Abstract
Background
The Mycobacterium tuberculosis (Mtb) proteasome has been established as a viable target for the development of anti-tuberculosis agents. In this study, the inhibitory activities of 100 plant-derived natural products on the Mtb proteasome were analyzed to identify novel potential inhibitors.
Methods
The fluorescent substrate Suc-Leu-Leu-Val-Tyr-AMC can be hydrolyzed by the proteasome to release free AMC, the fluorescence of which is proportional to the proteasome activity. The inhibitory activities of 100 natural products (each at a final concentration of 200 μM) were detected by this method using MG132 as a positive control.
Results
Twelve of these natural products (10 of which were flavonoids) inhibited the activity of the Mtb proteasome by more than 65%. Comparison of the structural differences between the flavonoids with good inhibitory activity and those without inhibitory activity revealed that the hydroxyl at the flavonoid C ring C-3 or the hydroxyl/methoxyl at the flavonoid A ring C-6 were critical for the inhibition of proteasomal activity.
Conclusions
These data indicate that flavonoids represent a basis for rational structural design in the process of novel anti-tuberculosis drug discovery.
Keywords: Mycobacterium tuberculosis, Proteasome inhibitor, Plant-derived natural products, Flavonoids
Background
Mycobacterium tuberculosis (Mtb) remains an important global pathogen that is considered to have infected one-third of the world’s population, causing the death of 1.3 million people, with 8.6 million new and relapsed infections in 2012 (World Health Organization Tuberculosis Data and Statistics, 2013) [1]. The development of new drugs is critical for the future control of tuberculosis (TB) and a number of promising compounds are currently in the pipeline at various stages of drug discovery and clinical development [2, 3]. New TB drugs are required to be active against both replicating and nonreplicating bacteria, and to penetrate tissues and granulomas to allow a reduced treatment duration [4].
The human immune system and classical antibacterial agents have the capacity to destroy Mtb in the proliferating state but not in the nonreplicating "drug tolerant" or "phenotypically drug resistant" state [5–7]. If the human immune system is compromised or medication is stopped, nonreplicating state Mtb quickly begin replicating [8]. The requirement for chemotherapy is prolonged for nonreplicating Mtb, which represents a major obstacle to the control of TB [9, 10]. Therefore, there is an urgent need to develop new drugs against nonreplicating Mtb to shorten the period of Mtb chemotherapy and to decrease the chances of treatment failure, Mtb relapse and the emergence of multidrug-resistant (MDR) strains [11, 12].
Mycobacteria are the only known bacterial pathogens with proteasomes are mycobacteria [13–15], which are essential for the degradation of certain proteins, survival of nitroxidative stress in vitro and maintenance of the nonreplicating state in vivo [16, 17]. Therefore, the Mtb proteasome may be required for virulence by mediating resistance to damage by reactive oxygen and nitrogen intermediates (ROI/RNI), and the acidic environment of the macrophage [18]. The Mtb proteasome removes proteins damaged by ROI or RNI, and shear detoxification proteins (such as repressors of genes that synthesize DNA repair enzymes). It also degrades transcription factors that regulate the expression of genes that counteract these host defenses [19, 20]. The Mtb proteasome has been established as a viable target for the development of anti-TB agents and proteasome inhibitors are implicated as potential drug candidates for the treatment of nonreplicating Mtb [21, 22]. In the current study, 100 natural products derived from plants were screened for inhibitory activity against the Mtb proteasome.
Methods
Plant-derived natural products and Mtb strain
One hundred natural products purified from plants were purchased from Shanghai Tauto Biotech (China) with purity >98% by HPLC. Suc-LLVY-7-amido-4-methylcoumarin (Suc-LLVY-AMC) was purchased from Boston Biochem (Cambridge, MA, USA). Stock solutions were prepared in DMSO.
Mtb H37Ra was cultivated on Middlebrook 7H9 medium (0.2% glycerol, 0.5% BSA, 0.2% dextrose, 0.085% NaCl and 0.05% Tween 80; pH 6.6) or on Middlebrook 7H11 plates.
Screening of 100 natural products
Screening methods were performed as described previously with minor modifications [17, 23]. Mtb H37Ra mid-log phase (OD580 1.0) cultures were centrifuged and the cell pellets were washed twice in phosphate buffered saline (PBS). The cell suspension was sonicated in a lysis buffer (50 mM Tris–HCl, pH 7.6, 50 mM NaCl, 5 mM MgCl2, 0.05% Triton X-100, 1 mM DTT and 0.5 mM EDTA). Lysates were centrifuged at 16,000 × g for 10 minutes and the supernatants were removed. The protein concentration of the supernatants was estimated with the Bradford assay. Proteasome activity of the supernatants was assessed. MG132 (a well-known proteasome inhibitor) was tested as a positive control. Reaction buffer containing substrate was added. Final concentrations were as follows: MG132 100 μM; test natural product 200 μM; Suc-LLVY-AMC 64 μM; protein concentration of Mtb lysates (supernatant) 25 μg/ml, HEPES 20 mM; EDTA 0.5 mM; SDS 0.34 mg/ml; pH 7.5. Each sample was tested in three duplicates. Plates were placed on an orbital shaker in an incubator at 37°C for 30 min and the fluorescence intensity of the free AMC was recorded using a luminescence microplate reader (Synergy-2, BioTek, USA) at excitation and emission wavelengths of 360 nm and 460 nm, respectively.
IC50 assay
One hundred natural products were screened to identify those with an inhibitory activity exceeding 65%. The inhibitory activity was calculated as the concentration of inhibitor resulting in a percentage of reduction in fluorescent units (FU) compared to that of the control. The fluorescence intensity of the chosen products was analyzed using the above method with a series of different concentrations (400 μM, 200 μM, 100 μM, 50 μM, 25 μM, 12.5 μM and 6.25 μM) and the corresponding inhibition activities were calculated. The IC50 values of natural products with good inhibitory activities were calculated by dose response curve. The IC50 values were calculated by fitting with the four parameter logistic (4-PL) model, y = A2 + (A1-A2)/(1 + (×/IC50)^p), with OriginPro 8.1 (OriginLab, Inc.), where y is percent inhibition, x is inhibitor concentration, p is the slope of the concentration–response curve, A1 is the minimal inhibition ratio from three independent assays, and A2 is the maximal inhibition ratio from three independent assays.
Results
Inhibitory activities of 100 natural products
The one hundred selected natural products represent 12 categories including terpanoid (27), flavonoid (27), alkaloid (14), coumarin (8), quinone (6), phenol (5), organic acid (4), lignan (3), nucleoside (1), glycoside (2), steroid sapogenin (2), and stilbene (1).
The Mtb proteasome inhibitory activity of MG132 was 79.66% at 100 μM, and the proteasome inhibitory activities of 12 of the 100 natural products (at 200 μM) were more than 65%. Specifically, these 12 products were hispidulin, baicalein, pectolinarin, myricetin, quercetin, curcumin, kaempferol, isoliquiritigenin, icariin, baicalin, celastrol and emodin (Table 1 and Figure 1). In addition to emodin (quinones) and tripterine (terpenoids), the remaining 10 natural products belonged to the flavonoids group.
Table 1.
Mtb proteasome inhibitory activities of 100 natural products and chemical categories
| Categories | Name | IA | Name | IA | Name | IA | Name | IA |
|---|---|---|---|---|---|---|---|---|
| Terpenoids | tripterine# | 69.56% | costunolide | 41.75% | ginkgolide | 39.90% | curcumol | 34.66% |
| dehydroandrographolide | 32.67% | dehydrocostuslactone | 32.15% | alantolactone | 32.12% | evodine | 31.77% | |
| tanshinone | 25.63% | astragaloside A | 23.59% | bilobalide | 18.56% | swertiamain | 14.39% | |
| artesunate | 13.83% | betulinol | 6.52% | ginsenoside | 2.62% | limonin | 1.61% | |
| glycyrrhetinic Acid | 0.00% | hemslecin A | 0.00% | madecassoside | 0.00% | andrographolide | 0.00% | |
| dipsacoside | 0.00% | geniposide | 0.00% | gentiopicroside | 0.00% | tubeimoside A | 0.00% | |
| notoginsenoside | 0.00% | asiaticoside | 0.00% | paclitaxel | 0.00% | |||
| Flavonoids | pectolinarin# | 88.69% | myricetin# | 84.50% | baicalein# | 84.03% | isoliquiritigenin# | 83.60% |
| hispidulin# | 82.06% | baicalin# | 78.47% | quercetin# | 74.40% | icarrin# | 74.34% | |
| curcumin# | 69.53% | kaempferol# | 68.29% | alpha mangostin | 64.16% | wogonoside | 59.01% | |
| wogonin | 53.97% | nevadensin | 48.01% | genistein | 45.74% | epigallocatechin gallate | 45.21% | |
| mangiferin | 31.88% | forsythin | 20.02% | paeoniflorin | 3.05% | tectoridin | 0.00% | |
| luteolin | 0.00% | rutin | 0.00% | silmyarin | 0.00% | puerarin | 0.00% | |
| apigenin | 0.00% | daidzein | 0.00% | diosmetin | 0.00% | |||
| Alkaloids | indirubin | 45.99% | martin hydrochloride | 43.66% | matrine | 23.53% | sophoridine | 11.85% |
| sinomenine | 3.77% | bergenin | 3.67% | peimine | 3.05% | berberine | 0.00% | |
| lycorine hydrochloride | 0.00% | hanfangchin B | 0.00% | hanfangchin A | 0.00% | camptothecin | 0.00% | |
| strychnine | 0.00% | cepharanthine | 0.00% | |||||
| Coumarins | imperatorin | 15.90% | scoparone | 0.00% | praeruptorin A | 0.00% | umbelliferone | 0.00% |
| coumarin | 0.00% | daphnetin | 0.00% | fraxin | 0.00% | osthole | 0.00% | |
| Quinones | emodin# | 73.09% | alkannin | 52.36% | rubimaillin | 46.37% | 1,8-dihydroxyanthraquinone | 36.30% |
| aloe-emodin | 34.30% | alizarin | 0.00% | |||||
| Phenols | gallogen | 62.50% | salidroside | 34.04% | paeonol | 0.00% | resveratrol | 0.00% |
| salicin | 0.00% | |||||||
| Organic acid | diffractic acid | 0.00% | syringic acid | 0.00% | shikimicacid | 0.00% | chlorogenic acid | 0.00% |
| Lignans | arctigenin | 27.10% | deoxyschizandrin | 23.86% | magnolol | 0.00% | ||
| Nucleoside | cordycepin | 7.77% | ||||||
| Glycoside | amygdalin | 1.87% | 2,3,5,4′-tetrahydroxy stilbene-2-O-β-D-glucoside | 0.00% | ||||
| Stilbenes | polydatin | 0.00% | ||||||
| Steroid sapogenin | sarsasapogenin | 0.00% | diosgenin | 0.00% | ||||
| Positive control | MG132 | 79.66% |
IA: inhibitory activity; #natural products with inhibitory activity of Mtb proteasome more than 65%. Different categories of natural products were highlighted using bold texts in the table. The concentration of natural products was 200 μM with the exception of MG132 (100 μM). Data are shown as mean of three independent experiments.
Figure 1.
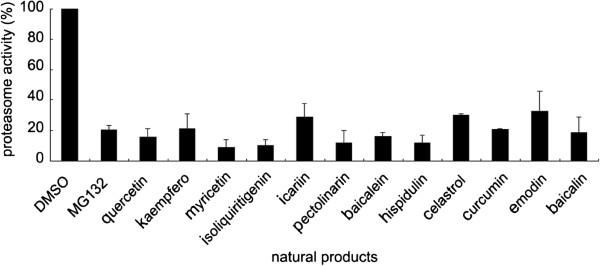
Mtb proteasome inhibitory activities of 12 natural products (200 μM). The inhibitory activities of 12 natural products (200 μM) were: hispidulin 82.06%, baicalein 84.30%, pectolinarin 88.69%, myricetin 84.50%, quercetin 74.40%, kaempferol 68.29%, isoliquiritigenin 83.60%, icariin 74.30%, baicalin 78.47%, curcumin 69.53%, celastrol 69.50% and emodin 73.09%, the inhibitory activity of MG132 (100 μM) is 79.66%. Data are shown as mean ± SD of three independent experiments.
IC50 values of 12 natural products against the Mtb proteasome
Twelve natural products showed concentration-dependent proteasomal inhibitory activities. IC50 values were calculated by fitting with the four parameter logistic (4-PL) model (Figures 2, 3 and Table 2) and the minimal IC50 of the 12 natural products was 45.65 μM.
Figure 2.
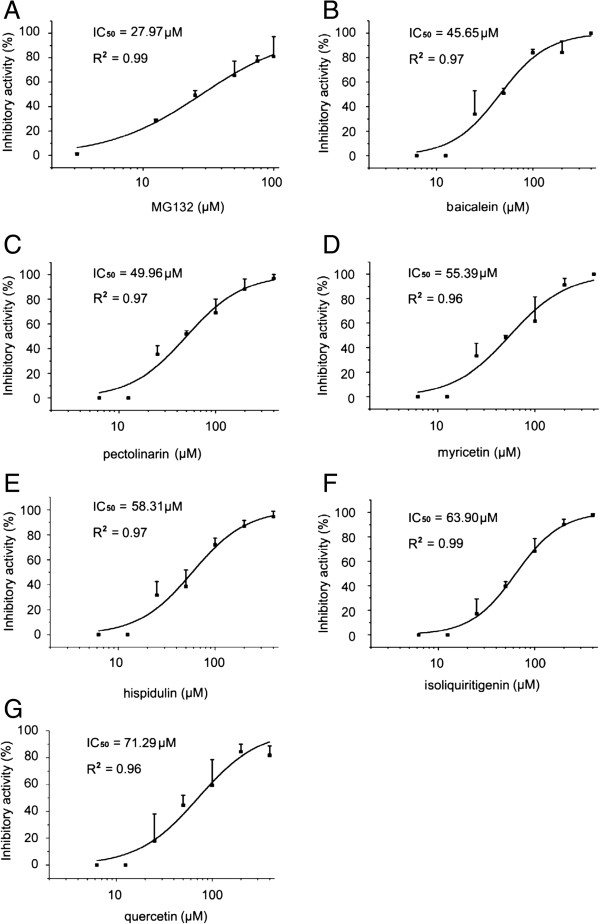
Dose response curves of inhibitory activities of MG132, baicalein, pectolinarin, myricetin, hispidulin, isoliquiritigenin and quercetin. The IC50 values were calculated by fitting with the four parameter logistic (4-PL) model: A. MG132 IC50=27.97 μM, R2=0.99. B. baicalein IC50=45.65 μM, R2=0.97. C. pectolinarin IC50=49.96 μM, R2=0.97. D. myricetin IC50=55.39 μM, R2=0.96. E. hispidulin IC50=58.31 μM, R2=0.97. F. isoliquiritigenin IC50=63.90 μM, R2=0.99. G. quercetin IC50=71.29 μM, R2=0.96. R2, adjust R square values. Dose response curves of inhibitory activity were presented. Data are shown as mean ± SD of three independent experiments.
Figure 3.
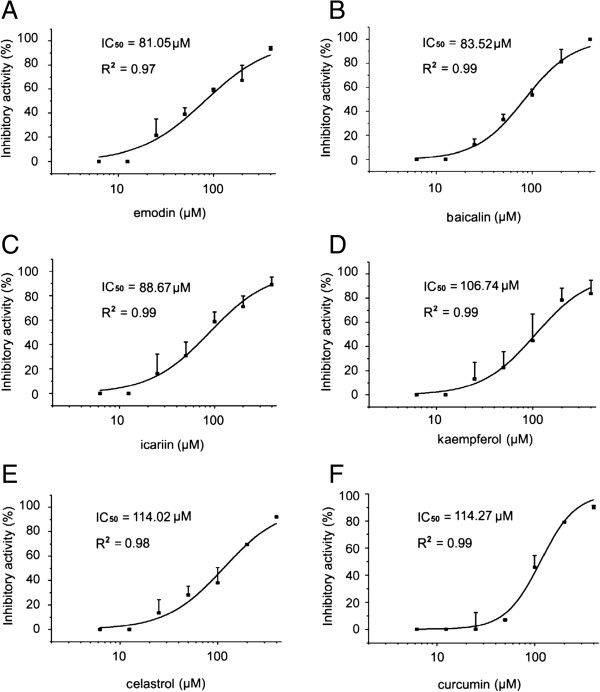
Dose response curves of inhibitory activities of emodin, baicalin, icariin, kaempferol, celastrol and curcumin. The IC50 values were calculated by fitting with the four parameter logistic (4-PL) model: A. emodin IC50=81.05 μM, R2=0.97. B. baicalin IC50=83.52 μM, R2=0.99. C. icariin IC50=88.67 μM, R2=0.99. D. kaempferol IC50=106.74 μM, R2=0.99. E. celastrol IC50=114.02 μM, R2=0.98. F. curcumin IC50=114.27 μM, R2=0.99. R2, adjust R square values. Dose response curves of inhibitory activity were presented. Data are shown as mean ± SD of three independent experiments.
Table 2.
IC 50 of 12 natural products
| Natural products | IC 50 (μM) | Natural products | IC 50 (μM) |
|---|---|---|---|
| baicalein | 45.65 | emodin | 81.05 |
| pectolinarin | 49.96 | icariin | 88.67 |
| quercetin | 71.29 | kaempfero | 106.74 |
| hispidulin | 58.31 | baicalin | 83.52 |
| myricetin | 55.39 | celastrol | 114.02 |
| isoliquiritigenin | 63.90 | curcumin | 114.27 |
| MG 132(control) | 27.97 |
The IC50 values of 12 natural products were calculated by dose response curve.
Discussion
Natural products, as similar as secondary metabolites, exhibit diversity in structures and biological activities. These compounds play an important role in the discovery of lead compounds and more than 50% of FDA-approved drugs are either natural products or natural product derivatives [24, 25]. Furthermore, natural products have specific selectivity for cellular targets [26], with biologically active natural products providing selective ligands for disease-related targets [27]. The in vitro inhibitory activity of crude extracts and/or pure active compounds extracted from plants against Mtb has been extensively reported [28–31].
Twelve of the 100 natural products selected for investigation in this study exhibited inhibitory activities against the proteasome exceeding 65%. Among these, 10 natural products were flavonoids. Thus, in our study, flavonoids showed better inhibitory activities against the Mtb proteasome than other categories, implicating flavonoids as potential proteasome inhibitors. Although some flavonoids showed inhibitory activities against the Mtb proteasome, the lowest IC50 (baicalein, 45.65 μM) was relatively higher than that of the positive control (MG132, 27.97 μM). Our study provides an insight into the unique features of potential proteasome inhibitors.
Natural flavonoids, which exist widely in nature, are secondary metabolites of plants and perform a wide range of functions. The chemical structure of flavonoids is based on a fifteen-carbon skeleton consisting of two benzene rings (A and B) linked by a heterocyclic pyrane ring (C), showing a common three ring structure (C6–C3–C6) (Figure 4) [32]. In this study, despite the finding that the inhibitory activities of flavonoids against the Mtb proteasome was better than that of other categories, there was a wide variation in the inhibitory activities between different flavonoid products. The structures of flavonoids with good inhibitory activity were shown in Figure 5 and those with little inhibitory activity were shown in Figure 6. The common structures of the flavonoids with effective inhibitory activity against the Mtb proteasome are hydroxyl residue at the C ring C-3, or hydroxyl/methoxyl residue at the A ring C-6. This indicates some key functional structures of flavonoids with inhibitory activity against the Mtb proteasome.
Figure 4.

Basic structures of flavonoids.
Figure 5.
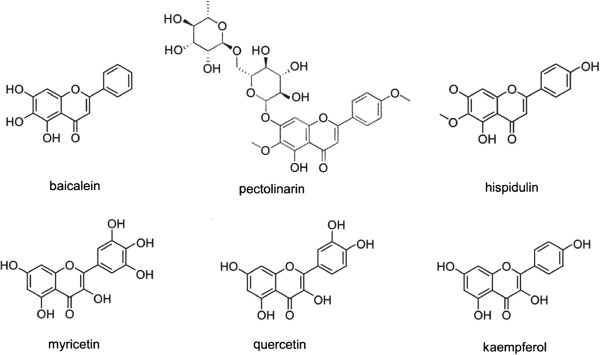
Six natural products of the flavonoids with good inhibitory activity on the Mtb proteasome. Inhibitory activities of six flavonoids (each 200 μM) were as follows: baicalein 84.30%, pectolinarin 88.69%, hispidulin 82.06%, myricetin 84.50%, quercetin 74.40% and kaempferol 68.29%. The common structural features are the hydroxyl at the C ring C-3 (myricetin, quercetin and kaempferol), or the hydroxyl (baicalein) or the methoxyl (pectolinarin and hispidulin) at the A ring C-6.
Figure 6.
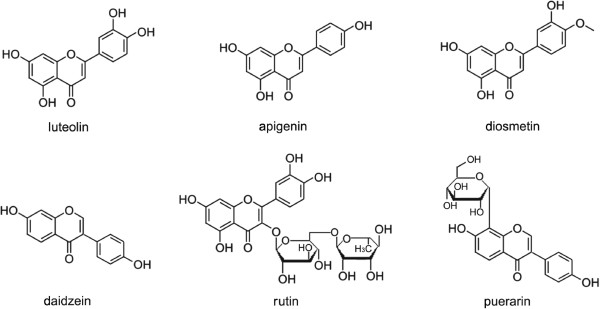
Six natural products of the flavonoids with little inhibitory activity on the Mtb proteasome. Inhibitory activities of six flavonoids (at 200 μM) were zero. Compared with Figure 5, these compounds are without the hydroxyl at the C ring C-3 and the hydroxyl or methoxyl at the A ring C-6.
The well-established free-radical scavenging ability of flavonoids depends on electron-donating hydroxyl group substitutions of the aromatic A ring and the heterocyclic C ring [33]. The C-2–C-3 double-bond conjugated to a C-4 carbonyl group on the C ring is responsible for antioxidant activity [23]. Comprehensive analysis of the correlations between functional groups in the flavonoid structures and their proteasome inhibitory activities are necessary to evaluate the effects of modifications at the C ring C-3 and the A ring C-6.
Conclusions
The flavonoid structure has been shown to be a viable template for the design of potent Mtb proteasome inhibitors. This information forms the basis of the rational design of flavonoid analogs retaining the key functional groups, which will focus direct screening and will be helpful to accelerate the discovery of potent inhibitors of the Mtb proteasome.
Acknowledgements
We thank Weigang He for excellent technical assistance in data analysis. This work was funded by the National Natural Science Foundation of China (81102869, 31100619, 81471537), Chen-guang Plan Project of Shanghai Educational Municipal Education Commission (11CG48), Shanghai Rising-Star Program (14QA1403400), and Specialized Research Fund for the Doctoral Program of Higher Education (20113107120014).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All authors have read and approved the final manuscript. YZ and XJ designed the experiment analyzed the data and prepared the manuscript. FG, JS, LW, XS, ZL and HZ performed the experiments.
Contributor Information
Yuejuan Zheng, Email: 13641776412@163.com.
Xin Jiang, Email: jiangxingao@163.com.
Feng Gao, Email: gaof891@aliyun.com.
Junxiang Song, Email: songjunxiang_ok@126.com.
Jinxia Sun, Email: jinxia0526@163.com.
Lixin Wang, Email: wlx831208@126.com.
Xiaoxia Sun, Email: 632539156@qq.com.
Zhenhui Lu, Email: tcmdoctorlu@163.com.
Huiyong Zhang, Email: zhanghuiyong1963@hotmail.com.
References
- 1.WHO . Global Tuberculosis Report. Geneva, Switzerland: WHO; 2012. [Google Scholar]
- 2.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469(7331):483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 3.Zumla A, Nahid P, Cole ST. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov. 2013;12(5):388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 4.Lechartier B, Rybniker J, Zumla A, Cole ST. Tuberculosis drug discovery in the post-post-genomic era. EMBO Mol Med. 2014;6(2):158–168. doi: 10.1002/emmm.201201772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff HI, Barry CE. Tuberculosis: metabolismand respiration in the absence of growth. Nat Rev Microbiol. 2005;3(1):70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 7.Dick T. Dormant tubercle bacilli: the key to more effective TB chemotherapy? J Antimicrob Chemother. 2001;47(1):117–118. doi: 10.1093/jac/47.1.117. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen L, Pieters J. Mycobacterial subversion of chemotherapeutic reagents and host defense tactics: challenges in tuberculosis drug development. Annu Rev Pharmacol Toxicol. 2009;49:427–453. doi: 10.1146/annurev-pharmtox-061008-103123. [DOI] [PubMed] [Google Scholar]
- 9.Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105(33):11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchison DA. Shortening the treatment of tuberculosis. Nat Biotechnol. 2005;23(2):187–188. doi: 10.1038/nbt0205-187. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y, Pieters J. Novel proteasome inhibitors as potential drugs to combat tuberculosis. J Mol Cell Biol. 2010;2(4):173–175. doi: 10.1093/jmcb/mjp053. [DOI] [PubMed] [Google Scholar]
- 12.Kim JH, O’Brien KM, Sharma R, Boshoff HI, Rehren G, Chakraborty S, Wallach JB, Monteleone M, Wilson DJ, Aldrich CC, Barry CE, 3rd, Rhee KY, Ehrt S, Schnappinger D. A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc Natl Acad Sci U S A. 2013;110(47):19095–19100. doi: 10.1073/pnas.1315860110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302(5652):1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 14.Lin G, Li D, de Carvalho LP, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H, Nathan C. Inhibitors selective for mycobacterial versus human proteasomes. Nature. 2009;461(7264):621–626. doi: 10.1038/nature08357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupas A, Zuhl F, Tamura T, Wolf S, Nagy I, De Mot R, Baumeister W. Eubacterial proteasomes. Mol Biol Rep. 1997;24(1–2):125–131. doi: 10.1023/A:1006803512761. [DOI] [PubMed] [Google Scholar]
- 16.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13(12):1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandotra S, Lebron MB, Ehrt S. The Mycobacterium tuberculosis proteasome active site threonine is essential for persistence yet dispensable for replication and resistance to nitric oxide. PLoS Pathog. 2010;6(8):e1001040. doi: 10.1371/journal.ppat.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerda-Maira F, Darwin KH. The Mycobacterium tuberculosis proteasome: more than just a barrel-shaped protease. Microbes Infect. 2009;11(14–15):1150–1155. doi: 10.1016/j.micinf.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322(5904):1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd Proteasomal protein degradation in mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284(5):3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nathan C, Gold B, Lin G, Stegman M, de Carvalho LP, Vandal O, Venugopal A, Bryk R. A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis (Edinb) 2008;88(Suppl):S25–S33. doi: 10.1016/S1472-9792(08)70034-9. [DOI] [PubMed] [Google Scholar]
- 22.Striebel F, Imkamp F, Özcelik D, Weber-Ban E. Pupylation as a signal for proteasomal degradation in bacteria. Biochim Biophys Acta. 2014;1843(1):103–113. doi: 10.1016/j.bbamcr.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 23.Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, Weich N, Nathan C. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol. 2006;59(5):1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 24.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kingston DG. Modern natural products drug discovery and its relevance to biodiversity conservation. J Nat Prod. 2011;74(3):496–511. doi: 10.1021/np100550t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagunin A, Filimonov D, Poroikov V. Multi-targeted natural products evaluation based on biological activity prediction with PASS. Curr Pharm Des. 2010;16(15):1703–1717. doi: 10.2174/138161210791164063. [DOI] [PubMed] [Google Scholar]
- 27.Gu J, Gui Y, Chen L, Yuan G, Lu HZ, Xu X. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS One. 2013;8(4):e62839. doi: 10.1371/journal.pone.0062839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta P, Bhatter P, D’souza D, Tolani M, Daswani P, Tetali P, Birdi T. Evaluating the anti Mycobacterium tuberculosis activity of Alpinia galanga (L.) Willd. axenically under reducing oxygen conditions and in intracellular assays. BMC Complement Altern Med. 2014;14:84. doi: 10.1186/1472-6882-14-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen JJ, Lin WJ, Shieh PC, Chen IS, Peng CF, Sung PJ. A new long-chain alkene and antituberculosis constituents from the leaves of Pourthiaea lucida. Chem Biodivers. 2010;7(3):717–721. doi: 10.1002/cbdv.200900198. [DOI] [PubMed] [Google Scholar]
- 30.Negi AS, Kumar JK, Luqman S, Saikia D, Khanuja SP. Antitubercular potential of plants: a brief account of some important molecules. Med Res Rev. 2010;30(4):603–645. doi: 10.1002/med.20170. [DOI] [PubMed] [Google Scholar]
- 31.Askun T, Tumen G, Satil F, Ates M. In vitro activity of methanol extracts of plants used as spices against Mycobacterium tuberculosis and other bacteria. Food Chem. 2009;116(1):289–294. doi: 10.1016/j.foodchem.2009.02.048. [DOI] [Google Scholar]
- 32.Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. Sci World J. 2013;2013:162750. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright B, Spencer JP, Lovegrove JA, Gibbins JM. Insights into dietary flavonoids as molecular templates for the design of anti-platelet drugs. Cardiovasc Res. 2013;97(1):13–22. doi: 10.1093/cvr/cvs304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1472-6882/14/400/prepub


