Abstract
The leucine biosynthetic pathway is essential for the growth of Mycobacterium tuberculosis and is a potential target for the design of new anti-tuberculosis drugs. The crystal structure of α-isopropylmalate synthase, which catalyzes the first committed step in this pathway, has been determined by multiwavelength anomalous dispersion methods and refined at 2.0-Å resolution in complex with its substrate α-ketoisovalerate. The structure reveals a tightly associated, domain-swapped dimer in which each monomer comprises an (α/β)8 TIM barrel catalytic domain, a helical linker domain, and a regulatory domain of novel fold. Mutational and crystallographic data indicate the latter as the site for leucine feedback inhibition of activity. Domain swapping enables the linker domain of one monomer to sit over the catalytic domain of the other, inserting residues into the active site that may be important in catalysis. The α-ketoisovalerate substrate binds to an active site zinc ion, adjacent to a cavity that can accommodate acetyl-CoA. Sequence and structural similarities point to a catalytic mechanism similar to that of malate synthase and an evolutionary relationship with an aldolase that catalyzes the reverse reaction on a similar substrate.
Mycobacterium tuberculosis remains one of mankind's deadliest pathogens, responsible for approximately 2 million deaths worldwide every year and estimated to infect one-third of the world's population (World Health Organization, www.who.int/gtb/). Although effective drugs exist, current therapy requires prolonged treatment with three to four drugs, leading to compliance problems and the emergence of multidrug resistance (1). Two features of the organism combine to make it one of the most serious disease-causing agents. First, it has a thick, waxy cell wall that is rich in novel lipids, glycolipids, and polysaccharides and provides a challenging barrier to drugs and other small molecules (2). Second, it can survive for many years in a dormant or persistent state within activated macrophages, to be reactivated as active tuberculosis (TB) later in life (3). The latter phenomenon has led to a deadly synergy with HIV/AIDS.
Large-scale transposon mutagenesis and in vitro growth studies have identified many essential biosynthetic pathways in M. tuberculosis (4). Among them are those for the synthesis of the branched-chain amino acids leucine, isoleucine, and valine. These pathways, which share common intermediates, are present in plants and microorganisms, but not humans, and are likely to be particularly important for the survival of M. tuberculosis inside macrophages, in the absence of an exogenous source of nutrients. In support of this notion, leucine auxotrophy (by insertional mutation of leuD) has been shown to attenuate both Mycobacterium bovis and M. tuberculosis, reducing their capacity to establish infections in mice, in vivo, and in macrophages, in vitro (5–7). M. tuberculosis also has shown sensitivity to inhibitors of acetolactate synthase (8), which catalyzes reactions required for the biosynthesis of all three branched-chain amino acids.
Leucine biosynthesis occurs via the isopropylmalate (IPM) pathway, starting with the formation of α-IPM from acetyl-CoA and α-ketoisovalerate (α-KIV), which is also the immediate precursor for valine biosynthesis. This aldol condensation-type reaction is catalyzed by α-IPM synthase (α-IPMS), encoded by leuA. Subsequent reactions in this pathway are catalyzed by α-IPM isomerase, which is encoded by leuC/D, and β-IPM dehydrogenase, which is encoded by leuB. Biochemical studies of α-IPMS have been limited to a few microorganisms, notably Salmonella typhimurium (9), Corynebacterium glutamicum (10), and Saccharomyces cerevisiae (11). The enzyme is dimeric (S. cerevisiae) or tetrameric (S. typhimurium), with a monomer molecular mass of 60–70 kDa, a divalent metal ion dependence, and an alkaline pH optimum. As is common for enzymes that catalyze the first committed step in a biosynthetic pathway (12, 13), it is subject to feedback inhibition by the end-product leucine (14). Despite its key role in this important pathway, no 3D structure is yet available for α-IPMS.
In this article we describe the crystal structure for α-IPMS from M. tuberculosis. The observation of bound substrate (α-KIV) and metal cofactor (Zn2+) identifies the active site and key residues within it, and structural similarities to several other enzymes that act on analogous substrates indicate previously unrecognized functional and evolutionary relationships. From these comparisons, we suggest a possible biochemical mechanism. The 3D structure also allows us to address other aspects of α-IPMS function, including the leucine feedback inhibition site, and provides a basis for inhibitor design directed toward the development of new anti-TB drugs.
Materials and Methods
Crystallization and Data Collection. M. tuberculosis α-IPMS, the gene product of ORF Rv3710, was cloned, expressed in Escherichia coli, and prepared and crystallized with incorporated selenomethionine (SeMet) as described (15). Crystals were dipped briefly in a cryoprotectant solution comprising 85% reservoir solution and 15% glycerol and flash-frozen in liquid nitrogen before data collection. Crystals of a fully occupied α-KIV complex were obtained by adding a solution containing 50 mM ZnCl2 and 50 mM α-KIV directly to drops containing SeMet α-IPMS crystals, to a final volume ratio of 1:10. After a 5-min incubation, the crystals were flash-frozen in a cryoprotectant comprising 75% reservoir solution, 10% substrate solution, and 15% glycerol. Crystals of the leucine complex were grown as for the native enzyme (15) but in the presence of KCl and leucine (5 mM each) and were frozen for data collection as for α-KIV complex crystals.
All x-ray data were collected at 113 K. SeMet multiwavelength anomalous diffraction (Se-MAD) data first were collected from the nonsubstrate bound crystal at the X8-C beamline at the Brookhaven National Laboratory (Upton, NY). These data were used to determine experimental phases, as outlined below. Additional high-resolution data sets were collected from substrate-bound and leucine-bound crystals on a Rigaku (Tokyo) RU-H3R rotating anode generator equipped with a Mar345 image plate detector. The crystals belong to space group P21 with unit cell dimensions a = 54.25, b = 154.73, and c = 68.82 Å, β = 98.05°, and a solvent content of 39%, assuming a dimer in the asymmetric unit (VM = 2.0 Å3/Da). All data were integrated and scaled with the hkl package (16). Data statistics are shown in Table 1.
Table 1. Data collection, processing, refinement, and phasing statistics.
| Statistics | SeMet λ = 0.9806 Å | SeMet λ = 0.9799 Å | SeMet λ = 0.9683 Å | α-KIV complex λ = 1.5418 Å | Leucine complex λ = 1.5418 Å |
|---|---|---|---|---|---|
| Data collection and processing | |||||
| Resolution, Å | 50-2.6 | 50-2.6 | 50-2.7 | 30-2.0 | 30-2.3 |
| Total reflections | 246,934 | 247,857 | 137,560 | 718,275 | 185,034 |
| Unique reflections | 34,054 | 34,207 | 33,783 | 74,305 | 49,393 |
| Completeness (final shell), % | 99.2 (98.4) | 99.3 (98.5) | 99.0 (98.1) | 98.4 (96.0) | 99.8 (100.0) |
| I/σ (final shell) | 14.8 (4.2) | 16.9 (5.0) | 11.1 (3.3) | 34.6 (3.0) | 16.0 (3.1) |
| Rmerge (final shell), % | 9.2 (44) | 8.5 (37) | 8.7 (38) | 7.2 (65) | 10.9 (45) |
| Rano | 0.074 | 0.063 | 0.073 | ||
| Refinement | |||||
| No. of reflections, working/test | 66,888/3,740 | 44,419/2,467 | |||
| R/Rfree | 0.169/0.197 | 0.191/0.244 | |||
| rms deviations, bonds/angles | 0.016/1.47 | 0.015/1.46 | |||
| Average B factors, Å2 | 39.2 | 29.6 | |||
| Protein atoms | 8,670 | 8,610 | |||
| Substrate atoms | 16 | 18 | |||
| Ions | 3 | 0 | |||
| Water molecules | 466 | 281 | |||
| Phasing | |||||
| solve figure of merit (22× Se) | 0.43 | ||||
| solveZ score | 72.09 | ||||
| resolve figure of merit | 0.53 |
Structure Determination and Refinement. The unliganded structure was solved by using the three-wavelength multiwavelength anomalous diffraction data and the program solve (17). solve identified and refined 22 selenium sites, giving a figure of merit of 0.43 and a Z score of 72.09. The solve map was subsequently input to resolve (18) for density modification and automatic model building. resolve gave an overall figure of merit of 0.53 and built ≈80% of the peptide backbone in the asymmetric unit including ≈50% of the side chains. A second model was built into the resolve map by using maid (19), and the starting model for refinement then was constructed by using a combination of the resolve and maid models and manual building in o (20). The model was refined by using iterative cycles of cns (21) simulated annealing, conjugate gradient minimization, and B-factor refinement. Throughout refinement, 2Fo - Fc maps (including experimental phases) and the multiwavelength anomalous diffraction-phased map were used for additional model building and structure validation. A large spherical peak of electron density was identified as a bound zinc ion from an x-ray fluorescence scan at Brookhaven National Laboratory and was refined with partial occupancy.
The liganded structures (containing either α-KIV or leucine) were solved by molecular replacement of the unliganded structure into the liganded structure unit cell. The structures were refined with cns as for the unliganded structure. Omit maps showed clear density for the α-KIV, Zn2+ ions, and leucine in their respective structures. These entities were included in refinement, as were solvent molecules, all treated as water. Each refinement was completed by subjecting the model to final cycles of refmac and cgmat refinement (22). The α-KIV complex structure has been deposited in the Protein Data Bank under PDB ID code 1sr9.
An acetyl-CoA molecule was modeled into a cavity adjacent to the α-KIV by using sybyl 6.9 (23) and gold 2.1 (24). Water molecules were removed, charges were assigned to the remaining atoms (including α-KIV) by using sybyl (Gasteiger method), and acetyl-CoA was docked into the putative binding pocket by using gold. gold assigned atom types by default, and two distance restraints were included in the docking: C51 acetyl-CoA (terminal acetyl carbon) to C2 α-KIV and C51 acetyl-CoA to His-379 Nε2 (both minimum 2.4 Å, maximum 3.2 Å, spring constant 5.0). For the docking process, an atom within the binding site was used as a starting position for the gold cavity search algorithm (15-Å radius). The gold genetic algorithm parameters included population size, 100; selection pressure, 1.1; number of operations, 100,000; number of islands, 5; niche size, 2; migrate, 10; mutate, 95; and crossover, 95.
Results and Discussion
Structure Determination and Model Quality. The structure of M. tuberculosis α-IPMS was solved at 2.6-Å resolution by using multiwavelength anomalous diffraction phases derived from the SeMet-substituted protein. This structure revealed a bound metal ion, identified as Zn2+ from an x-ray fluorescence scan, and associated density that resembled the natural substrate α-KIV, albeit partly occupied. We therefore prepared crystals of a fully occupied substrate complex and refined this structure at 2.0-Å resolution. The final R factor was 0.169 and Rfree was 0.197 (Table 1). The final model comprises the two molecules in the crystal asymmetric unit, which form the biological dimer, together with one molecule of α-KIV and one Zn2+ ion per monomer, and 466 water molecules. Molecule A comprises residues 18–643 of α-IPMS but with residues 427–433, 465–473, and 578–614 missing and assumed to be disordered. Molecule B also comprises residues 18–643, with residues 430–433, 464–474, and 580–613 missing. The main-chain torsion angles for both molecules correspond well with allowed values [91.3% in the most favored regions of the Ramachandran plot, as defined by procheck (25)], with only residues Arg-318 and Tyr-466, both located in β-turns, falling in disallowed regions.
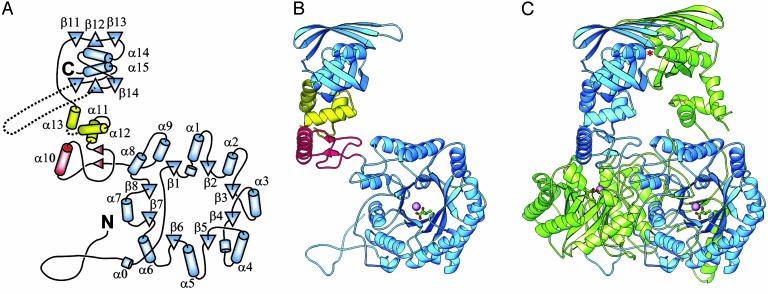
Overall Structure. M. tuberculosis α-IPMS forms an intimately associated, extended dimer (Fig. 1), which is assumed to represent the biological unit. The 70-kDa (644-residue) monomer is folded into two major domains, separated by two small subdomains that are joined by a flexible hinge (Fig. 1 A and B). The larger N-terminal domain, residues 51–368, forms an (α/β)8 TIM barrel that has N- and C-terminal extensions that contribute significantly to dimerization. Before the first β-strand, the polypeptide meanders in extended fashion over the domain surface and then extends as an N-terminal arm that wraps around the (α/β)8 barrel of the other monomer. After the last β-strand, two helices, α8 and α9, pack against the barrel surface before the polypeptide chain again extends outwards to form the first part of the linker domain, sitting on the surface of the second monomer. As is typical of (α/β)8 TIM barrel domains, the active site is located at the C-terminal end of the barrel, defined as the end that contains the C-termini of the eight β-strands. This active site is indicated clearly by the presence of the bound Zn2+ ion and its associated α-KIV substrate.
Fig. 1.
Structure of M. tuberculosis α-IPMS. (A) Topology of the monomer. β-Strands are shown as triangles, and helices are shown as cylinders. The helix numbering follows standard practice for TIM barrel domains. The barrel and regulatory domains are colored blue, and subdomains I and II of the linker are colored red and yellow, respectively. (B) Ribbon diagram of the monomer. The bound Zn2+ ion in the catalytic domain is shown as a large magenta sphere, and the α-KIV substrate is depicted as a ball-and-stick model. (C) Ribbon diagram of the dimer. The individual monomers are colored blue and green. An asterisk marks one leucine regulatory binding site (the other is obscured). B and C were drawn with ribbons (26).
After the TIM barrel domain, the linker domain is divided into two small subdomains. Subdomain I, residues 369–424, includes helix α10 and two short parallel β-strands, and subdomain II, residues 434–490, comprises a three-helix bundle that packs against the C-terminal domain. Between the two subdomains, the disordered residues 427–433 are the site of a flexible hinge that allows the two monomers to display quite different relative orientations of their N- and C-terminal halves (Fig. 1C). The structures on either side of this hinge match extremely well between the two monomers (rms difference in Cα positions ≈ 0.4 Å in each case), indicating that they are coherent structures separated by a single hinge.
The C-terminal regulatory (R) domain, residues 491–644, is an internally duplicated structure with a novel fold. It comprises two similar βββα units that are arranged such that the two α-helices pack together in the center, crossing at an angle of ≈34°, sandwiched between the two three-stranded, antiparallel β-sheets. The overall domain is thus constructed as a β-α-β three-layer sandwich. This domain is also the location of a curious feature of the M. tuberculosis α-IPMS enzyme, a variable number tandem repeat (VNTR) in the coding region of the leuA gene. Different strains of M. tuberculosis have different numbers of this 57-nt repeat, which translates to an exact 19-residue repeat in the α-IPMS enzyme (27). In the H37Rv strain, the subject of our study, two tandem repeats are present, corresponding to residues 575–612; these residues, which link β-strands β15 and β16, would project from the surface but are disordered in both monomers.
The dimer interface covers both major domains but not the intermediate linker domains, which make no contact with each other. The two (α/β)8 barrels pack against one another with an interface formed mainly by interactions involving helices α6, α7, and α8 and are tied together by N- and C-terminal extensions. The N-terminal residues 18–50 from one barrel wind as an extended arm over the surface of the other, covering the opening of its N-terminal end. Similarly, at the C-terminal end of the barrel, the chain crosses to the other monomer in a domainswapping interaction such that subdomain I sits over the end of the other barrel, with the N terminus of helix α10 over its active site (Fig. 1C). The R domains also dimerize such that two six-stranded, antiparallel β-sheets extending over both monomers are formed and sandwich the four helices. The N-termini of the helices from the two R-domain monomers are directed toward each other at the interface with the N terminus of helix α14 facing that of α15′ and α15 facing α14′.
Dimerization buries 7,650 Å2 of the monomer surface area, representing one quarter of the total monomer surface. Approximately 80% of the total buried surface area is contributed by the dimerization of the barrel, including the N- and C-terminal extensions. Without these extensions the barrel would contribute only 12% to the total buried surface. The very large buried surface and, in particular, the domain swapping barrel extensions must give a significant stabilization of the biological unit. The hinge in the linker region, however, allows adjustment of the R domain dimer orientation relative to that of the TIM barrel dimer. In the present structure, this flexibility manifests as an unusual asymmetry in the dimer in which the orientation of the R domain relative to its respective barrel differs for each monomer.
Sequence and Structure Comparisons. Alignment of α-IPMS sequences from the current database shows that the M. tuberculosis enzyme most closely resembles the yeast α-IPMS, with which it shares ≈45% sequence identity over the full length; the yeast enzyme differs substantially only in the absence of the TB-specific variable number tandem repeat motifs. In contrast, almost all of the bacterial enzymes lack the N-terminal extension on the TIM barrel domain, and several other loops, making them substantially smaller; also some archaeal enzymes appear to lack the C-terminal R domain completely. Some 20 residues are completely invariant over 30 sequences we compared, almost all in the TIM barrel domain, where several conserved sequence motifs can be identified.
Searches of the Protein Data Bank with the secondary structure matching algorithm ssm (www.ebi.ac.uk/msd-srv/ssm) gave a very large number of structural matches for the (α/β)8 TIM barrel domain but no significant match for the protein as a whole. Most of the matches simply reflect the ubiquitous occurrence of the TIM barrel fold, which covers many functionally diverse enzymes, grouped in scop (28) into 26 superfamilies. The top hit with ssm, however, revealed a striking similarity with the aldolase portion, DmpG, of the bifunctional 4-hydroxy-2-ketovalerate aldolase/acylating acetaldehyde dehydrogenase (29). Although the DmpG aldolase has no N-terminal extension, its TIM barrel portion, up to and including helix α9, aligns extremely well to the barrel of α-IPMS; 240 residues are aligned with an rms difference in Cα atomic positions of 2.1 Å, a quality (Q) score of 0.28, and a Z score of 9.6. The two interacting aldolase monomers also dimerize in an identical fashion to those of α-IPMS, and the active sites of both enzymes share identical or similar residues, disposed similarly in space. The DmpG aldolase also has a “communication domain” with four helices that follows the TIM barrel domain, like the linker domain in α-IPMS, but in DmpG this sits on its own TIM barrel domain instead of domain swapping as in α-IPMS. Sequence identity in the common portions of α-IPMS and DmpG is ≈15%, and we conclude that these two proteins share a common evolutionary ancestry. Among many other hits to the TIM barrel domain of α-IPMS, we find a number of aldolases, but these are not metal-dependent and appear to fit better into other (α/β)8 families.
Searches with the C-terminal R domain of α-IPMS, by contrast, gave only one weakly significant hit: the solution structure of an E. coli ribosome-associated cold-shock response protein that contains a double-stranded RNA binding domain fold [PDB ID code 1n3g (30)]. Approximately 68 residues from the R domain of α-IPMS could be matched to the RNA binding domain (an αβββα fold) with an rms difference in Cα positions of 2.65 Å.
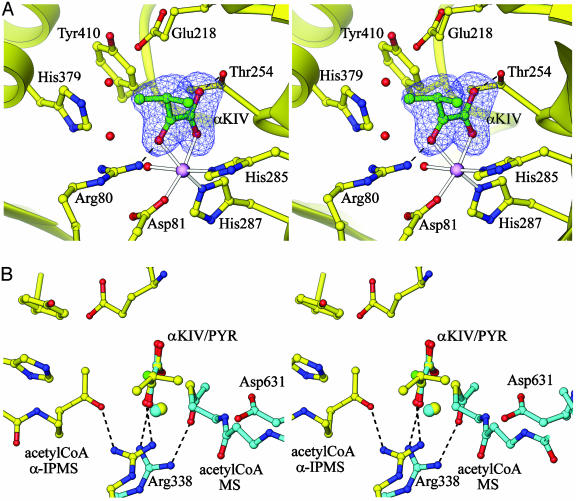
Active Site and Substrate Binding. The active site (Fig. 2A) is located at the C-terminal end of each (α/β)8 barrel, clearly identified by the presence of the bound Zn2+ ion. The metal ion is coordinated by three protein ligands: Asp-81 from helix α1′, following strand β1, and His-285 and His-287 from the C-terminal end of strand β7. The two His ligands are part of a HxH(D/N)D sequence motif that is conserved in all α-IPMS enzymes. Each ligand is stabilized by hydrogen bonding, Asp-81 with Asn-321 Nδ2, His-285 with Glu-309 Oε2, and His-287 with the carbonyl oxygen of Gly-320. Three octahedral coordination sites remain and are filled in the substrate complex by a water molecule and two carbonyl oxygens of the substrate, α-KIV (Fig. 2 A). Hydrogen bonds to Arg-80 and Thr-254, both completely invariant in all α-IPMS enzymes, further stabilize substrate binding. Analogs of all of these residues are present in the DmpG aldolase, where Asp-18, His-200, and His-202 coordinate the active site Mn2+ ion and are hydrogen-bonded to Asn-236, Asp-224, and the carbonyl oxygen of Gly-235, and Arg-17 and Ser-171 hydrogen-bond to the substrate.
Fig. 2.
Active site of α-IPMS, shown in stereoview. (A) The bound α-KIV substrate is shown built into its electron density (from a 2Fo - Fc omit map, contoured at 1σ), adjacent to the Zn2+ ion (magenta sphere). Two water molecules in the adjacent acetyl-CoA cavity are shown with red spheres. His-379 and Tyr-410 contribute to the active site from the other monomer. Zn-ligand bonds are shown as outlined rods, and hydrogen bonds are shown as broken lines. (B) Comparison of the active sites of α-IPMS (yellow) and MS (cyan). Superposition of the Zn2+ ion and α-KIV substrate of α-IPMS on to the Mg2+ and pyruvate of the E. coli MS/pyruvate/CoA ternary complex (31) shows that the key residues Arg-338 and Asp-631 in MS match Arg-80 and either Glu-218 or His-379 in α-IPMS. Acetyl-CoA (modeled for α-IPMS, experimentally located for MS) approaches from opposite sides of the substrate in the two cases. Binding on the same side as for MS is precluded in α-IPMS by the presence of side chains (not shown, for clarity). Images were drawn with ribbons (26).
A cavity adjacent to the bound α-KIV, opening to a groove on the enzyme surface, is of appropriate size (≈10-Å long and total volume ≈200 Å3) to allow acetyl-CoA to approach the substrate. This cavity is surrounded by further invariant residues, Glu-218, Glu-317, and Arg-318 from a conserved GxGERxG sequence motif (residues 314–320), and His-379 and Tyr-410 from the other monomer; His-379 belongs to the N terminus of helix α10, which sits over the C-terminal end of the barrel, and Tyr-410 belongs to a finger of polypeptide that projects into the active site. CoA can adopt widely varying conformations in different enzymes (31), making its mode of binding difficult to model in detail. The thioacetyl moiety, however, can be modeled convincingly at the end of the cavity, adjacent to the α-KIV, with its methyl carbon and acetyl oxygen atoms displacing bound water molecules (Fig. 2B); the methyl carbon sits between His-379 Nε2, Asp-218 Oδ1, and the C2 atom of α-KIV, 3.0–3.5 Å from each, and the acetyl oxygen is hydrogen-bonded to Arg-80 Nη1.
Catalytic Mechanism. The arrangement of the conserved active site residues relative to the substrate, metal, and modeled acetyl-CoA suggests a potential mechanism for the aldolcondensation reaction that forms α-IPM from α-KIV. This spatial arrangement is similar to that of malate synthase (MS), which catalyzes a very similar reaction, an acetyl-CoA-dependent Claisen condensation of glyoxylate to malate (31–33). Both α-IPMS and MS have (α/β)8 barrel catalytic domains but appear otherwise structurally unrelated. They share no significant sequence identity, the metal ion (Zn2+ for α-IPMS, Mg2+ for MS) is located at a different position around the barrel, and when the metal ions and bound substrate molecules are superimposed, the binding sites for acetyl-CoA are found to be on opposite sides of the substrate (Fig. 2B). What they do appear to share is convergence to common chemistry, by using very similar functional groups acting on similar substrates.
In both reactions the first step is hydrogen abstraction from the methyl group of acetyl-CoA. In α-IPMS, two invariant residues, His-379 and Glu-218, are in position to act as the essential base, analogous to Asp-631 in E. coli MS (Fig. 2B). His-379 is oriented by stacking with Tyr-410, also invariant; both these residues are contributed by the domain-swapped linker subdomain I of the other monomer, giving a mechanism for allosteric regulation. Hydrogen abstraction promotes enolization of the acetyl group, the enol formation stabilized by interaction with the positively charged side chain of Arg-80, which also binds a carbonyl oxygen of the substrate. In MS, Arg-338 has the same structural and electrostatic role (Fig. 2B). Subsequent electronic rearrangement promotes nucleophilic attack on the α-KIV substrate by the enol. The zinc ion not only positions α-KIV appropriately but also polarizes the C2 carbonyl bond to induce positive charge on the carbon, making it susceptible to nucleophilic attack. Further, the zinc-bound water is in a good position to participate in the subsequent hydrolytic attack on the acetyl-CoA/α-KIV intermediate, which leads to formation of the product and release of CoA. A similar role of the water ligand has been proposed for MS (33) and would be favored in α-IPMS by the ability of Zn2+ to adopt a variety of coordination geometries. Given its role in substrate binding, the Zn2+ ion also provides an attractive template for the design of potential inhibitors for α-IPMS.
The relationship between α-IPMS and the bifunctional aldolase/dehydrogenase DmpG/DmpF is intriguing. The catalytic domain of α-IPMS is clearly homologous with the aldolase domain DmpG; they share the same catalytic machinery and bind similar substrates (α-KIV and 4-hydroxy-2-ketovalerate, respectively). In α-IPMS, however, an acetyl group is transferred directly from acetyl-CoA to α-KIV, in an aldol condensation, whereas in DmpG, an acetyl group is removed from 4-hydroxy-2-ketovalerate, in an aldol cleavage, and transferred by means of a long channel to CoA on the DmpF domain, effectively the reverse reaction separated in space.
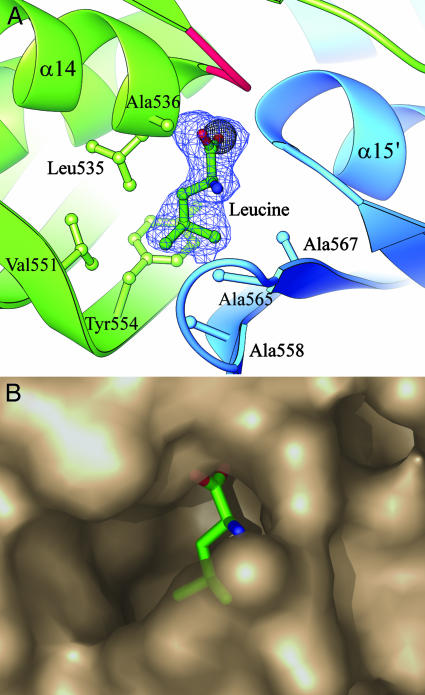
Role of the C-Terminal Domain. The C-terminal domain has a novel fold that is a duplication of a βββα structure previously seen only in an αβββα double-stranded RNA binding domain (30). This structure gives no clue to its function in α-IPMS, however, because the double-stranded RNA binding site corresponds to the N-terminal end of the helices, which in α-IPMS are buried in the dimer interface. What, then, is the role of the C-terminal domain in α-IPMS? Evidence for a functional role comes from characterization of seven natural mutants of S. cerevisiae α-IPMS that give resistance to 5,5,5-trifluoro-dl-leucine (34). These mutations all map to the C-terminal domain and affect the regulatory control of α-IPMS activity; we therefore now identify this as a regulatory (R) domain. Three mutations that abolish leucine feedback inhibition, G514D, G516D, and S519T in S. cerevisiae α-IPMS, map to residues Gly-531, Gly-533, and Ala-536 in the M. tuberculosis enzyme, which are clustered at the dimer interface adjacent to a small cavity between the N-termini of opposing helices α14 and α15′ (Fig. 3A). This cavity is occupied in the substrate structure by a partial-occupancy Cl- ion (Fig. 3A).
Fig. 3.
Leucine binding site for feedback inhibition. (A) Leucine is shown built into its electron density (2Fo - Fc omit map, contoured at 1σ) in the leucine complex structure. The carboxyl group displaces a chloride ion (gray sphere) seen in the leucine-free substrate structure, between the N-termini of helices α14 and α15′. Mutation of residues equivalent to Gly-531, Gly-533 (in the loop colored red), and Ala-536 abolish feedback inhibition in S. cerevisiae α-IPMS. Also shown are the residues that form the hydrophobic cavity for the isopropyl side chain. (B) Semitransparent PYMOL (DeLano Scientific, www.pymol.org) surface diagram of the leucine binding pocket.
To test our hypothesis that this could be the allosteric leucine binding site, we cocrystallized α-IPMS with leucine. We found that leucine is indeed bound in this site, displacing the chloride ion. There are two equivalent sites per dimer, with leucine bound in the same fashion in both. Its carboxyl group sits between the two helix N-termini, neutralizing their positive charge and hydrogen-bonding to peptide NH groups, and its side chain is in an adjacent hydrophobic pocket bounded by Leu-535, Ala-536, Val-551, Tyr-554, Ala-558, Ala-565, and Ala-567 (Fig. 3). An adjacent loop folds slightly over the binding site, and becomes better ordered, on leucine binding, but we do not see any larger changes that would lead to inhibition of catalysis.
Other examples of enzymes that are subject to end-product regulation, and bind their product inhibitors on domains remote from their catalytic domains, include ATP phosphoribosyl transferase (HisG), phosphoglycerate dehydrogenase (PGD), and threonine deaminase (TD), which are subject to regulation by histidine, serine, and isoleucine/valine, respectively. In HisF, histidine binding causes a major conformational change and a dimer-to-hexamer transition (35). No such change occurs in α-IPMS, so PGD and TD may be better analogs. In PGD, paired regulatory domains bind serine at their dimer interface and are believed to communicate this binding to the distant catalytic site by means of flexible domain connections (12). A similar mechanism is believed to operate in TD, where again the regulatory domains dimerize (13). The most important common theme may be the concept of binding at a dimer interface being translated to a distant catalytic domain by means of flexible domain linkages. Although the exact details of the feedback mechanism in α-IPMS are not apparent from our crystal structures, some of the mutations that affect leucine feedback inhibition or regulation by CoA in S. cerevisiae α-IPMS correspond to residues Glu-556, His-557, and Ala-568, which are located on the lower β-sheet, in contact with the linker subdomain II. Mutations here could affect communication with the catalytic domain by altering the orientations of the linker subdomains and thence the residues (His-379 and Tyr-410) that project from them into the active site.
Acknowledgments
We thank Dr. Li-Wei Hung for the synchrotron data collection at Brookhaven National Laboratory. This work was carried out as part of the TB Structural Genomics Consortium (www.doe-mbi.ucla.edu/TB) and received financial support from the Health Research Council of New Zealand.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TB, tuberculosis; IPM, isopropylmalate; α-IPMS, α-IPM synthase; α-KIV, α-ketoisovalerate; SeMet, selenomethionine; MS, malate synthase; R domain, regulatory domain.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1sr9).
References
- 1.Rattan, A., Kalia, A. & Ahmad, N. (1998) Emerg. Infect. Dis. 4, 195-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan, P. J. & Nikaido, H. (1995) Annu. Rev. Biochem. 64, 29-63. [DOI] [PubMed] [Google Scholar]
- 3.O'Regan, A. & Joyce-Brady, M. (2001) Brit. Med. J. 323, 635 (lett.). [PMC free article] [PubMed] [Google Scholar]
- 4.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2003) Mol. Microbiol. 48, 77-84. [DOI] [PubMed] [Google Scholar]
- 5.McAdam, R. A., Weisbrod, T. R., Martin, J., Scuderi, J. D., Brown, A. M., Cirillo, J. D., Bloom, B. R. & Jacobs, W. R., Jr. (1995) Infect. Immun. 63, 1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bange, F. C., Brown, A. M. & Jacobs, W. R., Jr. (1996) Infect. Immun. 64, 1794-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hondalus, M. K., Bardarov, S., Russell, R., Chan, J., Jacobs, W. R., Jr., & Bloom, B. R. (2000) Infect. Immun. 68, 2888-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grandoni, J. A., Marta, P. T. & Schloss, J. V. (1998) J. Antimicrob. Chemoth. 42, 475-482. [DOI] [PubMed] [Google Scholar]
- 9.Kohlhaw, B., Leary, T. R. & Umbarger, H. E. (1969) J. Biol. Chem. 244, 2218-2225. [PubMed] [Google Scholar]
- 10.Patek, M., Krumbach, K., Eggeling, L. & Sahm, H. (1994) Appl. Environ. Microbiol. 60, 133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohlhaw, G. B. (1988) Methods Enzymol. 166, 414-423. [DOI] [PubMed] [Google Scholar]
- 12.Schuller, D. J., Grant, G. A. & Banaszak, L. J. (1995) Nat. Struct. Biol. 2, 69-76. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher, D. T., Gilliland, G. L., Xiao, G., Zondlo, J., Fisher, K. E., Chinchilla, D. & Eisenstein, E. (1998) Structure (London) 6, 465-475. [DOI] [PubMed] [Google Scholar]
- 14.Ulm, E. H., Böhme, R. & Kohlhaw, G. (1972) J. Bacteriol. 110, 1118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koon, N., Squire, C. J. & Baker, E. N. (2004) Acta Crystallogr. D 60, in press. [DOI] [PubMed]
- 16.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 17.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitt, D. G. (2001) Acta Crystallogr. D 57, 1013-1019. [DOI] [PubMed] [Google Scholar]
- 20.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 21.Brünger, A. T., Adams, P. T., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 23.Blanco, M. (1991) J. Comput. Chem. 12, 237-247. [Google Scholar]
- 24.Jones, G., Willet, P. & Glen, R. C. (1995) J. Mol. Biol. 245, 45-53. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski, R., MacArthur, M., Moss, D. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 26.Carson, M. (1997) Methods Enzymol. 277, 493-505. [PubMed] [Google Scholar]
- 27.Chanchaem, W. & Palittapongarnpim, P. (2002) Tuberculosis 82, 1-6. [DOI] [PubMed] [Google Scholar]
- 28.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536-540. [DOI] [PubMed] [Google Scholar]
- 29.Manjasetty, B. A., Powlowski, J. & Vrielink, A. (2003) Proc. Natl. Acad. Sci. USA 100, 6992-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rak, A., Kalinin, A., Shcherbakov, D. & Bayer, P. (2002) Biochem. Biophys. Res. Comm. 299, 710-714. [DOI] [PubMed] [Google Scholar]
- 31.Anstrom, D. M., Kallio, K. & Remington, S. J. (2003) Protein Sci. 12, 1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard, B. R., Endrizzi, J. A. & Remington, S. J. (2000) Biochemistry 39, 3156-3168. [DOI] [PubMed] [Google Scholar]
- 33.Smith, C. V., Huang, C.-C., Miczak, A., Russell, D. G., Sacchettini, J. C. & Honer zu Bentrup, K. (2003) J. Biol. Chem. 278, 1735-1743. [DOI] [PubMed] [Google Scholar]
- 34.Cavalieri, D., Casalone, E., Bendoni, B., Fia, G., Polsinelli, M. & Barberio, C. (1999) Mol. Gen. Genet. 261, 152-160. [DOI] [PubMed] [Google Scholar]
- 35.Cho, Y., Sharma, V. & Sacchettini, J. C. (2003) J. Biol. Chem. 278, 8333-8339. [DOI] [PubMed] [Google Scholar]