Abstract
The pancreas is both an exocrine and endocrine endodermal organ involved in digestion and glucose homeostasis. During embryogenesis, the anlagen of the pancreas arise from dorsal and ventral evaginations of the foregut that later fuse to form a single organ. In order to better understand the molecular genetics of early pancreas development, we sought to isolate markers that are uniquely expressed in this tissue. Microarray analysis was performed comparing dissected pancreatic buds, liver buds, and the stomach region of tadpole stage Xenopus embryos. A total of 912 genes were found to be differentially expressed between these organs during early stages of organogenesis. K-means clustering analysis predicted 120 of these genes to be specifically enriched in the pancreas. Of these, we report on the novel expression patterns of 24 genes. Our analyses implicate the involvement of previously unsuspected signaling pathways during early pancreas development.
INTRODUCTION
The embryonic pancreas in vertebrates forms from dorsal and ventral protrusions of the primitive gut epithelium. These pancreatic buds grow, fuse, and branch to eventually form the definitive pancreas. Like the mammalian pancreas, the pancreas of the frog Xenopus is composed of both endocrine and exocrine tissues and can be readily studied due to easy access to large numbers of externally developing embryos (Blitz et al. 2006). The exocrine tissue of both mammals and amphibians is organized into grape-like clusters of cells called acini and their associated ducts, which produce and secrete enzymes into the duodenum that function in digestion of food. Embedded within this exocrine tissue are five types of endocrine cells, which secrete hormones into the bloodstream to regulate somatic tissue uptake and hepatic release of glucose as metabolic conditions require. Endocrine cells are organized into the so-called Islets of Langerhans that produce insulin (β cells), amylin (β cells), glucagon (α cells), somatostatin (δ cells), ghrelin (ε cells), and pancreatic polypeptide (PP cells). Malfunction of the pancreas is the root cause of several diseases including diabetes mellitus, pancreatitis and pancreatic cancers. Diabetes mellitus is a metabolic disorder characterized by hyperglycemia (high blood sugar) and can be considered to break down into two subtypes. Type 1 diabetes is caused by autoimmune destruction of the insulin-producing β cells (reviewed in Devendra et al., 2004), while Type 2 diabetes is characterized by a system-wide resistance to insulin that sometimes progresses to the loss of β cell function. Type 1 diabetes is an incurable chronic condition, which can only be remedied by insulin supplementation. Thus, it has been argued that considerable benefit to patients with type 1 diabetes (and severe forms of type II diabetes) could be derived via an increased understanding of the molecular and developmental underpinning of pancreas differentiation that fosters the development of β cell replacement and/or regeneration therapies.
In recent years rapid progress has been made in understanding the molecular genetic basis of pancreas formation. The pancreas undergoes a series of developmental processes: 1) the formation of the endoderm germ layer, 2) regional specification of the endoderm to form a “pre-pancreatic” domains, 3) the formation/outgrowth of pancreatic buds and subsequent fusion of the dorsal and ventral buds, and 4) the differentiation and expansion of pancreatic tissues with endocrine, exocrine and duct cells (Murtaugh and Melton, 2003). In both zebrafish and Xenopus (teleost and tetrapod), there exists a conserved gene regulatory network controlling early endodermal development (Stainier, 2002; Koide et al., 2005; Wardle and Smith, 2006). Coordinated expression of transcription factors such as VEGT and SOX17, and the members of the MIX and GATA family genes are required for the early endoderm formation (Zhang et al., 1998; Weber et al., 2000; Xanthos et al., 2001; Lee et al., 2001; Latinkic et al., 2003; Kofron et al., 2004; Sinner et al., 2004). Signals emanating from the mesoderm control the establishment of anteroposterior and dorsoventral patterning of the endoderm. Retinoic acid, which is secreted from the anterior paraxial mesoderm is required for the proper development of pancreas in Xenopus, zebrafish and mice (Stafford and Prince 2002; Stafford et al., 2004; 2006; Martin et al., 2005; Pan et al., 2007). Signals from notochord promote the induction of dorsal pancreas (Kim et al., 1997), and purified activin βB and FGF2 mimic the notochord function (Hebrok et al., 2000). Growth factors including sonic hedgehog, Wnts, and Notch are also implicated in various processes of pancreatic development (Hebrok et al., 2000; reviewed in Kim & MacDonald, 2002).
Pancreatic cell fates become irreversibly determined through the interactions of various intracellular transcription factors. During embryogenesis, endoderm is specified by GATAs and SOX17, and subsequently the foregut endoderm, which gives rise to the liver, duodenum and pancreas, is established by the expression of FOXA2 and HNF6 (Jacquemin et al., 2000; Lee et al., 2005). The homeodomain-containing transcription factors such as HLXB9, PDX1, PTF1A participate in pancreatic bud formation and growth (Li et al., 1999; Harrison et al., 1999; Ahlgren et al., 1996; Offield et al., 1996; Kim et al., 2002, reviewed in Schwitzgebel, 2001, Kawaguchi et al, 2002). Pancreatic progenitors are specified for endocrine and non-endocrine fates by the differential expression of the transcription factor NGN3. NEUROD1 and PAX6 promote the development of endocrine progenitor differentiation. PDX1, NKX2.2 and NKX.6.1 are expressed in cells that are destined to become specific β-endocrine cells (Sander et al., 2000). The goal of the present study is to further identify signaling components and transcription factors of potential important for pancreas development.
One approach to uncover the processes of pancreas development is to establish a comprehensive transcription profile using “genomics” methodologies. Using DNA microarrays, novel regulators of β cell development and signature marker genes representing specific pancreatic cells were identified in mice (Gu et al., 2004; Chiang and Melton, 2003). In Xenopus, we have previously reported on the isolation and characterization of various organ specific endodermal markers using our homemade Xenopus neurula stage (stage 15) and tailbud stage (stage 25) cDNA microarrays (Shin et al., 2005; Park et al., 2007). In this current study, we instead utilized Xenopus Affymetrix GeneChip arrays representing over 14,400 transcripts and compared the gene expression profiles of the developing pancreas, liver, and stomach of tadpole stage (stage 42) embryos. The stage was chosen for the analyses because the pancreatic and liver buds are easily identifiable by visual inspection and thus can be isolated with minimum contamination of other nearby organs. Additionally, the pancreas of stage 42 embryos expresses genes such as Pdx1, Ptf1a, Sox9 that are critical for pancreas development, suggesting that other genes involved in pancreas development can be still isolated. Here we report on the identification and expression analysis of 24 novel Xenopus genes that are enriched in the pancreas. Our analyses implicate the involvement of previously unsuspected signaling pathways during early pancreas development.
RESULTS AND DISCUSSION
Identification of genes expressed in embryonic pancreatic buds using microarrays
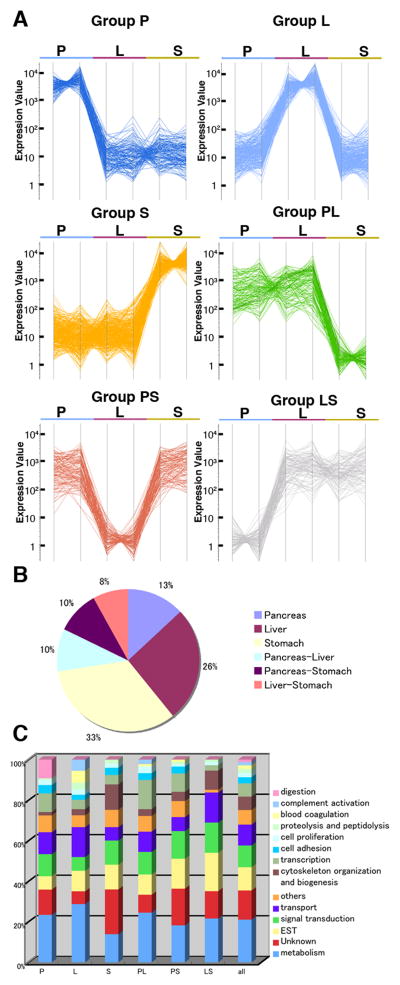
We wished to identify genes that are specifically expressed in the developing pancreas of early Xenopus embryos to better understand the process of endodermal organ differentiation. During embryonic gut formation, the liver and pancreatic rudiments bud out from the epithelium of the developing digestive tube posterior to the future stomach (Kelly and Melton 2000; Chalmers and Slack, 1998). Like the situation in mammals, the dorsal pancreatic bud originates from the gut tube underneath a portion of the notochord and the ventral pancreatic buds derive from the ventral gut near the heart and liver. These pancreatic primordia are independently induced and later fuse to form a single pancreas. We isolated the pancreas, liver, and stomach buds from stage 42 embryos (swimming tadpole stage shortly after the dorsal and ventral pancreatic buds fuse), and performed Affymetrix gene expression analyses using Xenopus laevis genome arrays. These organ buds mostly consist of the endoderm component, but also include the associated mesoderm. Experiments were performed twice, using two independently dissected tissue samples. Expressionist software (GeneData) was used to refine microarray data, including gene expression profiling analyses (see Materials and Methods). To identify genes that are differentially expressed between the pancreas, liver, and stomach, we first selected genes that passed the analysis of variance (ANOVA) with a criterion of p values of less than 0.05. Four thousand two hundred two genes satisfied this standard. Next, the expression levels of each gene in the pancreas, liver and stomach, were compared. Genes with average expression difference ratios of greater than three-fold were selected for further analyses. After manually removing genes that are redundantly represented in the Affymetrix chips, 912 genes were subjected to subsequent analyses (see Supplementary Table 1). These genes were separated into six expression groups based on K-means clustering analysis (Fig. 1A). Group P contains 120 genes that are specifically enriched in the pancreas. Group L contains 237 genes preferentially expressed in the liver, and 303 genes in Group S are enriched in the stomach. Groups PL, PS and LS contain the genes that are enriched in both the pancreas and liver but not stomach (90 genes), pancreas and stomach but not liver (88 genes), and liver and stomach but not pancreas (74 genes), respectively (Fig. 1A and B). To identify the most closely related human orthologs we performed BLAST searches, followed by reciprocal BLAST analyses. The homologous genes thus represent the best bidirectional BLAST hit between Xenopus laevis and human and were then assigned official gene symbols and full names of their human counterparts (Supplementary Table 1). Whenever a Xenopus gene encodes a putative protein but fails to identify a human ortholog, “not available” (“N/A”) was assigned. Other remaining Xenopus genes were simply listed by EST identification numbers.
Fig. 1.
Clustering analysis of genes expressed in the embryonic pancreas, liver, and stomach. (A) Six expression groups generated by K-means clustering analysis. Individual line represents the expression level of a given gene in the pancreas (P), liver (L) or stomach (S). Abbreviations: PL, Pancreas and liver; PS, pancreas and stomach; LS, liver and stomach. (B) A chart indicating the percentages of six groups, which are represented by a total of 912. (C) Percentages of highly represented GO terms for each cluster.
We examined the biological properties of the six clustering groups by examining Gene Ontology (GO) terms associated with genes in each cluster (Suppl Table 1, Fig. 1C). Among the 912 genes, the most frequently observed GO term is linked to metabolism (21%), followed by signal transduction (11%), transport (10%), cytoskeletal organization and biogenesis (7%), transcription (7%), cell adhesion (3%), cell proliferation (2%), proteolysis and peptidolysis (2%), blood coagulation (2%), complement activation (2%), and digestion (1%). A GO term of “digestion” was represented at 9% in Group P, in contrast to 1% among the 912 pancreas/liver/stomach genes. Since the exocrine pancreas secretes numerous digestive enzymes into the duodenum, this finding is consistent with a known role of the pancreas. In the embryonic liver (Group L), “blood coagulation” and “complement activation” represented 6% and 5%, respectively. This is consistent with the liver’s important role in hematopoiesis and as the major producer of complement activation proteins to regulate inflammatory processes. Group S is enriched in genes involved in “cytoskeleton organization and biogenesis” (13%) including actin and myosin, in contrast to the other two groups (2% for Group P and 3% for Group L), which may be linked to the contractile action of the stomach, requiring enhanced musculature.
Based on this annotation, we first compared the gene lists in each clustered subgroup against the published literature to corroborate that both our organ bud dissections did indeed enrich for organ-specific mRNAs, and our K-means clustering analysis correctly assigned these into appropriate subgroups (see Table 1). Group P genes included AMY2A, CEL, CLDN6, CPA1, DNASE1, ELA1, ELA2A, HLXB9, INS, PDIA2, and PRSS2, which are known to be expressed in the pancreas (Afelik et al., 2004; Sogame et al., 2003, Horb and Slack, 2002; Kelly and Melton, 2000; Chen et al., 2004; Park et al., 2007). Similarly in Group L, we found genes associated with liver development, which are A2M, ALB, AMBP, BF, FGG, HHEX, KNG1, TBX2 and TF (Zorn and Mason, 2001, Park et al., 2007). Group S included ATP4B, CTSE, SOX2, and STARD10, which are expressed in the stomach (Park et al, 2007). XBP1 is expressed in both the liver and pancreas (Park et al., 2007) and belongs to Group PL. PAX6 is expressed in both the pancreas and intestine of stage 45 embryos (Kelly and Melton, 2000) and is assigned to Group PS. Based on these results, we concluded that our dissections/microarray data are of high quality and that the clustering analysis successfully separated the 912 genes into appropriate expression groups.
Table 1.
Partial list of known marker genes identified in this study.
| Gene symbol | Gene description | Biological process | Group | Xenopus Unigene ID | References |
|---|---|---|---|---|---|
| AMY2A | amylase, alpha 2A; pancreatic | digestion | P | Xl.21603 | Horb et al., 2002 |
| CEL | carboxyl ester lipase (bile salt-stimulated lipase) | digestion | P | Xl.24591 | Sogame et al., 2003 |
| CLDN6 | claudin 6 | cell adhesion | P | Xl.4725 | Park et al., 2007 |
| CPA1 | carboxypeptidase A1 (pancreatic) | digestion | P | Xl.26019 | Kelly and |
| DNASE1 | deoxyribonuclease I | digestion | P | Xl.932 | Sogame et al., 2003 |
| ELA1 | elastase 1, pancreatic | digestion | P | Xl.24478 | Horb et al., 2002 |
| ELA2A | elastase 2A | digestion | P | Xl.23779 | Sogame et al., 2003 |
| HLXB9 | homeo box HB9 | transcription | P | Xl.286 | Chen et al., 2004 |
| INS | insulin | signal transduction | P | Xl.817 | Kelly and Melton, 2000; |
| PDIA2 | protein disulfide isomerase-associated 2 | P | Xl.24424 | Sogame et a | |
| PRSS2 | protease, serine, 2 (trypsin 2) | digestion | P | Xl.119 | Horb et al., 2002 |
| A2M | alpha-2-macroglobulin | transport | L | Xl.24285 | Park et al., 2007 |
| ALB | albumin | transport | L | Xl.395 | Zorn and Mason, 2001 |
| AMBP | alpha-1-microglobulin/bikunin precursor | anti-inflammatory response | L | Xl.26215 | Zorn and Mason, 2001 |
| BF | B-factor, properdin | complement activation | L | Xl.5152 | Park et al., 2007 |
| FGG | fibrinogen, gamma polypeptide | blood coagulation | L | Xl.5168 | Zorn and Mason, 2001 |
| HHEX | hematopoietically expressed homeobox | transcription | L | Xl.225 | Zorn and Mason, 2001 |
| KNG1 | kininogen 1 | blood coagulation | L | Xl.10766 | Park et al., 2007 |
| TBX2 | T-box 2 | transcription | L | Xl.931 | Park et al., 2007 |
| TF | transferrin | transport | L | Xl.534 | Zorn and Mason, 2001 |
| ATP4B | ATPase, H+/K+ exchanging, beta polypeptide | transport | S | Xl.241 | unpublished |
| CTSE | cathepsin E | proteolysis and peptidolysis | S | Xl.35410 | unpublished |
| STARD10 | START domain containing 10 | transport | S | Xl.2843 | Park et al., 2007 |
| SOX2 | SRY (sex determining region Y)-box 2 | transcription | S | Xl.188 | Park et al., 2007 |
| XBP1 | X-box binding protein 1 | transcription | PL | Xl.4624 | Park et al., 2007 |
| PAX6 | paired box gene 6 (aniridia, keratitis) | transcription | PS | Xl.647 | Kelly and Melton, 2000 |
Overview of our findings
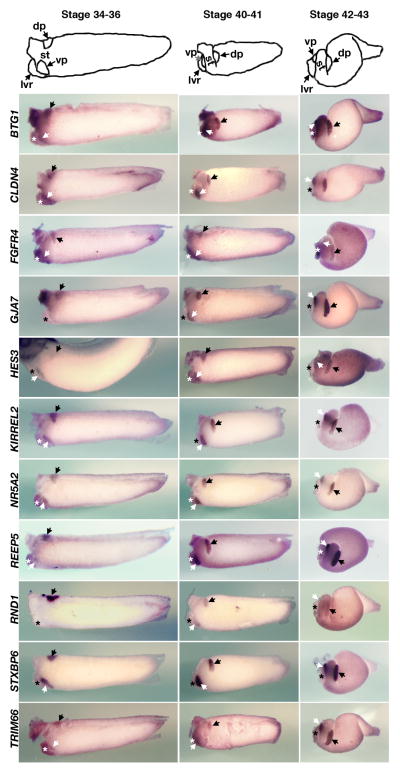
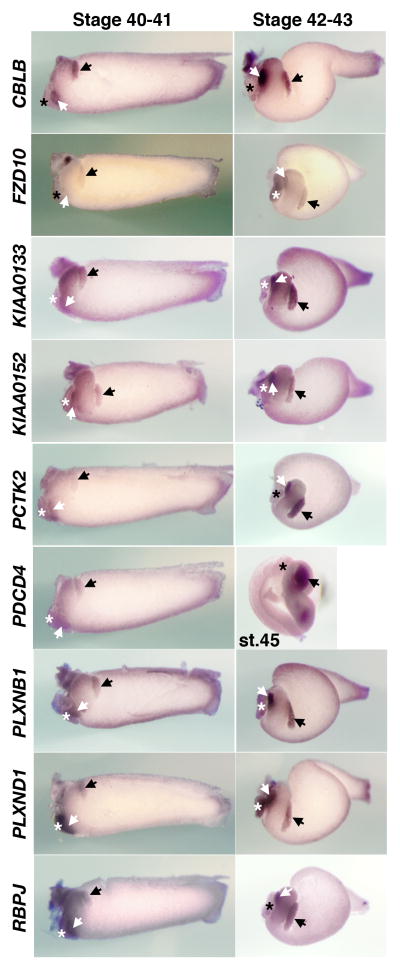
Our current microarray analyses show that many of the Xenopus genes found to be expressed in the stomach, liver, and pancreas have clear orthologs in human and other species, although their specific roles in each organ have not been recognized previously. We also wished to confirm the findings of the microarray data by whole mount in situ hybridization. Among the 120 pancreatic (group P) genes, 24 previously uncharacterized candidate genes were selected based on the following criteria. Eighteen genes were selected based on three GO terms, “transcription”, “signal transduction”, and “cell adhesion”. In addition, based on the array results 6 genes expressing at high levels in the pancreas were also chosen from the PL and PS groups because they may serve as useful marker genes in the future. The results of whole mount in situ hybridizations performed on dissected guts of tadpole stage embryos are shown in Figures 2 through 4, and information on genes previously not reported to be expressed in the pancreas is listed in Table 2. We also report on the expression patterns of these genes in whole embryos (Suppl Figures 1–5). We note that while most of the in situ hybridization patterns are consistent with the predicted patterns based on the K-means clustering analysis, in some cases in situ patterns do not match with the clustering classification. This discrepancy may be attributed to the fact that while we isolated RNA from dissections performed at a single stage (stage 42), we examined gene expression patterns by whole mount in situ hybridization at several different stages.
Fig. 2.
Expression of novel pancreatic genes in dissected guts of tadpole stage embryos. Gene expression patterns were revealed by whole mount in situ hybridization at three representative stages, stage 34–36, stage 40–41, and stage 42–43. These genes are expressed in the pancreatic rudiments of stage 34–36 embryos. The dorsal and ventral pancreatic buds are indicated with the black and white arrows, respectively and the liver bud is marked by an asterisk. A whole embryo was shown for HES3. While the Xenopus laevis gene designated here as HES3 (also known as Xenopus ESR2) has a best blastP match to human HES5, blastN to X. tropicalis and synteny relations identifies it as a gene distinct from tropicalis hes5 called hes3. Anterior to the left, dorsal to the top.
Fig. 4.
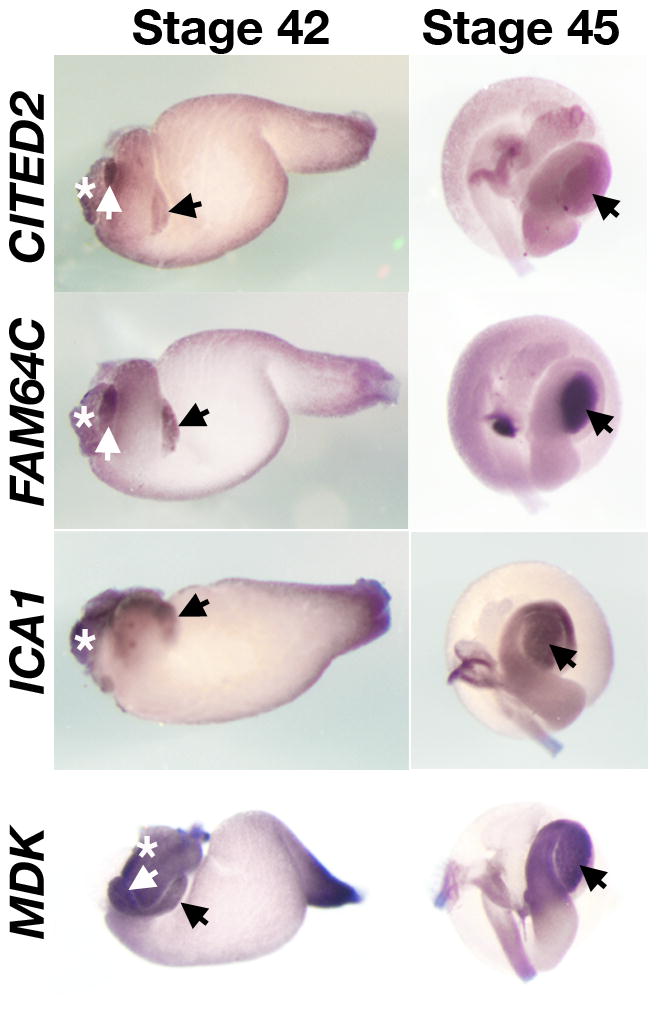
Expression of pancreatic genes in the gut of stage 42 and 45 embryos. The black arrows at stage 45 indicate the pancreas after the fusion of dorsal and ventral pancreas and the liver bud is marked by an asterisk.
Table 2.
List of novel Xenopus pancreatic genes identified in this study
| Gene symbol | Gene description | Biological process | Group | Xenopus Unigene ID | Clone |
|---|---|---|---|---|---|
| BTG1 | B-cell translocation gene 1, anti-proliferative | transcription | P | Xl.25750 | XL005n04 |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | protein ubiquitination | P | Xl.17445 | XL013i01 |
| CITED2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | transcription | PL | Xl.25598 | XL019i09 |
| CLDN4 | claudin 4 | cell adhesion | P | Xl.23306 | XL001g02 |
| FAM46C | family with sequence similarity 46, member C | unknown | PL | Xl.4393 | XL197e22 |
| FGFR4 | fibroblast growth factor receptor 4 | signal transduction | PL | Xl.1016 | XL070o16 |
| FZD10 | frizzled homolog 10 (Drosophila) | signal transduction | P | Xl.559 | XL048i11 |
| GJA7 | gap junction protein, alpha 7, 45kDa (connexin 45) | transporter | P | Xl.3337 | XL038c12 |
| HES3 | hairy and enhancer of split 3 (Drosophila) | transcription | P | Xl.12067 | XL031a05 |
| ICA1 | islet cell autoantigen 1, 69kDa | transporter | PS | Xl.70385 | XL183c20 |
| KIAA0133 | KIAA0133 | unknown | P | Xl.13330 | XL045a04 |
| KIAA0152 | KIAA0152 | unknown | P | Xl.2971 | XL075f01 |
| KIRREL2 | kin of IRRE like 2 (Drosophila) | cell adhesion | P | Xl.76492 | XL088m05 |
| MDK | midkine (neurite growth-promoting factor 2) | signal transduction | PS | Xl.897 | XL091d20 |
| NR5A2 | nuclear receptor subfamily 5, group A, member 2 | signal transduction | P | Xl.12129 | XL106i11 |
| PCTK2 | PCTAIRE protein kinase 2 | cell cycle | P | Xl.55323 | XL058l19 |
| PDCD4 | programmed cell death 4 (neoplastic transformation inhibitor) | apoptosis | P | Xl.14956 | XL214b12 |
| PLXNB1 | plexin B1 | signal transduction | P | Xl.72590 | XL084f22 |
| PLXND1 | plexin D1 | signal transduction | PL | Xl.18576 | XL073a19 |
| RBPJ | recombining binding protein suppressor of hairless (Drosophila) | transcription | P | Xl.12006 | XL202i12 |
| REEP5 | receptor accessory protein 5 | signal transduction | P | Xl.8593 | XL174p24 |
| RND1 | Rho family GTPase 1 | signal transduction | P | Xl.517 | XL001c12 |
| STXBP6 | syntaxin binding protein 6 (amisyn) | transporter | P | Xl.5823 | XL004n21 |
| TRIM66 | tripartite motif-containing 66 | transcription | P | Xl.24102 | XL084d03 |
The in situ data are presented in Figures 2–4 according to the timing of earliest onset of each genes expression. Among the 24 genes selected based on the above criteria, we find that ten genes (BTG1, CLDN4, FGFR4, GJA7, HES3, KIRREL2, NR5A2, REEF5, STXBP6, and TRIM66) are initially expressed in stage 35 embryos in both dorsal and ventral pancreatic buds, with the exception of RND1, which is first detected in the dorsal pancreas and later becomes expressed in the ventral pancreas (Figure 2). We originally identified FGFR4 to be expressed in both the pancreas and the liver based on microarray data. However whole mount in situ hybridization of stage 35 embryos revealed that this gene is initially detected in the dorsal and ventral pancreatic buds, but later is expressed in other tissues. During subsequent stages, morphogenetic movements of the endoderm rotate the dorsal and ventral pancreatic buds toward one another on the left side of the trunk (compare stage 35 to stages 40–43). The pancreatic buds fuse with one another medial to the stomach by stage 40 (Kelly and Melton, 2000). Figure 3 shows expression of nine other genes (CBLB, FZD10, KIAA0133, KIAA0152, PCTK2, PDCD4, PLXNB1, PLXND1, and RBPJ) that are initially detected in the dorsal and ventral pancreas of stage 40 embryos. Additionally, several genes belonging to the PL group (CITED2, FAM46C) and the PS group (ICA1, MDK) are detected subsequently (st 42, Figure 4).
Fig. 3.
Expression of novel pancreatic genes in the gut of stage 40–43 embryos. The dorsal and ventral pancreatic buds are indicated with the black and white arrows, respectively and the liver bud is marked by an asterisk. PDCD4 expression shown is a stage 45 embryo,
Genes expressed in the pancreas
RND1- and PLXNB1-mediated signaling
We detected specific expression of RND1 and PLXNB1 in the developing pancreas of stage 35 and stage 40 embryos, respectively (Figures 2 and 3). Since both PLXNB1 and RND1 physically interact to regulate cell contraction in cell culture assays (Oinuma et al., 2003), it is tempting to propose the involvement of a RND1-PLXNB1 signaling pathway in pancreas morphogenesis. Recently, RND1 was also shown to interact with FLRT3 (Fibronectin Leucine-rich Repeat Transmembrane 3), a type I transmembrane protein containing extracellular leucine-rich repeats (Ogata et al., 2007). FLRT3 and RND1 physically interact to modulate cell adhesion during early embryogenesis by controlling the cell surface levels of cadherin through regulated endocytosis. RND1- and PLXNB1-mediated signaling may play roles in coordinating pancreatic morphogenesis.
KIRREL2 in pancreas development
The KIRREL gene family encodes transmembrane proteins with 5 Immunoglobulin (Ig) -like domains (Sun et al., 2003), suggesting possible roles as receptors or cell adhesion molecules. At stage 35 and afterward, KIRREL2 is expressed both in the ventral and dorsal pancreas (Figure 2 and suppl Figure 3) but not before. At stage 25 KIRREL expression is also detected in the pronephros, the brain, eyes, and the posterior tip of the notochord of early tailbud stage embryos (suppl Figure 3) and the expression persists until stage 40. In human, there are three KIRREL genes (KIRREL, KIRREL2, and KIRREL3), and KIRREL2 is expressed specifically in β cells of human adult pancreas (Sun et al., 2003). While the Xenopus KIRREL family member discussed here is the ortholog to human KIRREL2, it is yet to be determined whether Xenopus KIRREL2 is similarly expressed in the β cells of adult Xenopus pancreas.
FGF signaling in pancreas development
FGF signaling has been implicated in mediating epithelial-mesenchymal interactions in the developing pancreas (Miralles et al., 1999) and promotes the expansion of pancreatic progenitor cells (Papadopoulou and Edlund, 2005). We identified FGFR4 expression in both the liver and pancreas of developing Xenopus (Figure 2). This finding is consistent with a previous report that FGFR4 is expressed in the liver and the endodermal epithelium of the E12–14 mouse pancreas (Stark et al., 1991). In mice, FGF10 has been implicated in the normal growth of early pancreas buds (Bhushan et al., 2001), and the maintenance of PTF1A expression in the dorsal pancreatic bud (Jacquemin et al., 2006). Misexpression of FGF4 in the early pancreas using the PDX1 promoter was shown to disrupt pancreas development (Dichmann et al., 2003). In zebrafish, FGF10 and FGF24 are shown to display redundant activity in patterning the pancreatic lateral plate mesoderm (Manfroid et al., 2007).
FZD10 and WNT signaling in pancreas development
While numerous studies have demonstrated roles for Wnt signaling in pancreas development, the precise functions of this pathway are yet to be determined. Gene targeting of β-catenin in the pancreas using PDX1-Cre recombinase suggests that s-catenin is required for development of the exocrine pancreas but not the endocrine cells (Dessimoz et al., 2005; Murtaugh et al., 2005; Wells et al., 2007). In a more recent study, Wnt signaling has been implicated in islet β cell proliferation (Rulifson et al., 2007). Our clustering analysis (Suppl Table 1) shows that FZD10 belongs to Group P. However, whole mount in situ hybridization revealed that FZD10 is weakly expressed in the liver as well (Figure 3). FZD10 was previously shown to mediate canonical Wnt signaling in the presence of Wnt7b and LRP5 (Wang et al., 2005).
Hedgehog signaling in pancreas development
Hedgehog (Hh) signaling likely plays multiple roles in pancreas development. Inhibition of sonic hedgehog (Shn) signaling in the endoderm is crucial for specification of the dorsal pancreas domain (Hebrok et al., 1998; Kim and Melton, 1998). Furthermore, appropriate levels of Hh signaling also appear to regulate cell proliferation/organ size, morphogenesis, and the function of pancreatic tissue at the later stages of development (Hebrok et al., 2000). Our clustering analysis (Suppl Table 1) has grouped the Hh ligands (Shh and Dhh) into Group S and grouped the Hh receptors (PTCH2, and SMO) into Group LS, and therefore these signaling components are lacking in the early pancreas, consistent with the observation that the inhibition of Hh signaling is important for early pancreas development.
RBPJ and HES3: Notch signaling
Loss-of-function experiments have shown that the Notch signaling pathway promotes the self-renewal of pancreatic progenitor cells and/orexocrine cell lineage commitment (Apelqvist et al., 1999, Jensen et al., 2000). We find that notch pathway genes such as HES3 (a Xenopus gene closely related to human HES5; Figure 2) and RBPJ (a nuclear component of the Notch signaling pathway; Figure 3), are expressed in the Xenopus pancreas, whereas DLL1 (a Notch/Delta ligand) is expressed in the stomach (Suppl Table 1). ESR10, another enhancer of split related gene closely related to HES3 and HES5, was previously shown to be expressed in the Xenopus pancreas (Chen et al., 2004). These data are consistent with the notion that Notch/Delta signaling and its downstream components such as HES3/5 and ESR10 play important roles in pancreas development.
Other novel genes expressed in developing pancreas
CITED2
CBP/p300-interacting transactivator has a Glu/Asp-rich carboxy-terminal domain 2 and encodes a transcriptional modulator. Knockout of the CITED2 gene results in embryonic lethality with heart and neural tube defects (Yin et al., 2002; Weninger et al., 2005; Chen et al., 2007). CITED2 is also required for mouse fetal liver development (Qu et al., 2007) and our microarray data suggest that it is expressed in the developing embryonic liver. While expression of CITED2 has been reported in the heart and the kidney of mouse and chick embryos (Dunwoodie et al., 1998; Schlange et al., 2000; Boyle et al., 2007), our finding of its expression in the pancreas (Figure 4) is novel.
PCTK2
PCTAIRE protein kinase 2 is a Cdc2-related protein kinase and is known to be expressed in terminally-differentiated neurons (Hirose et al., 1997). However, its expression in the pancreas (Figure 3) has not yet been reported. Since a related protein, PCTK1, was shown to play a role in membrane trafficking from endoplasmic reticulum to Golgi (Palmer et al., 2005), we speculate that PCTK2 might be involved in the similar secretory pathway during pancreas development.
REEP5
Receptor accessory protein 5 is a member of the Receptor Expression Enhancing Protein (REEP) family. REEP1 is involved in cell surface expression of odorant receptors that belong to the G-protein coupled receptor (GPCR) superfamily (Saito et al., 2004). Specific expression of REEP5 in the pancreas (Figure 2) has not been reported previously.
TRIM66
Tripartite motif containing 66, also known as transcriptional intermediary factor 1 delta (TIF1delta), has been implicated in heterochromatin-mediated gene silencing (Khetchoumian et al., 2004). TRIM66 was reported to be expressed in testes (Khetchoumian et al., 2004), but its expression in the pancreas (Figure 2) has not been described.
STXBP6
Syntaxin binding protein 6, also known as amisyn, was isolated as a syntaxin-binding protein enriched in brain (Scales et al., 2002) and is thought to act as an inhibitor of exocytosis (Constable et al., 2005). STXBP6 is also expressed in the cement gland, an amphibian-specific transient structure that is notable for being highly secretory. STXBP6 is also expressed in cranial ganglia before mid-tailbud stage (Suppl Figure 5). At the mid-tailbud stage (stage 30–31), STXBP6 expression is first detected in the ventral pancreas (Suppl Figure 5), and later (stage 35) in both pancreatic buds (Figures 2 and Suppl Figure 5). At stage 40, STXBP6 expression is maintained in the cement gland, cranial ganglia, and pancreas (Figure 2 and Suppl Figure 5).
BTG1
B-cell translocation gene 1 is a member of the family of antiproliferative genes. BTG1 expression is maximal at the G0/G1 phases of the cell cycle and decreases when cells progress through G1 (Rouault et al., 1992). It interacts with a variety of transcription factors including retinoic acid and estrogen receptors, MyoD, and CAF1 (CCR4 associated factor 1), to regulate cellular differentiation (Busson et al., 2005; Prevot et al., 2001). While BTG1 is expressed in the adult pancreas, and weakly in the stomach and the liver (Prevot et al., 2001), the function of BTG1 in the embryonic pancreas (Figure 2) is unknown.
CBLB
Cas-Br-M (murine) ecotropic retroviral transforming sequence b is an E3 ubiquitin ligase and is expressed in the embryonic pancreas (Figure 3). CBLB is involved in the negative regulation of autoimmunity (Bachmaier et al., 2000; Chiang et al., 2000) and has been suggested as a susceptibility gene for Type 1 diabetes based on its association with autoimmunity in animal models and its role in T-cell co-stimulatory signaling (Yokoi et al., 2002). However, no naturally occurring mutations of CBLB have so far been linked to type 1 diabetes in human (Kosoy et al., 2004).
GJA7 and PLXND1
Gap junction protein alpha 7, also known as connexin 45, is a component of gap junctions. Its expression has been described in vascular cells of pancreatic islets (Theis et al., 2004) and is expressed in the embryonic Xenopus pancreas (Figure 2). GJA7-deficient mice have vascular defects and die between E9.5 and E10.5 (Kruger et al., 2000). Plexin D1 (PLXND1) is also involved in vascular patterning (Torres-Vazquez et al., 2004), and we find that it is expressed in both the developing liver and pancreas (Figure 3). GJA7 and PLXND1 may have their roles in vascular patterning during pancreas development.
ICA1
Islet Cell Autoantigen 1 is an arfaptin-related protein associated with membrane trafficking at the Golgi complex and immature secretory granules in neurosecretory cells (Spitzenberger et al., 2003). ICA1 is also known as ICA69, and is a candidate gene in autoimmunity associated with type I diabetes (Pietropaolo et al., 1993). The expression is particularly strong in adult pancreatic tissues (Mally et al., 1996), but we also find that 1CA1 is expressed during embryonic pancreas development (Figure 4).
MDK
Midkine is a heparin-binding growth factor involved in many biological processes including angiogenesis, cancer, and neural development (reviewed in Kadomatsu and Muramatsu, 2004). MDK has also been implicated in diabeticnephropathy by accelerating the intracellular signaling network evoked by hyperglycemia (Kosugi et al., 2006). We find that MDK is expressed in the pancreas and the stomach but not in the liver of Xenopus (Figure 4), consistent with the previous observations in mouse embryos (Kadomatsu et al., 1990). While the expression of MDK is up-regulated in pancreatic cancer (Aridome et al., 1995), its role during pancreas development is unclear.
NR5A2
Nuclear Receptor subfamily 5, group A, member 2 is a Ftz-F1-related nuclear orphan receptor B and expressed in the pancreas and the liver (Figure 2), and the similar expression patterns were observed in mice (Annicotte et al. 2003; Fayard et al., 2003). PDX1 controls the expression of NR5A2, which in turn controls the expression of CEL (Carboxyl Ester Lipase) (Fayard et al., 2003). In Xenopus, NR5A2 has been suggested to be a transcriptional activator (Ellinger-Ziegelbauer et al., 1994), but its exact function and spatiotemporal expression studies during organogenesis have not been examined. While NR5A2 deficient mice have been generated, its in vivo role during pancreas development has not been reported (Pare et al., 2004).
PDCD4
Programmed Cell Death protein 4 is a recently discovered tumorsuppressor protein that inhibits protein synthesis by interfering with the assembly of the cap-dependent translation initiation complex. (Cmarik et al., 1999). In connection with the pancreas, PDCD4 was shown to suppress the expression of carbonic anhydrase type II. Since tumor cells require a high bicarbonate flux for their growth, carbonic anhydrase suppression results in growth inhibition of endocrine tumors such as insulinomas and glucagonomas (Lankat-Buttgereit et al., 2004). PDCD4 expression during pancreas development (Figure 3) has not been reported in other animals, though PDCD4 seems to be expressed in a wide variety of tissues.
Other genes with unknown function
FAM46C (Family with sequence similarity 46, member C), KIAA0133, and KIAA0152 (Figures 3 and 4) are evolutionarily conserved genes, although their functions are yet to be determined. KIAA0152 is expressed in human pancreatic islets (Ylipaasto et al., 2005).
Why were some other known pancreas markers not in the list?
In Xenopus, transcription factors such as SOX9, PTF1A/P48, PDX1 (XLHBOX8) are all expressed in the developing pancreas, but these are not included in our gene lists. These genes were inaccessible to our microarray analysis as PTF1A and PDX1 were not included in the Xenopus Affymetrix version 1.0 microarray chip design. In the case of SOX9, the expression level in the pancreas was high, however its expression values in the liver and the stomach varied significantly between replicate experiments. Therefore, these data did not pass the standards we set for our ANOVA analysis (our cutoff was a p value ≤ 0.05).
Synexpression group genes
The term “synexpression groups” was coined to describe functionally related genes that share similar spatial expression patterns (Niehrs and Pollet, 1999). Thus, it was proposed that tissue specific patterns of coexpression of genes may be a predictor for genes that work together in a common pathway or toward a common goal. Interestingly, many genes involved in steps of early neural patterning (e.g., NKX2.2, NKX6.1, NGN3, NEUROD, PAX6, PAX4) are also expressed in the pancreas, and this observation has led to the notion that neural and pancreatic genes/tissues may share a functional similarity, perhaps in regulating a group of genes controlling secretory function. Our current Xenopus situ hybridization study agrees with this view as a majority of the genes expressed in the Xenopus pancreas are also expressed in the brain (71 %, n=24). Along this theme, we also report a synexpression group of pancreas genes (BTG1, CLDN4, FZD10, GJA7, KIRREL2, RBPJ, and RND1) that are coexpressed at the tip of growing tail buds (Suppl Figures 1–3, and Suppl Figure 5).
Usefulness of the Xenopus system to study organ development
Xenopus has so far been under-utilized as a means of studying the processes of organogenesis (Blitz et al., 2006). However, among the advantages offered by Xenopus that could contribute significantly to a better understanding of the development of organs such as the pancreas is the provision of a fresh set of molecular markers. Availability of such markers would be useful to better define the developmental processes of differentiation, in this case, leading to the formation of a functional pancreas during embryogenesis. More significantly, identification of such markers could aid in the development of novel diagnostic tools for the detection of pancreatic diseases in humans and in manipulating the growth and differentiation of human and mouse stem cells to develop cures for pancreatic disorders such as diabetes mellitus. Since this strategy was successful in identifying genes expressed in the pancreas, we predict that in situ hybridizations for genes belonging to Groups L and S will similarly reveal new markers for liver and stomach development.
EXPERIMENTAL PROCEDURES
Isolation of endodermal organ buds
Xenopus laevis embryos were fertilized in vitro, dejellied in 2% cysteine hydrochloride (pH 7.8) and maintained in 0.1X Modified Barth’s Saline (MBS) (pH 7.4). At stage 42, embryos were anesthetized in 0.01% ethyl 3-aminobenzoate methanesulfonate (Sigma, A5040) in 0.1X MBS. Then, the embryos were transferred to 1X MBS containing 0.01% ethyl 3-aminobenzoate methanesulfonate. Using watchmakers forceps the gut was removed from the rest of the tadpole tissues, and the pancreas, liver, and stomach buds were isolated and subjected to RNA isolation using TRIZOL reagent (Invitrogen).
GeneChip analyses
Total RNAs isolated from the endodermal organs were subjected to microarray analyses using Xenopus laevis Genome Arrays (Affymetrix, version 1.0). Two biologically independent experiments were carried out. Microarray data were refined and analyzed using Expressionist software (GeneData, Inc.). Analysis of variance (ANOVA) was performed to identify statistically significant genes (with p values less than 0.05) that are expressed differentially between the pancreas, liver, and stomach. Six gene groups representing enrichment in pancreas (P), liver (L), stomach (S), pancreas and liver (PL), pancreas and stomach (PS), liver and stomach (LS) were generated by k-means clustering.
Whole mount In situ hybridization
Whole mount in situ hybridization was carried out essentially as described previously (Harland, 1991). Embryos at various stages were fixed in MEMFA, washed in methanol, and stored in ethanol at −20°C. For the preparation of gut tissue, the anterior and posterior regions of embryos were removed by cutting transversely at the levels of heart and proctodeum, from which the ectodermal layer, notochord and somites were removed. cDNA clones were obtained from a Xenopus EST cDNA library (kind gift of Dr. Naoto Ueno, NIBB). To generate anti-sense in situ hybridization probes, cDNA inserts were PCR-amplified using M13 forward and reverse primers, and DIG-labeled RNA probes were directly generated using T7 RNA polymerase.
Supplementary Material
Supplementary Figures 1–5. Expression patterns of genes identified in this study in whole embryos at tailbud and tadpole stages.
Supplementary Table 1. Microarray data for genes described in this study. Column headings are described further in the Excel file data sheet named Explanation of Columns.
Acknowledgments
We thank Dr. Naoto Ueno for providing us with Xenopus EST cDNA library. This work was supported by NIH grants to K.W.Y.C.
References
- Afelik S, Chen Y, Pieler T. Pancreatic protein disulfide isomerase (XPDIp) is an early marker for the exocrine lineage of the developing pancreas in Xenopus laevis embryos. Gene Expr Patterns. 2004;4:71–76. doi: 10.1016/s1567-133x(03)00150-9. [DOI] [PubMed] [Google Scholar]
- Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Annicotte JS, Fayard E, Swift GH, Selander L, Edlund H, Tanaka T, Kodama T, Schoonjans K, Auwerx J. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol Cell Biol. 2003;23:6713–6724. doi: 10.1128/MCB.23.19.6713-6724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Aridome K, Tsutsui J, Takao S, Kadomatsu K, Ozawa M, Aikou T, Muramatsu T. Increased midkine gene expression in human gastrointestinal cancers. Jpn J Cancer Res. 1995;86:655–661. doi: 10.1111/j.1349-7006.1995.tb02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmaier K, Krawczyk C, Kozieradzki I, Kong YY, Sasaki T, Oliveira-dos-Santos A, Mariathasan S, Bouchard D, Wakeham A, Itie A, Le J, Ohashi PS, Sarosi I, Nishina H, Lipkowitz S, Penninger JM. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Andelfinger G, Horb ME. Germ layers to organs: Using Xenopus to study “later” development. Seminar in Cell Dev Biol. 2006;17:133–145. doi: 10.1016/j.semcdb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Boyle S, Shioda T, Perantoni AO, de Caestecker M. Cited1 and Cited2 are differentially expressed in the developing kidney but are not required for nephrogenesis. Dev Dyn. 2007;236:2321–2330. doi: 10.1002/dvdy.21242. [DOI] [PubMed] [Google Scholar]
- Busson M, Carazo A, Seyer P, Grandemange S, Casas F, Pessemesse L, Rouault JP, Wrutniak-Cabello C, Cabello G. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene. 2005;24:1698–1710. doi: 10.1038/sj.onc.1208373. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Slack JM. Development of the gut in Xenopus laevis. Dev Dyn. 1998;212:509–521. doi: 10.1002/(SICI)1097-0177(199808)212:4<509::AID-AJA4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pan FC, Brandes N, Afelik S, Solter M, Pieler T. Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Dev Biol. 2004;271:144–160. doi: 10.1016/j.ydbio.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Chen Y, Haviernik P, Bunting KD, Yang YC. Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood. 2007;110:2889–2898. doi: 10.1182/blood-2007-01-066316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ, Jang IK, Gutkind JS, Shevach E, Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- Chiang MK, Melton DA. Single-cell transcript analysis of pancreas development. Dev Cell. 2003;4:383–93. doi: 10.1016/s1534-5807(03)00035-2. [DOI] [PubMed] [Google Scholar]
- Cmarik JL, Min H, Hegamyer G, Zhan S, Kulesz-Martin M, Yoshinaga H, Matsuhashi S, Colburn NH. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc Natl Acad Sci U S A. 1999;96:14037–14042. doi: 10.1073/pnas.96.24.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constable JR, Graham ME, Morgan A, Burgoyne RD. Amisyn regulates exocytosis and fusion pore stability by both syntaxin-dependent and syntaxin-independent mechanisms. J Biol Chem. 2005;280:31615–31623. doi: 10.1074/jbc.M505858200. [DOI] [PubMed] [Google Scholar]
- Dessimoz J, Bonnard C, Huelsken J, Grapin-Botton A. Pancreas-specific deletion of beta-catenin reveals Wnt-dependent and Wnt-independent functions during development. Curr Biol. 2005;15:1677–1683. doi: 10.1016/j.cub.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Devendra D, Liu E, Eisenbarth GS. Type 1 diabetes: recent developments. BMJ. 2004;328:750–754. doi: 10.1136/bmj.328.7442.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichmann DS, Miller CP, Jensen J, Scott Heller R, Serup P. Expression and misexpression of members of the FGF and TGFbeta families of growth factors in the developing mouse pancreas. Dev Dyn. 2003;226:663–674. doi: 10.1002/dvdy.10270. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Rodriguez TA, Beddington RS. Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Hihi AK, Laudet V, Keller H, Wahli W, Dreyer C. FTZ-F1-related orphan receptors in Xenopus laevis: transcriptional regulators differentially expressed during early embryogenesis. Mol Cell Biol. 1994;14:2786–2797. doi: 10.1128/mcb.14.4.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayard E, Schoonjans K, Annicotte JS, Auwerx J. Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J Biol Chem. 2003;278:35725–35731. doi: 10.1074/jbc.M302370200. [DOI] [PubMed] [Google Scholar]
- Gu G, Wells JM, Dombkowski D, Preffer F, Aronow B, Melton DA. Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development. 2004;131:165–179. doi: 10.1242/dev.00921. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, St Jacques B, McMahon AP, Melton DA. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- Hirose T, Tamaru T, Okumura N, Nagai K, Okada M. PCTAIRE 2, a Cdc2-related serine/threonine kinase, is predominantly expressed in terminally differentiated neurons. Eur J Biochem. 1997;249:481–488. doi: 10.1111/j.1432-1033.1997.t01-1-00481.x. [DOI] [PubMed] [Google Scholar]
- Horb ME, Slack JM. Expression of amylase and other pancreatic genes in Xenopus. Mech Dev. 2002;113:153–157. doi: 10.1016/s0925-4773(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, Rousseau GG, Lemaigre FP. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Kadomatsu K, Huang RP, Suganuma T, Murata F, Muramatsu T. A retinoic acid responsive gene MK found in the teratocarcinoma system is expressed in spatially and temporally controlled manner during mouse embryogenesis. J Cell Biol. 1990;110:607–616. doi: 10.1083/jcb.110.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadomatsu K, Muramatsu T. Midkine and pleiotrophin in neural development and cancer. Cancer Lett. 2004;204:127–143. doi: 10.1016/S0304-3835(03)00450-6. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kelly OG, Melton DA. Development of the pancreas in Xenopus laevis. Dev Dyn. 2000;218:615–627. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1027>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Khetchoumian K, Teletin M, Mark M, Lerouge T, Cervino M, Oulad-Abdelghani M, Chambon P, Losson R. TIF1delta, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J Biol Chem. 2004;279:48329–48341. doi: 10.1074/jbc.M404779200. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kim SK, MacDonald RJ. Signaling and transcriptional control of pancreatic organogenesis. Curr Opin Genet Dev. 2002;12:540–547. doi: 10.1016/s0959-437x(02)00338-6. [DOI] [PubMed] [Google Scholar]
- Kofron M, Wylie C, Heasman J. The role of Mixer in patterning the early Xenopus embryo. Development. 2004;131:2431–2441. doi: 10.1242/dev.01132. [DOI] [PubMed] [Google Scholar]
- Koide T, Hayata T, Cho KW. Xenopus as a model system to study transcriptional regulatory networks. Proc Natl Acad Sci U S A. 2005;102:4943–4948. doi: 10.1073/pnas.0408125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy R, Yokoi N, Seino S, Concannon P. Polymorphic variation in the CBLB gene in human type 1 diabetes. Genes Immun. 2004;5:232–235. doi: 10.1038/sj.gene.6364057. [DOI] [PubMed] [Google Scholar]
- Kosugi T, Yuzawa Y, Sato W, Kawai H, Matsuo S, Takei Y, Muramatsu T, Kadomatsu K. Growth factor midkine is involved in the pathogenesis of diabetic nephropathy. Am J Pathol. 2006;168:9–19. doi: 10.2353/ajpath.2006.050488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger O, Plum A, Kim JS, Winterhager E, Maxeiner S, Hallas G, Kirchhoff S, Traub O, Lamers WH, Willecke K. Defective vascular development in connexin 45-deficient mice. Development. 2000;127:4179–4193. doi: 10.1242/dev.127.19.4179. [DOI] [PubMed] [Google Scholar]
- Lankat-Buttgereit B, Gregel C, Knolle A, Hasilik A, Arnold R, Göke R. Pdcd4 inhibits growth of tumor cells by suppression of carbonic anhydrase type II. Mol Cell Endocrinol. 2004;214:149–153. doi: 10.1016/j.mce.2003.10.058. [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- Lee MA, Heasman J, Whitman M. Timing of endogenous activin–like signals and regional specification of the Xenopus embryo. Development. 2001;128:2939–52. doi: 10.1242/dev.128.15.2939. [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Li H, Arber S, Jessell TM, Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat Genet. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- Mally MI, Cirulli V, Hayek A, Otonkoski T. ICA69 is expressed equally in the human endocrine and exocrine pancreas. Diabetologia. 1996;39:474–480. doi: 10.1007/BF00400680. [DOI] [PubMed] [Google Scholar]
- Manfroid I, Delporte F, Baudhuin A, Motte P, Neumann CJ, Voz ML, Martial JA, Peers B. Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development. 2007;134:4011–4021. doi: 10.1242/dev.007823. [DOI] [PubMed] [Google Scholar]
- Martin M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, Dolle P, Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Miralles F, Czernichow P, Ozaki K, Itoh N, Scharfmann R. Signaling through fibroblast growth factor receptor 2b plays a key role in the development of the exocrine pancreas. Proc Natl Acad Sci U S A. 1999;96:6267–72. doi: 10.1073/pnas.96.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Melton DA. Genes, signals, and lineages in pancreas development. Annu Rev Cell Dev Biol. 2003;19:71–89. doi: 10.1146/annurev.cellbio.19.111301.144752. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Law AC, Dor Y, Melton DA. Beta-catenin is essential for pancreatic acinar but not islet development. Development. 2005;132:4663–4674. doi: 10.1242/dev.02063. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ogata S, Morokuma J, Hayata T, Kolle G, Niehrs C, Ueno N, Cho KW. TGF-beta signaling-mediated morphogenesis: modulation of cell adhesion via cadherin endocytosis. Genes Dev. 2007;21:1817–1831. doi: 10.1101/gad.1541807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I, Katoh H, Harada A, Negishi M. Direct interaction of Rnd1 with Plexin-B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin-B1 and induces cell contraction in COS-7 cells. J Biol Chem. 2003;278:25671–25677. doi: 10.1074/jbc.M303047200. [DOI] [PubMed] [Google Scholar]
- Palmer KJ, Konkel JE, Stephens DJ. PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J Cell Sci. 2005;118:3839–3847. doi: 10.1242/jcs.02496. [DOI] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Bayha E, Pieler T. Retinoic acid-mediated patterning of the pre-pancreatic endoderm in Xenopus operates via direct and indirect mechanisms. Mech Dev. 2007;124:518–31. doi: 10.1016/j.mod.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Papadopoulou S, Edlund H. Attenuated Wnt signaling perturbs pancreatic growth but not pancreatic function. Diabtes. 2005;54:2844–51. doi: 10.2337/diabetes.54.10.2844. [DOI] [PubMed] [Google Scholar]
- Pare JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Belanger L. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J Biol Chem. 2004;279:21206–21216. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- Park EC, Hayata T, Cho KW, Han JK. Xenopus cDNA microarray identification of genes with endodermal organ expression. Dev Dyn. 2007;236:1633–1649. doi: 10.1002/dvdy.21167. [DOI] [PubMed] [Google Scholar]
- Pietropaolo M, Castaño L, Babu S, Buelow R, Kuo YL, Martin S, Martin A, Powers AC, Prochazka M, Naggert J, Leiter EH, Eisenbarth GS. Islet cell autoantigen 69 kD (ICA69). Molecular cloning and characterization of a novel diabetes-associated autoantigen. J Clin Invest. 1993;92:359–371. doi: 10.1172/JCI116574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevot D, Morel AP, Voeltzel T, Rostan MC, Rimokh R, Magaud JP, Corbo L. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J Biol Chem. 2001;276:9640–9648. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]
- Qu X, Lam E, Doughman YQ, Chen Y, Chou YT, Lam M, Turakhia M, Dunwoodie SL, Watanabe M, Xu B, Duncan SA, Yang YC. Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J. 2007;26:4445–4456. doi: 10.1038/sj.emboj.7601883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault JP, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud JP. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson IC, Karnik SK, Heiser PW, ten Berge D, Chen H, Gu X, Taketo MM, Nusse R, Hebrok M, Kim SK. Wnt signaling regulates pancreatic beta cell proliferation. Proc Natl Acad Sci U S A. 2007;104:6247–6252. doi: 10.1073/pnas.0701509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Hesser BA, Masuda ES, Scheller RH. Amisyn, a novel syntaxin-binding protein that may regulate SNARE complex assembly. J Biol Chem. 2002;277:28271–28279. doi: 10.1074/jbc.M204929200. [DOI] [PubMed] [Google Scholar]
- Schlange T, Andree B, Arnold H, Brand T. Expression analysis of the chicken homologue of CITED2 during early stages of embryonic development. Mech Dev. 2000;98:157–160. doi: 10.1016/s0925-4773(00)00454-8. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel VM. Programming of the pancreas. Mol Cell Endocrinol. 2001;185:99–108. doi: 10.1016/s0303-7207(01)00628-1. [DOI] [PubMed] [Google Scholar]
- Shin Y, Kitayama A, Koide T, Peiffer DA, Mochii M, Liao A, Ueno N, Cho KW. Identification of neural genes using Xenopus DNA microarrays. Dev Dyn. 2005;232:432–444. doi: 10.1002/dvdy.20229. [DOI] [PubMed] [Google Scholar]
- Sinner D, Rankin S, Lee M, Zorn AM. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004;131:3069–3080. doi: 10.1242/dev.01176. [DOI] [PubMed] [Google Scholar]
- Sogame A, Hayata T, Asashima M. Screening for novel pancreatic genes from in vitro-induced pancreas in Xenopus. Dev Growth Differ. 2003;45:143–152. doi: 10.1034/j.1600-0854.2004.00683.x. [DOI] [PubMed] [Google Scholar]
- Spitzenberger F, Pietropaolo S, Verkade P, Habermann B, Lacas-Gervais S, Mziaut H, Pietropaolo M, Solimena M. Islet cell autoantigen of 69 kDa is an arfaptin-related protein associated with the Golgi complex of insulinoma INS-1 cells. J Biol Chem. 2003;278:26166–26173. doi: 10.1074/jbc.M213222200. [DOI] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–20. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Stafford D, Hornbruch A, Mueller PR, Prince VE. A conserved role for retinoid signaling in vertebrate pancreas development. Dev Genes Evol. 2004;214:432–441. doi: 10.1007/s00427-004-0420-6. [DOI] [PubMed] [Google Scholar]
- Stainier DY. A glimpse into the molecular entrails of endoderm formation. Genes Dev. 2002;16:893–907. doi: 10.1101/gad.974902. [DOI] [PubMed] [Google Scholar]
- Stark KL, McMahon JA, McMahon AP. FGFR-4, a new member of the fibroblast growth factor receptor family, expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development. 1991;113:641–651. doi: 10.1242/dev.113.2.641. [DOI] [PubMed] [Google Scholar]
- Sun C, Kilburn D, Lukashin A, Crowell T, Gardner H, Brundiers R, Diefenbach B, Carulli JP. Kirrel2, a novel immunoglobulin superfamily gene expressed primarily in beta cells of the pancreatic islets. Genomics. 2003;82:130–142. doi: 10.1016/s0888-7543(03)00110-1. [DOI] [PubMed] [Google Scholar]
- Theis M, Mas C, Doring B, Degen J, Brink C, Caille D, Charollais A, Kruger O, Plum A, Nepote V, Herrera P, Meda P, Willecke K. Replacement by a lacZ reporter gene assigns mouse connexin36, 45 and 43 to distinct cell types in pancreatic islets. Exp Cell Res. 2004;294:18–29. doi: 10.1016/j.yexcr.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Torres-Vazquez J, Gitler AD, Fraser SD, Berk JD, Van N, Pham Fishman MC, Childs S, Epstein JA, Weinstein BM. Semaphorin-plexin signaling guides patterning of the developing vasculature. Dev Cell. 2004;7:117–123. doi: 10.1016/j.devcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shu W, Lu MM, Morrisey EE. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol Cell Biol. 2005;25:5022–5030. doi: 10.1128/MCB.25.12.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle FC, Smith JC. Transcriptional regulation of mesendoderm formation in. Xenopus Semin Cell Dev Biol. 2006;17:99–109. doi: 10.1016/j.semcdb.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Weber H, Symes CE, Walmsley ME, Rodaway AR, Patient RK. A role for GATA5 in Xenopus endoderm specification. Development. 2000;127:4345–4360. doi: 10.1242/dev.127.20.4345. [DOI] [PubMed] [Google Scholar]
- Wells JM, Esni F, Boivin GP, Aronow BJ, Stuart W, Combs C, Sklenka A, Leach SD, Lowy AM. Wnt/beta-catenin signaling is required for development of the exocrine pancreas. BMC Dev Biol. 2007;7:4. doi: 10.1186/1471-213X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger WJ, Floro KL, Bennett MB, Withington SL, Preis JI, Barbera JP, Mohun TJ, Dunwoodie SL. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development. 2005;132:1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- Xanthos JB, Kofron M, Wylie C, Heasman J. Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development. 2001;128:167–180. doi: 10.1242/dev.128.2.167. [DOI] [PubMed] [Google Scholar]
- Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, Yuan S, Prabhakar NR, Herrup K, Conlon RA, Hoit BD, Watanabe M, Yang YC. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci U S A. 2002;99:10488–10493. doi: 10.1073/pnas.162371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi N, Komeda K, Wang HY, Yano H, Kitada K, Saitoh Y, Seino Y, Yasuda K, Serikawa T, Seino S. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat Genet. 2002;31:391–394. doi: 10.1038/ng927. [DOI] [PubMed] [Google Scholar]
- Ylipaasto P, Kutlu B, Rasilainen S, Rasschaert J, Salmela K, Teerijoki H, Korsgren O, Lahesmaa R, Hovi T, Eizirik DL, Otonkoski T, Roivainen M. Global profiling of coxsackievirus-and cytokine-induced gene expression in human pancreatic islets. Diabetologia. 2005;48:1510–1522. doi: 10.1007/s00125-005-1839-7. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Mason J. Gene expression in the embryonic Xenopus liver. 2001. Mech Dev. 103:153–157. doi: 10.1016/s0925-4773(01)00341-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–5. Expression patterns of genes identified in this study in whole embryos at tailbud and tadpole stages.
Supplementary Table 1. Microarray data for genes described in this study. Column headings are described further in the Excel file data sheet named Explanation of Columns.