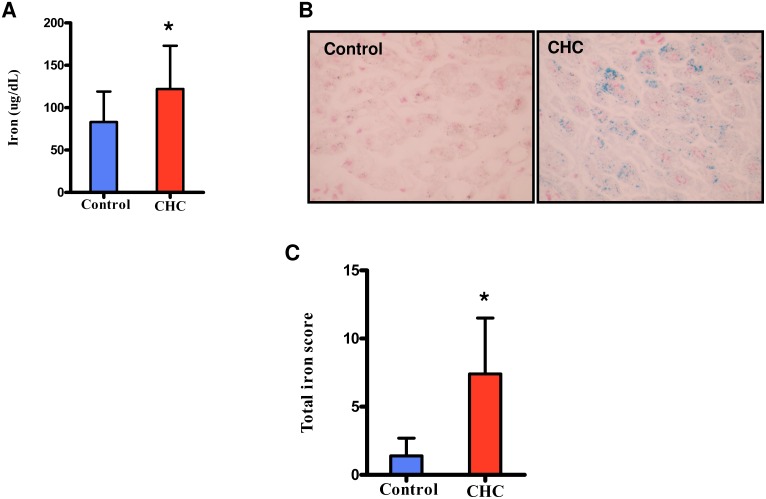
It has been shown that patients with CHC have higher levels of serum iron [9], [11]. Consistent with these observations, we found that serum levels of iron in patients with CHC in this study were markedly higher than those in the control group (Fig. 1A). Meanwhile, serum levels of transferrin saturation, and ferritin levels in patients with CHC were significantly higher than those in the control group (Table 1). In addition, moderate iron deposition in the liver was observed in patients with CHC (Fig. 1B), and TIS scores were much higher in patients with CHC than in the control group (Fig. 1C).
Figure 1. Iron concentrations in serum, and iron localization in the liver.
CHC (N = 15) and Control (N = 12). (A) Serum levels of iron. (B) Iron in liver sections with Prussian blue staining. (C) Total iron score. *P<0.05 relative to control group.
The expression of hepcidin in liver is reduced in patients with CHC
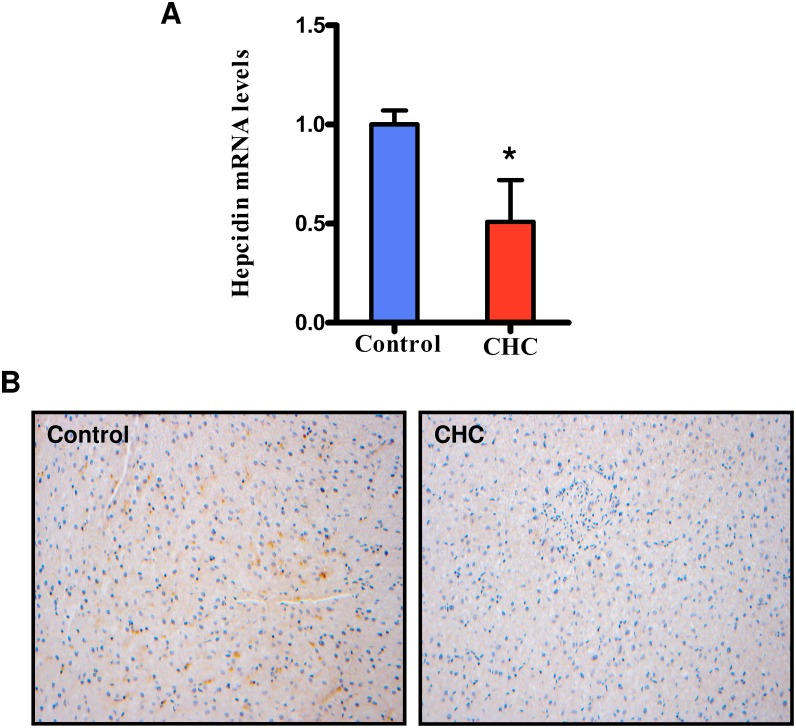
Next, we determined the mRNA levels of hepcidin by qPCR in all subjects. As shown in Fig. 2A, the mRNA levels of hepcidin in the patients with CHC were significantly lower than those in the control group. Meanwhile, we measured the protein levels of prohepcidin in the liver by immunohistochemical staining. Patients with CHC exhibited a decrease in the expression of hepatic prohepicidin, compared with the control group.
Figure 2. The expression of hepcidin in the liver.
CHC (N = 15) and Control (N = 12). (A) mRNA levels of hepcidin determined by qPCR. (B) The protein expression of prohepcidin determined by immunohistochemistry. *P<0.05 relative to control group.
Expression of duodenal Fpn1 is up-regulation in patients with CHC
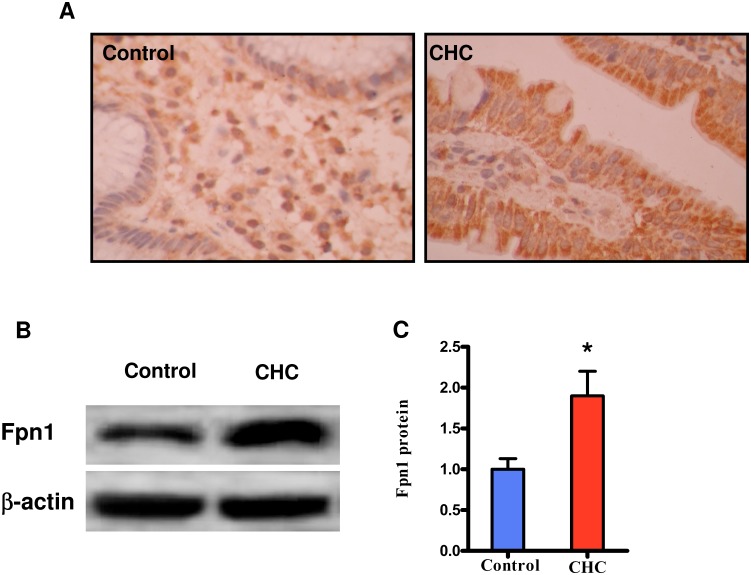
Hepcidin binds to the iron exporter ferroportin, leading to its internalization and degradation [5]. Thus, reduced hepcidin levels probably results in an increase in the levels of Fpn1 in the duodenum. To test this hypothesis, we measured the expression of Fpn1 in the duodenum by immunohistochemical analysis. We found that the protein expression of Fpn1 in the duodenum was significantly higher in patients with CHC than in the control group (Fig. 3A). To further confirm these results, the protein expression of Fpn1 in the duodenum was detected by western blotting. As shown in Fig. 3B and C, the protein levels of duodenal Fpn1 were markedly up-regulated in patients with CHC, compared with those in the control group. In contrast, the mRNA levels of Fpn1 the patients with CHC were comparable to those in the control group (data not shown).
Figure 3. The protein expression of duodenal Fpn1.
CHC (N = 15) and Control (N = 12). The protein expression of duodenal Fpn1 determined by immunohistochemistry (A), and Western blot (B), respectively. (C) Quantification of protein levels by Western blot in the two groups. *P<0.05 relative to control group.
Correlation between duodenal Fpn1 levels and clinical parameters
We next examined correlation of duodenal Fpn1 protein with inflammatory factors and iron parameters (Table 2). We found that hemoglobin concentrations, red blood cell counts, hematocrit, and the serum levels of ALT and AST, were not correlated with duodenal Fpn1 protein levels. As a potent stimulator of prohepcidin expression in liver [7], [24], IL-6 in serum was significantly increased in patients with CHC (Table 1). However, a decrease in the expression of hepatic prohepicidin was observed in patients with CHC. Thus, it is unlikely that the expression of prohepcidin is regulated by inflammation in CHC. Furthermore, we found that there were no significant correlations between duodenal Fpn1 protein and inflammation factors (IL-6, and TNFα) in patients with CHC. In contrast, Fpn1 protein levels were negatively correlated with hepcidin mRNA levels. Finally, we found that there were significant correlations between Fpn1 protein levels and serum iron levels. Taken together, these results suggest that elevated duodenal Fpn1 protein levels are probably due to reduced hepcidin expression, leading to an increase in serum levels of iron.
Table 2. Correlations between clinical findings and Fpn1 protein levels in patients with CHC.
| r | P value | |
| ALT (IU/L) | 0.153 | 0.0912 |
| AST (IU/L) | 0.168 | 0.1340 |
| Hematocrit (%) | 0.112 | 0.1781 |
| RBC (104/mm3) | 0.154 | 0.0898 |
| Hemoglobin (g/L) | 0.177 | 0.0612 |
| Serum ferritin (ng/ml) | 0.528 | <0.0001 |
| Transferrin saturation (%) | 0.368 | 0.0015 |
| Serum iron (µg/dL) | 0.288 | 0.0236 |
| TIS | 0.465 | <0.0001 |
| Hepcidin RNA levels | −0.381 | 0.0011 |
| IL-6 (pg/ml) | 0.179 | 0.0781 |
| TNFα (pg/ml) | 0.118 | 0.1390 |
Iron overload in blood does not increase the risk of bacterial infection in patients with CHC
Many species of pathogenic bacteria require iron for growth as iron acquisition proteins are virulence factors for these pathogens [16]–[18]. As iron overload in blood occurs in patients with CHC, we hypothesized that increased iron levels in blood could increase the infection rate of pathogenic bacteria in patients with CHC. To test this idea, we determined the composition of bacterial communities in blood using high-throughput 454 pyrosequencing. In total, we collected blood samples from 12 control patients and 15 patients with CHC, for analysis of the bacterial 16S ribosomal RNA (rRNA) genes. The variable regions (V1–V2) of the bacterial rRNA 16S genes were amplified by PCR using a primer set with a unique 10-nt barcode. Three blood samples from the CHC group and three blood samples from the control group were excluded from the following analysis due to sequences less than 500, which possibly resulted from sequencing bias. For the remaining 21 samples, we obtained a dataset consisting of 31, 205 high-quality 16S rRNA gene sequences with an average of 1486±188 (S.E.) sequences per sample (Table 3). In this study, the most notable contamination was derived from the reagents for DNA extract and PCR. But our PCR-based bacteria detections for all these reagents were negative.
Table 3. 454 data summary.
| CHC Group (n = 12) | Control Group (n = 9) | Total (n = 21) | |
| Sequence number | 1960±248 | 854±79 | 1486±188 |
| OTUs a | 73±7 | 64±6 | 69±5 |
| Shannon b | 2.21±0.13 | 2.57±0.14 | 2.37±0.10 |
| Simpson b | 0.53±0.03 | 0.60±0.02 | 0.56±0.02 |
OTU was equal to bacterial species level according to 97% sequence identity.
Bacterial diversity Index.
From the dataset, we identified a total of 265 operational taxonomic units (OTUs) (detected from at least two blood samples) based on the conventional criterion of 97% sequence similarity (equal to species level), with 64±6 (n = 9) OTUs per sample in blood from the control group, 73±7 (n = 12) OTUs per sample in blood from the CHC group (Table 3). Furthermore, we found 14 of 16 dominant OTUs can be matched to known bacterial species according to 97% sequence similarity, which accounted to about 90% of all bacterial 16S rDNA sequences (Table 4).
Table 4. Identification of 14 dominant bacterial species and their abundance distribution in CHC and control groups.
| OTUs ID | CHC Group#(n = 12) | Control Group#(n = 9) | Similar Species | Genbank Accession | Identity |
| OTU01 | 66.10±2.97% | 60.70±2.16% | Pseudomonas aeruginosa DK2 | NC_018080 | 99% |
| OTU02 *** | 7.13±1.15% | 12.80±0.65% | Delftia acidovorans SPH-1 | NC_010002 | 99% |
| OTU03 | 5.52±2.29% | 2.15±1.75% | Pseudomonas fluorescens A506 | NC_017911 | 99% |
| OTU04 | 3.48±0.22% | 3.80±0.33% | Pseudomonas aeruginosa DK2 | NC_018080 | 99% |
| OTU05 | 2.49±0.21% | 2.89±0.13% | Pseudomonas aeruginosa DK2 | NC_018080 | 98% |
| OTU06 | 1.75±0.52% | 0.81±0.10% | Escherichia coli str. K-12 substr. W3110 | NC_007779 | 99% |
| OTU07 | 1.11±0.10% | 1.16±0.09% | Pseudomonas aeruginosa DK2 | NC_018080 | 99% |
| OTU08 | 1.36±0.60% | 0.42±0.42% | Rhodococcus qingshengii strain djl-6 | NR_043535 | 99% |
| OTU09 | 0.51±0.06% | 0.68±0.10% | Pseudomonas aeruginosa DK2 | NC_018080 | 99% |
| OTU10 | 0.50±0.06% | 0.68±0.07% | Pseudomonas aeruginosa DK2 | NC_018080 | 99% |
| OTU11 | 0.60±0.19% | 0.26±0.06% | Escherichia coli O26:H11 str. 11368 | NC_013361 | 99% |
| OTU12 | 0.46±0.20% | 0.25±0.21% | Phyllobacterium myrsinacearum | AB681132 | 99% |
| OTU13 | 0.33±0.11% | 0.41±0.13% | Propionibacterium acnes 6609 | NC_017535 | 99% |
| OTU14 | 0.36±0.07% | 0.36±0.07% | Pseudomonas aeruginosa DK2 | NC_018080 | 99% |
Relative abundance given by mean ± SE.
***P = <0.001 (t-test).
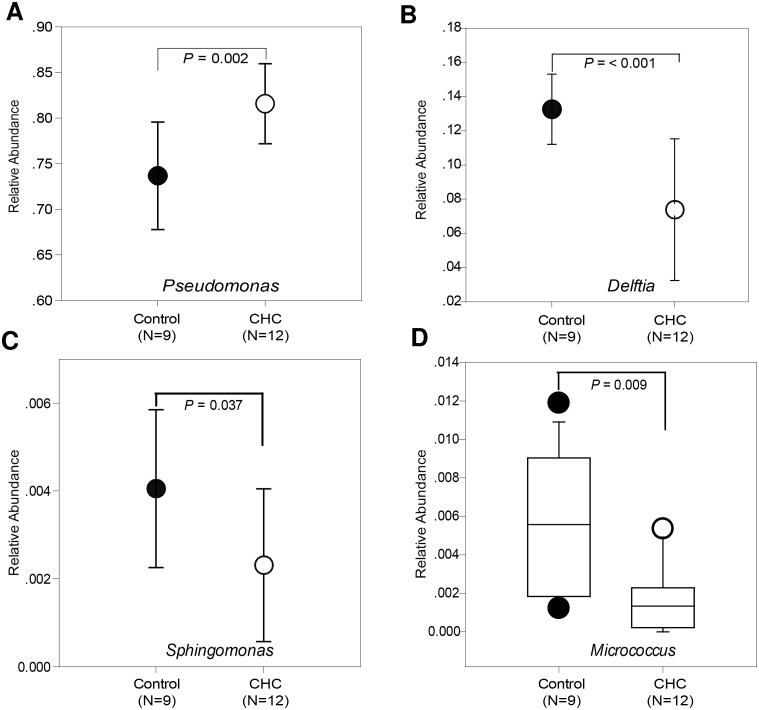
We observed no difference in bacterial community diversities between the control and CHC groups (evenness by Shannon and Simpson indexes in Table 3). However, we found that the bacteria genus Pseudomonas in the CHC group was significantly higher than that in control group (81.60±4.38% versus 73.70±5.90%) (Fig. 4A). We also discovered a significant decrease in the abundance of three rare bacterial genera Delftia, Sphingomonas, and Micrococcus in the CHC group, compared with the control group (Fig. 4C–D).
Figure 4. Bacterial genera comparison of blood samples between the CHC (N = 12) and control (N = 9) groups.
(A) Pseudomonas; (B) Delftia; (C) Sphingomonas; (D) Micrococcus.
Discussion
It has been reported that the serum levels of hepcidin and mRNA levels of hepcidin in the liver in patients with CHC are significantly lower than normal subjects or patients without HCV infection [11]–[13]. Consistent with these observations, our results confirm that patients with CHC have low levels of hepcidin in serum and prohepicidin in the liver. The expression of hepcidin is significantly induced by iron overload and inflammation [6], [7] and downregulated in response to low iron levels, erythropoiesis, and hypoxia [8]. However, high serum levels of iron and IL-6, and iron deposit in liver does not result in high levels of hepcidin in patients with CHC. The fact that ROS induced by HCV proteins inhibits the expression of hepcidin [14], [15] strongly suggests that the ROS-mediated signaling overcomes inflammatory and iron-sensing pathways in hepcidin regulation in patients with CHC. Thus, elevated iron levels in serum are largely attributable to reduced hepcidin expression in liver.
As a peptide hormone, hepcidin function as a regulator of iron homeostasis by inhibiting the iron transporter ferroportin [5]. Nishina et al has previously reported that ferroportin expression in the duodenum and macrophages is significantly up-regulated in HCV transgenic mice [15]. However, the HCV transgenic mouse model used by Nishina et al is characterized by a lack of hepatic inflammation. Therefore, the animal model is different from patients with CHC whose serum levels of IL-6 are elevated [25]. In an attempt to shed light on the pathogenesis of iron overload in CHC, we determined the expression of duodenal ferroportin. Our results have demonstrated that the protein levels of Fpn1 in the duodenum increase in patients with CHC. This observation is consistent with presently elucidated hepcidin functions. When serum levels of hepcidin are reduced, Fpn1 in the duodenum is elevated. Increased intestinal iron absorption, at least in part, account for high levels of serum iron in patients with CHC. However, we do not exclude the possibility that high levels of serum iron are due to an increase in the protein expression of Fpn1 in reticuloendothelial macrophages in patients with CHC.
It has been well-established that CHC is associated with elevated serum iron indices and hepatic iron stores [9], [11]. Interestingly, in vitro experiments demonstrate that iron blocks HCV replication in hepatoma cell lines [26], [27]. These findings suggest that iron can exert antiviral effects. On the other hand, pathogenic bacteria require iron for survival [17], [18]. Thus, inflammatory hypoferremia has been regarded as an antibacterial defense mechanism to limit bacterial iron availability [16]–[18]. Currently, the origin of blood-associated bacteria remains unclear. Using multiple methods, such as dark-field microscopy, fluorescent in situ hybridization, and PCR-based analysis, several studies have demonstrated that a variety of bacteria exist in the normal human blood [28]–[30]. In this study, our results demonstrate that bacterial genera from control and CHC groups exhibit a similar pattern, suggesting that an overabundance of iron does not alter the bacterial communities in patients with CHC. It has been reported that Helicobacter 16S rDNA is detected in more than 60.0% of liver samples from patients with HCV using PCR methods [31]. However, we failed to detect the presence of Helicobacter species in the blood samples from patients with CHC. Thus, coinfection with Helicobacter species in patients with CHC does not seem to be common. Consistent with previous observations [28]–[30], our study indicate that Pseudomonas is the most abundant bacterial genus in blood. Although Pseudomonas genus in the CHC group are significantly higher than those in control group (Figure 4), all dominant species within this genus have no significant difference between the CHC and control groups (Table 4). These results indicate that the member of Pseudomonas genus may not have any clinical significance related to HCV infection. Consistent with significant decrease of Delftia genus in the CHC group, relative abundance of its representative species (Delftia acidovorans SPH-1) in the CHC group is significantly lower than that in the control group (Table 4). However, whether this bacterial species has the clinical significance related to HCV infection remains unclear. Taken together, this study suggests that iron overload in blood does not increase potential risk of bacterial infection in patients with CHC.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All raw 454 sequence data used in this study was deposited to the NCBI Sequence Read Archive (SRA) (accession number SRP042638) (www.ncbi.nlm.nih.gov/sra).
Funding Statement
The authors acknowledge funding from a grant (2010ZC126) from Yunnan Department of Science and Technology (to LM), and a grant (81170171) from the National Natural Science Foundation of China (to JY). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hentze MW, Muckenthaler MU, Andrews NC (2004) Balancing acts: molecular control of mammalian iron metabolism. Cell 117: 285–297. [DOI] [PubMed] [Google Scholar]
- 2. Viatte L, Lesbordes-Brion JC, Lou DQ, Bennoun M, Nicolas G, et al. (2005) Deregulation of proteins involved in iron metabolism in hepcidin-deficient mice. Blood 105: 4861–4864. [DOI] [PubMed] [Google Scholar]
- 3. Mackenzie B, Garrick MD (2005) Iron Imports. II. Iron uptake at the apical membrane in the intestine. Am J Physiol Gastrointest Liver Physiol 289: G981–986. [DOI] [PubMed] [Google Scholar]
- 4. Fraenkel PG, Traver D, Donovan A, Zahrieh D, Zon LI (2005) Ferroportin1 is required for normal iron cycling in zebrafish. J Clin Invest 115: 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, et al. (2004) Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306: 2090–2093. [DOI] [PubMed] [Google Scholar]
- 6. Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, et al. (2001) A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 276: 7811–7819. [DOI] [PubMed] [Google Scholar]
- 7. Lee P, Peng H, Gelbart T, Wang L, Beutler E (2005) Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc Natl Acad Sci U S A 102: 1906–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, et al. (2002) The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest 110: 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hezode C, Cazeneuve C, Coue O, Roudot-Thoraval F, Lonjon I, et al. (1999) Liver iron accumulation in patients with chronic active hepatitis C: prevalence and role of hemochromatosis gene mutations and relationship with hepatic histological lesions. J Hepatol 31: 979–984. [DOI] [PubMed] [Google Scholar]
- 10. Fujita N, Horiike S, Sugimoto R, Tanaka H, Iwasa M, et al. (2007) Hepatic oxidative DNA damage correlates with iron overload in chronic hepatitis C patients. Free Radic Biol Med 42: 353–362. [DOI] [PubMed] [Google Scholar]
- 11. Fujita N, Sugimoto R, Takeo M, Urawa N, Mifuji R, et al. (2007) Hepcidin expression in the liver: relatively low level in patients with chronic hepatitis C. Mol Med. 13: 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Girelli D, Pasino M, Goodnough JB, Nemeth E, Guido M, et al. (2009) Reduced serum hepcidin levels in patients with chronic hepatitis C. J Hepatol. 51: 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horl WH, Schmidt A (2013) Low hepcidin triggers hepatic iron accumulation in patients with hepatitis C. Nephrol Dial Transplant. [DOI] [PubMed]
- 14. Miura K, Taura K, Kodama Y, Schnabl B, Brenner DA (2008) Hepatitis C virus-induced oxidative stress suppresses hepcidin expression through increased histone deacetylase activity. Hepatology 48: 1420–1429. [DOI] [PubMed] [Google Scholar]
- 15. Nishina S, Hino K, Korenaga M, Vecchi C, Pietrangelo A, et al. (2008) Hepatitis C virus-induced reactive oxygen species raise hepatic iron level in mice by reducing hepcidin transcription. Gastroenterology 134: 226–238. [DOI] [PubMed] [Google Scholar]
- 16. Skaar EP (2010) The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog 6: e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Markel TA, Crisostomo PR, Wang M, Herring CM, Meldrum KK, et al. (2007) The struggle for iron: gastrointestinal microbes modulate the host immune response during infection. J Leukoc Biol 81: 393–400. [DOI] [PubMed] [Google Scholar]
- 18. Paradkar PN, De Domenico I, Durchfort N, Zohn I, Kaplan J, et al. (2008) Iron depletion limits intracellular bacterial growth in macrophages. Blood 112: 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ratziu V, Bellentani S, Cortez-Pinto H, Day C, Marchesini G (2010) A position statement on NAFLD/NASH based on the EASL 2009 special conference. J Hepatol 53: 372–384. [DOI] [PubMed] [Google Scholar]
- 20. Rapisarda C, Puppi J, Hughes RD, Dhawan A, Farnaud S, et al. (2010) Transferrin receptor 2 is crucial for iron sensing in human hepatocytes. Am J Physiol Gastrointest Liver Physiol 299: G778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Spandidos A, Wang H, Seed B (2012) PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res 40: D1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Z, Zhai H, Geng J, Yu R, Ren H, et al. (2013) Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. Am J Gastroenterol 108: 1601–1611. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Z, Geng J, Tang X, Fan H, Xu J, et al. (2013) Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J 8: 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, et al. (2004) IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest 113: 1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Migita K, Abiru S, Maeda Y, Daikoku M, Ohata K, et al. (2006) Serum levels of interleukin-6 and its soluble receptors in patients with hepatitis C virus infection. Hum Immunol 67: 27–32. [DOI] [PubMed] [Google Scholar]
- 26. Fillebeen C, Rivas-Estilla AM, Bisaillon M, Ponka P, Muckenthaler M, et al. (2005) Iron inactivates the RNA polymerase NS5B and suppresses subgenomic replication of hepatitis C Virus. J Biol Chem 280: 9049–9057. [DOI] [PubMed] [Google Scholar]
- 27. Fillebeen C, Pantopoulos K (2010) Iron inhibits replication of infectious hepatitis C virus in permissive Huh7.5.1 cells. J Hepatol 53: 995–999. [DOI] [PubMed] [Google Scholar]
- 28. Nikkari S, McLaughlin IJ, Bi W, Dodge DE, Relman DA (2001) Does blood of healthy subjects contain bacterial ribosomal DNA? J Clin Microbiol 39: 1956–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McLaughlin RW, Vali H, Lau PC, Palfree RG, De Ciccio A, et al. (2002) Are there naturally occurring pleomorphic bacteria in the blood of healthy humans? J Clin Microbiol 40: 4771–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moriyama K, Ando C, Tashiro K, Kuhara S, Okamura S, et al. (2008) Polymerase chain reaction detection of bacterial 16S rRNA gene in human blood. Microbiol Immunol 52: 375–382. [DOI] [PubMed] [Google Scholar]
- 31. Rocha M, Avenaud P, Menard A, Le Bail B, Balabaud C, et al. (2005) Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut 54: 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All raw 454 sequence data used in this study was deposited to the NCBI Sequence Read Archive (SRA) (accession number SRP042638) (www.ncbi.nlm.nih.gov/sra).