Abstract
Specific recognition of double-stranded RNA (dsRNA) by dsRNA-binding domains (dsRBDs) is involved in a large number of biological and regulatory processes. Although structures of dsRBDs in complex with dsRNA have revealed how they can bind to dsRNA in general, these do not explain how a dsRBD can recognize specific RNAs. Rnt1p, a member of the RNase III family of dsRNA endonucleases, is a key component of the Saccharomyces cerevisiae RNA-processing machinery. The Rnt1p dsRBD has been implicated in targeting this endonuclease to its RNA substrates, by recognizing hairpins closed by AGNN tetraloops. We report the solution structure of Rnt1p dsRBD complexed to the 5′ terminal hairpin of one of its small nucleolar RNA substrates, the snR47 precursor. The conserved AGNN tetraloop fold is retained in the protein-RNA complex. The dsRBD contacts the RNA at successive minor, major, and tetraloop minor grooves on one face of the helix. Surprisingly, neither the universally conserved G nor the highly conserved A are recognized by specific hydrogen bonds to the bases. Rather, the N-terminal helix fits snugly into the minor groove of the RNA tetraloop and top of the stem, interacting in a non-sequence-specific manner with the sugar-phosphate backbone and the two nonconserved tetraloop bases. Mutational analysis of residues that contact the tetraloop region show that they are functionally important for RNA processing in the context of the entire protein in vivo. These results show how a single dsRBD can convey specificity for particular RNA targets, by structure specific recognition of a conserved tetraloop fold.
Binding of double-stranded RNA (dsRNA) by proteins is mediated by dsRNA-binding domains (dsRBDs), a 65- to 75-aa domain with a conserved α1β1β2β3α2 fold (1, 2). dsRBD-containing proteins play essential roles in a wide variety of biological and regulatory processes (3, 4). dsRBDs are found in dsRNA-dependent protein kinases, in proteins involved in dsRNA editing, developmental regulation through RNA localization and translational regulation, as well as dsRNA endonucleases of the RNase III family (3). The latter include the Dicer and Drosha proteins that play key roles in RNA interference and in the processing of microRNAs (5, 6) which bind to and cleave dsRNA (7–9).
The identification of the mechanism by which dsRBDs bind dsRNA would provide a structural framework to study a number of regulatory processes based on dsRBD–dsRNA interactions. Two structures of dsRBDs, from Xenopus laevis RNA-binding protein A (Xlrbpa) and Drosophila melanogaster Staufen, in complex with model dsRNA have been described (10, 11). In these complexes, the conformation of the bound protein is essentially the same as that of the free protein, and the helix α1, the β3–α2 loop, and the β1–β2 loop (Fig. 1A) interact primarily with the sugar-phosphate backbone of successive minor, major, and minor grooves, respectively, on one face of the RNA. These complex structures explain how dsRBDs can recognize and bind dsRNA vs. ds or single-stranded (ss) DNA or ssRNA, but leave open the question of how dsRBDs recognize specific dsRNA targets. Most proteins containing dsRBDs have substrate specificity in vivo, and it has been shown that at least in some cases this specificity is conferred by the dsRBDs (12). Some proteins contain multiple dsRBDs, which may contribute to more specific interactions (3). Identifying the structural basis for the binding specificity of individual dsRBDs is a major challenge in our understanding of the biological roles and specificities of these ubiquitous RNA-binding domains.
Fig. 1.
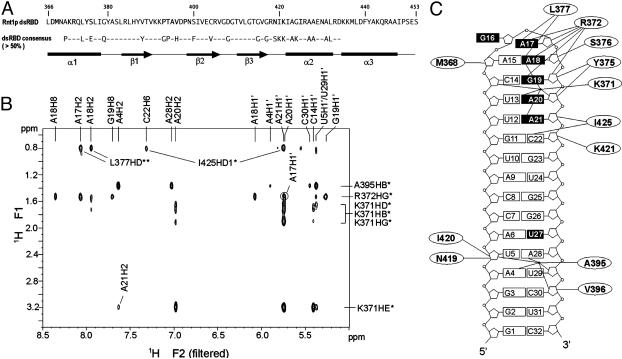
(A) Sequence of Rnt1p dsRBD (366–453) used in the NMR studies. Amino acids that contact the RNA in the complex are shown in bold. The consensus sequence (>50%) of dsRBDs (3) and a schematic of the secondary structure elements of Rnt1p dsRBD in the complex are indicated below. (B) Portion of an F1f NOESY (22) spectrum acquired on a 13C,15N-Rnt1p dsRBD/unlabeled snR47h sample in D2O. The intermolecular NOE assignments are indicated. (C) Sequence of snR47h and schematic overview of intermolecular NOEs observed between Rnt1p dsRBD and RNA. Bases of nucleotides that were affected in interference assays (15) are represented by black boxes.
Rnt1p, a key component of the yeast RNA processing machinery, is the only Saccharomyces cerevisiae representative of the RNase III family. Rnt1p is involved in the processing of the precursors of a large number of stable RNAs, including ribosomal RNAs, small nuclear RNAs, and small nucleolar RNAs (snoRNAs) (13), and in degradation pathways for unprocessed pre-mRNAs (14). All Rnt1p substrates contain stem–loop structures capped by tetraloops with the consensus sequence AGNN (13, 15). Although most bacterial and eukaryotic RNases III cleave dsRNA in a non-sequence-specific manner, the presence of these AGNN terminal tetraloops is a strong determinant for Rnt1p binding and cleavage (15, 16). The AGNN tetraloop present at one end of the dsRNA has been shown to dictate the position of the cleavage site, 14–16 bp away from the tetraloop (15). Structural studies have shown that AGNN tetraloops adopt a specific fold, independently from the binding of Rnt1p (17, 18). Deletion analyses have shown that the single dsRBD of Rnt1p is responsible for the specificity of Rnt1p for these terminal tetraloops (16). Thus, it is likely that the Rnt1p dsRBD recognizes and binds to the dsRNA substrate at the tetraloop, thereby positioning the nuclease domain at the cleavage site. In this model, the dsRBD would play a major function in the ruler-type mechanism of the enzyme.
Here we present the solution structure of the Rnt1p dsRBD complexed to a 14-bp RNA hairpin capped by an AGAA tetraloop, derived from one of the Rnt1p substrates, the snR47 precursor RNA. This is the only structure of a dsRBD in complex with a physiologically relevant RNA target reported to date. Surprisingly, the structure reveals that the dsRBD recognizes the fold of the terminal tetraloop rather than the conserved sequence, and only the nonconserved nucleotides of the tetraloop are contacted by the protein. The fold of the tetraloop and a bend at the adjacent base pairs provide a minor groove-binding pocket for the two helical turns of the N-terminal helix α1. Mutational analysis of residues of helix α1 that contact the tetraloop region show that these residues are functionally important for RNA processing in vivo. These results show how a single dsRBD can convey specificity for particular RNA targets, by fold recognition of a minimally conserved tetraloop sequence.
Materials and Methods
NMR Sample Preparation. A peptide consisting of residues 366–453 of Rnt1p (Rnt1p dsRBD) was overexpressed as a glutathione transferase (GST) fusion protein in BL21-Codon-plus (RIL) cell (Stratagene). The Rnt1p dsRBD was purified by glutathione affinity chromatography, thrombin cleavage, and gel filtration, and concentrated by using Centriprep (Millipore). NMR samples of a 32-nt RNA (snR47h) (Fig. 1C) derived from the 5′ extension of snR47 snoRNA (15) (nucleotides 12–45; U18-A21 deleted, U17 replaced by C) were prepared unlabeled, uniformly 13C,15N-labeled, and A-, U-, G-, and C- 13C,15N-labeled as described (17). All protein–RNA samples were prepared at 1:1 molar ratio, 1.0–1.5 mM in 20 mM NaPi buffer, pH 6.5, 150 mM NaCl in either 2H2O or 95% 1H2O/5% 2H2O.
NMR Spectroscopy. All NMR spectra were recorded at 30°C on Bruker DRX 500- and 600-MHz spectrometers. Protein resonances in the complex were assigned from five 3D NMR experiments: CBCANH, CBCA(CO)NH, HBHA(CO)NH, HCCH-TOCSY, and HCCH-COSY (19) recorded on 13C,15N-labeled Rnt1p dsRBD/unlabeled snR47h samples. The snR47h RNA in the complex was assigned from 2D NOESY, TOCSY, and 1H-13C HSQC and 3D HCCH-TOCSY and HCCH-COSY spectra on labeled and unlabeled samples as described (17, 20). 2D NOESY, 3D 13C-NOESY-HMQC (heteronuclear multiple quantum correlation) (21) and 3D 15N-NOESY-HSQC (heteronuclear single quantum correlation) (19) experiments were used to obtain NOEs for distance restraints. Additional RNA and all intermolecular NOEs were assigned by using a suite of four 2D filtered/edited NOESY experiments (22) on four samples of unlabeled Rnt1p dsRBD with 13C-,15N-A-, U-, C-, or G-labeled RNA and one 13C,15N-labeled Rnt1p dsRBD/unlabeled RNA sample. Residual dipolar couplings (RDCs) were measured from 1JHN differences in t1-coupled HSQCs in the presence and absence of C12E6/hexanol (23).
Structure Determination. A total of 2,359 experimental distance restraints were obtained from NOE intensities and categorized as strong (1.8–3.0 Å), medium (1.8–4.5 Å), and weak (1.8–6.0 Å). Hydrogen-bond restraints were used for the 80 amino acids that had slowly exchanging amides and for the 14 Watson–Crick base pairs. Ribose conformation and χ angles were analyzed as described (17). α, β, γ, and ε torsion angles (±30°) were included for nucleotides with NOE patterns consistent with A-form geometry (Table 1). The structures were calculated by using NIH x-plor (24) with standard simulated annealing protocols. First, 100 templates were generated from extended protein and RNA structures with randomized orientations. The protein and RNA were initially separated by 70 Å and were folded simultaneously during 80 ps of high-temperature dynamics followed by 75 ps of slow cooling from 2,000 K to 100 K. The structures were then subjected to two rounds of refinement from 1,000 K to 100 K. Forty-three RDC restraints (±2 Hz) were incorporated during the second round. The force constant for RDCs was slowly increased from 0.001 to 0.2 Kcal·mol-1·Hz-2. Axial (-35 Hz) and rhombic (0.66) components of the alignment tensor were derived from a grid search procedure (25). Fifteen structures were selected for analysis based on total energy. Experimental and structural statistics are summarized in Table 1. The structures were analyzed by using molmol. Hydrogen bonds between the protein and RNA were deduced based on heavy atom distances and angles (<3.4 Å and >120° for direct and 5.0 Å for water-mediated H-bonds).
Table 1. Structure calculation statistics.
| Distance and dihedral restraints | Protein | RNA |
| Total NOE restraints | 1,614 | 699 |
| Intraresidue | 606 | 229 |
| Sequential (i+2) | 405 | 340 |
| Medium (i+2 to i+4) | 303 | |
| Long range (> i+4) | 261 | 130 |
| Ambiguous | 39 | |
| Intermolecular NOE restraints | 46 | |
| Hydrogen bond restraints | 80 | 34 |
| RDC restraints | 43 | |
| Dihedral angle restraints | 225 | |
| Structure statistics (15 lowest energy structures) | ||
| No. of NOE violation > 0.2 Å | 1.5 ± 1.2 | |
| No. of NOE violation > 0.5 Å | 0 | |
| Maximum violation, Å | 0.378 | |
| Mean deviation of NOE violation, Å | 0.019 ± 0.001 | |
| rms deviation of RDC, Hz | 0.52 ± 0.01 | |
| Angle violations > 5° | 0 | |
| rms deviation from ideal covalent geometry | ||
| Bond length, Å | 0.0034 ± 0.00004 | |
| Angle, ° | 0.764 ± 0.004 | |
| Improper, ° | 0.36 ± 0.01 | |
| rms deviation from the mean structure (15 lowest energy structures) | Backbone | Heavy atoms |
| Protein (366—447), Å | 0.66 ± 0.12 | 1.10 ± 0.11 |
| RNA (1—32), Å | 0.87 ± 0.22 | 0.82 ± 0.18 |
| Complex (1—32, 366—447), Å | 0.87 ± 0.18 | 1.04 ± 0.14 |
The force constants used in the final refinement: 50 kcal·mol-1·Å-2 for distance restraints, 200 kcal·mol-1·Rad-2 for torsion angle restraints, and 4 kcal·mol-1·Å-2 for the quartic van der Waals repulsion terms.
Mutagenesis and in Vivo RNA Processing Assays. Mutations were introduced in the S. cerevisiae BMA64 strain (26) at the chromosomal RNT1 (YMR239C) locus by a two-step homologous recombination method (27). All mutations were confirmed by sequencing of 200 nucleotides of the region spanning the site of the mutation. Northern blot analysis of Rnt1p substrates was performed as described (14, 26). Western blot analysis was performed by using affinity purified polyclonal antibodies raised against a GST–Rnt1p fusion (Covance).
Results
Rnt1p dsRBD Forms a Specific Complex with the snR47 Precursor 5′ Terminal Hairpin. We chose to study a derivative of the 5′ terminal hairpin of the yeast snR47 snoRNA precursor, because the interaction of Rnt1p with this substrate has been well characterized biochemically (15, 17) and the structure of its AGAA tetraloop had been determined (17). The 32-nt hairpin contains the sequence immediately adjacent to the Rnt1p cleavage site, with the four-base bulge in the middle of the stem deleted because it has no effect on Rnt1p binding or substrate cleavage (G.C., unpublished data), and these internal loops are not conserved among Rnt1p substrates. Complex formation between Rnt1p dsRBD and snR47h was initially assayed by chemical shift changes observed in 1H-15N HSQC spectra on addition of unlabeled RNA to 15N-labeled Rnt1p dsRBD (Fig. 6, which is published as supporting information on the PNAS web site). A subset of the crosspeaks shifted at each step of the titration until a stoichiometric ratio of protein to RNA was reached, indicating that a specific complex in fast exchange on the NMR time scale was formed without significant rearrangement of the protein structure. 1H-13C HSQC spectra of the reverse titration of unlabeled protein with 13C,15N-labeled RNA showed significant chemical shift changes for nucleotides A5, C14-A21, U29, and C30. Formation of a specific complex was further confirmed by the observation of intermolecular NOEs in 2D filtered/edited NOESY spectra (22) (Fig. 1B).
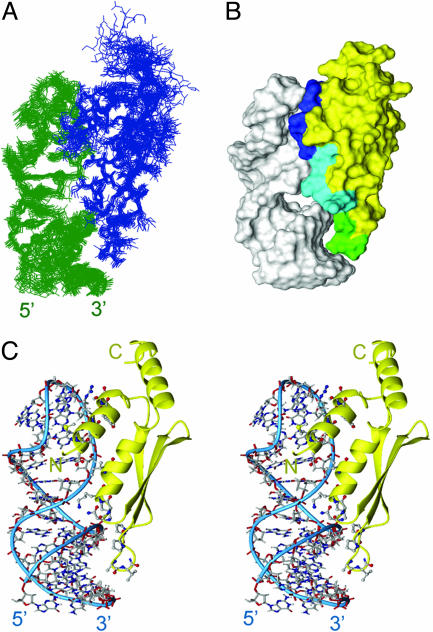
Overview of the Rnt1p dsRBD–snR47h RNA Complex. The structure of the Rnt1p dsRBD–snR47h complex (Fig. 2A) is well determined with an overall rms deviation of 1.04 Å (Table 1). The protein–RNA interface was defined by 46 unambiguously assigned intermolecular NOEs between the protein and the RNA tetraloop, major groove, and minor groove protons (Fig. 1 B and C). Rnt1p dsRBD adopts a fold similar to previously described αβββα dsRBD structures (28, 29) but with the addition of a third α-helix at the C terminus. The three amphipathic α-helices are packed against one another on one face of the three-stranded antiparallel β-sheet. The hydrophobic face of α3 interacts extensively with the C terminus of α1 and α1–β1 loop. A charged guanadinium group (R445) is located near the C terminus (negative end of the helix dipole) of α1.
Fig. 2.
Overview of the Rnt1p dsRBD–snR47h RNA complex. (A) Superposition of the 15 lowest energy structures. The protein is shown in blue, and RNA is shown in green. (B) Solvent-accessible surface of the lowest energy structure. Amino acids contacting successive minor, major, and tetraloop minor grooves on one face of the RNA helix are colored green, cyan, and blue, respectively. The rest of the protein is yellow, and the RNA is white. (C) Stereoview of the lowest energy structure. The RNA is shown in lines with the helical backbone indicated by thin blue cylinder and the protein in ribbons with amino acids at the protein–RNA interface shown as ball and sticks.
The RNA in the complex retains the A-form stem and the AGNN tetraloop fold present in the free RNA (17) (Fig. 2). The backbone turn of the tetraloop is between the second and third nucleotide, and is facilitated by the syn G, which has a hydrogen bond between its amino group and nonbridging phosphate oxygen. The first A and universally conserved G stack on the 5′ side of the helix and point into the major groove, whereas the last two As stack on the 3′ side of the helix and point into the minor groove. The tetraloop is closed by a sheared A15–A18 base pair. The one significant change in the RNA structure in the complex is a small bend in the top of the helix just below the tetraloop toward the minor groove. Both chemical shift changes and NOEs provide experimental support for the conformational change in this region of the RNA.
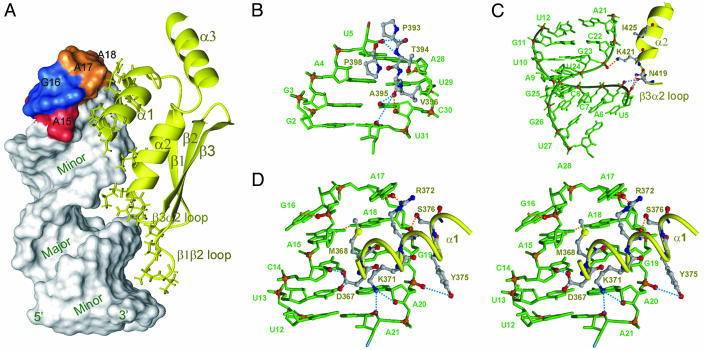
Rnt1p dsRBD binds on one face of the RNA over almost its entire length (13 bp plus tetraloop) and interacts primarily with the sugar-phosphate backbone in three regions (Fig. 2B). Residue side chains insert into successive minor and major grooves, with helix α1 lying in the minor groove of the tetraloop and the adjacent two base pairs, the N-terminal end of helix α2, and the β3–α2 loop contacting the sugar-phosphate backbone of the major groove, and the β1–β2 loop contacting the minor groove 10–13 bp away from the tetraloop, as described in detail below.
The dsRBD Binds Successive Minor and Major Grooves Along One Face of the RNA Helix. The interactions between Rnt1p dsRBD and the RNA stem are similar to those observed in the crystal structure of the Xlrbpa dsRBD–RNA complex (10). The spacing between the β1–β2 loop and the N-terminal end of helix α2 is ideal for interaction of these regions with adjacent minor and major grooves, respectively, of an A-form RNA helix (Fig. 3A). At the bottom of the RNA stem, four residues on the β1–β2 loop contact the minor groove of base pairs 2–5 (Fig. 3B). Three peptide backbone groups (P393 CO, T394 NH, and A395 CO) interact with three ribose groups (U5 2′OH, C30 2′OH, and U31 O4′) and one base (C30 O2) via direct and water-mediated hydrogen bonds. The aliphatic side chains of A395, V396, and P398 are in van der Waals contact with U29, C30, and U5, respectively. On the same face of the RNA helix, the β3–α2 loop and the N terminus of helix α2 interact with the phosphodiester backbone across the major groove from base pairs 5–12, contacting residues U5, A6, G23, and A21 (Fig. 3C). On the 5′ side of the major groove, the N419 backbone CO likely contacts the phosphate groups of U5 and A6 via water-mediated hydrogen bonds, and the side chain NH forms a direct hydrogen bond with the nonbridging oxygen of U5 phosphate. Across the major groove, the side chain of K421 hydrogen bonds directly with the G23 phosphate group, and I425 side chain is in van der Waals contact with A21. Most dsRBDs have several positively charged residues in β3–α2 loop and N-terminal end of α2 that interact with the major groove, whereas in Rnt1p there are only two, N419 and K421 (Fig. 1A).
Fig. 3.
Interactions between Rnt1p dsRBD and snR47h RNA. Lowest energy structure is shown. (A) Solvent-accessible surface of the RNA showing the major and minor grooves with the AGAA tetraloop nucleotides colored red, blue, and orange, and the protein in yellow ribbon. Protein side chains that interact with the RNA are shown as sticks. (B–D) Details of specific interactions of the protein with the minor groove (B), major groove (C), and tetraloop minor groove (stereoview) (D) are shown. Nucleotides are green, with phosphates and O2′ of interacting riboses in red. Protein side chains are sticks, with backbone as yellow ribbon. Direct and water-mediated protein–RNA H-bonds are indicated as orange and blue dashed lines, respectively.
Helix α1 Specifically Recognizes the Minor Groove of the Tetraloop. One full turn of the RNA helix away from the interaction of the β1–β2 loop with the minor groove of base pairs 3–5 and two helical turns of helix α1 lie in the minor groove of the tetraloop and adjacent base pairs (Fig. 3D). The N terminus of helix α1 contacts the 5′ side of the loop, whereas the C terminus interacts with 3′ side of the loop. R372 and S376 interact via the minor groove with A17 and A18, the two nonconserved 3′ nucleotides of the AGNN tetraloop. The R372 guanidinium group and S376 OH hydrogen bond to the 2′OH of A17 and A18, respectively, and the R372 aliphatic side chain is in van der Waals contact with the two bases. On the 5′ side of the tetraloop, the nonpolar side chain of M368 is stacked on the A15 ribose. Helix α1 also contacts three base pairs below the tetraloop by using D367, K371, and Y375. The D367 side chain carboxyl interacts with C14 2′OH through a potential water-mediated hydrogen bond. The side chain of K371 inserts into the minor groove, with its aliphatic side chain in van der Waals contact with A20 and its amino group hydrogen bonding to the A20 2′OH and A21 O4′. The phenyl ring of Y375 stacks next to the G19 ribose and hydrogen bonds to the A20 phosphate group.
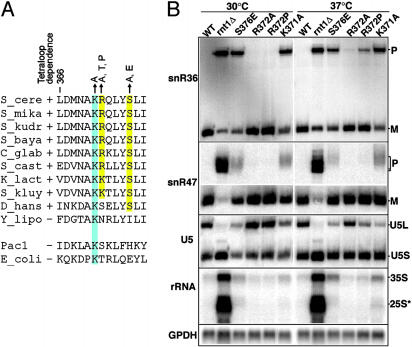
In Vivo Analysis of Helix α1 Mutants. We investigated whether residues within helix α1 that contact the tetraloop region are important for RNA processing in vivo. We focused on the three amino acids that directly interact with the tetraloop minor groove and are either universally conserved among RNases III (K371) or conserved only among RNases III that recognize AGNN tetraloops (R372 and S376) (Fig. 4A). The effect of mutation of these residues was tested in vivo by introducing the mutations into the chromosomal RNT1 gene by homologous recombination (27). K371 was mutated to Ala and S376 to Ala and Glu. This latter mutation was chosen because a Glu is often found in dsRBDs of bacterial RNases III, which do not rely on tetraloop recognition for cleavage (Fig. 4A). Thus, we hypothesized that introduction of a bacterium-specific sequence in the dsRBD may induce a negative effect on AGNN tetraloop recognition. Similarly, R372 was mutated to Thr in addition to Ala, because Thr is the most common amino acid at this position in bacteria. We also serendipitously recovered a Pro-372 mutation, which was kept for further analysis. None of these mutations would be expected to significantly destabilize Rnt1p, and consistent with this, mutant proteins were expressed at levels similar to the wild-type (data not shown).
Fig. 4.
(A) Sequence alignment for helix α1 from RNases III found in Hemiascomycetes species that require (+) or do not require (-) AGNN tetraloops for cleavage. Sequences for E. coli Rnc and Schizosaccharomyces pombe PacI are shown for comparison. The amino acid mutations studied are indicated by arrows above the sequence. (B) Northern blot analysis of RNA processing in wild-type and Rnt1p dsRBD mutants. M, mature RNA; P, precursor RNA; 35S, unprocessed primary ribosomal RNA precursor; 25S*, 3′ extended species of the 25S. GPDH is the glyceraldehyde phosphate dehydrogenase loading control.
Processing of Rnt1p substrates from the four major families, one H/ACA snoRNA (snR36), one C/D snoRNA (snR47), one spliceosomal small nuclear RNA (U5), and the 3′ external transcribed spacer (ETS) in the ribosomal RNA, was investigated by Northern analysis (Fig. 4B). The K371A and S376E mutations exhibited the strongest phenotypes, with a reduction of the mature snR36 snoRNA and a strong accumulation of unprocessed precursors. The R372P mutation showed a temperature-sensitive phenotype, with a partial accumulation of unprocessed precursor at 37°C. The R372A mutation showed a very subtle processing phenotype at 37°C, as shown by a small accumulation of unprocessed snR36 precursor at 37°C. The S376A and R372T mutations showed no obvious phenotypes at any temperature (data not shown). Analysis of the snR47 C/D snoRNA showed similar phenotypes, with the exception that the effects were more pronounced for the accumulation of unprocessed precursor than for the reduction of mature snoRNA.
The U5 small nuclear RNA is present in two forms: a long form (U5L), whose processing strictly relies on Rnt1p, and a short form (U5S) generated through a Rnt1p-independent pathway (30). Synthesis of these two forms is the result of a competition between these two pathways, and any perturbation of Rnt1p activity results in a decrease of the U5L/U5S ratio. As observed previously for the two snoRNAs substrates analyzed, the S376E and K371A mutants showed the strongest phenotypes, which were more pronounced at 37°C. The R372P mutant also exhibited a thermosensitive phenotype. No effect of R372A mutation was detected for this substrate.
Finally, we investigated the effect of the mutations on the processing of the precursor of ribosomal RNA. Rnt1p cleavage occurs in the 3′ external transcribed spacer of the pre-rRNA, and this cleavage generates the 35S pre-rRNA. In the absence of Rnt1p, extended forms of this precursor and of the 25S rRNA accumulate (31, 32) (Fig. 4B). The S376E and K371A mutants showed a significant accumulation of the 35S extended species. The 3′-extended forms of the 25S rRNA could also be observed in the K371A mutant, especially at 37°C. For the other mutants, the background level of accumulation of these extended species in the wild-type strain makes it difficult to interpret the results.
Discussion
Interaction of Helix α1 with the AGNN Tetraloop Minor Groove Determines the Binding Site Specificity of Rnt1p. We have solved the structure of the dsRBD domain of Rnt1p in complex with a target RNA derived from one of its precursor snoRNA substrates, snR47. Previous studies have shown that Rnt1p, unlike most other members of the RNase III family that cleave dsRNA with little regard to sequence specificity, specifically recognizes terminal AGNN tetraloops via its dsRBD domain and cleaves dsRNA at a fixed distance of 14–16 nt from the loop. Structural studies of RNA hairpins containing AGAA, AGUU, and AGUC tetraloops (17, 18) showed that they have a common fold with the conserved A and G stacking on each other on the 5′ side, the nonconserved nucleotides stacking on each other on the 3′ side, and a backbone turn after the G, which is in the syn conformation. The structure of the complex reveals that this conserved RNA tetraloop fold is retained in the protein–RNA complex and is specifically recognized by helix α1. Surprisingly, neither the universally conserved G nor the highly conserved A are recognized by specific hydrogen bonds to the bases. Rather, helix α1 fits snugly into the minor groove side of the tetraloop, interacting with the sugar-phosphate backbone and the nonconserved 3′ bases, and extending into the minor groove at the top of stem.
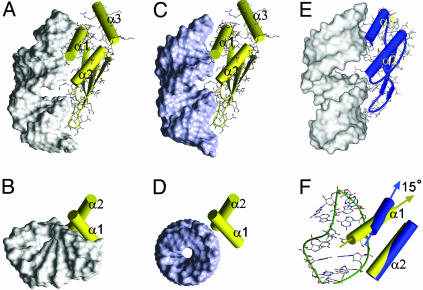
The interaction of helix α1 with the tetraloop provides an explanation for the 5-fold greater affinity of the Rnt1p dsRBD for AGNN hairpins compared to a GUGA hairpin (16). In the Xlrbpa dsRBD–dsRNA complex, helix α1 also contacts the minor groove (Fig. 5E), but only two amino acid residues interact with three nucleotides, whereas six amino acids of Rnt1p interact with seven nucelotides. The additional contacts seen in the Rnt1p dsRBD complex are caused by the fold of the tetraloop and the slight bend of the top of the stem toward the minor groove, which create a pocket in which the two turns of helix α1 fit (Fig. 5 A and B). In a modeled complex of Rnt1p dsRBD with A-form RNA, only the C terminus of helix α1 is positioned to contact the RNA (Fig. 5 C and D). A superposition of the Xlrpba and Rnt1p dsRBDs over the β-sheets and α2 (backbone rms deviation = 1.0 Å) reveals a 15° difference in the orientations of the α1 helices (Fig. 5F). This difference in the angle of α1 positions it to lie perfectly along the minor groove of the AGNN tetraloop without changing the spacing of contacts to the minor groove one turn of the helix away and the intervening major groove. The orientation of helix α1 is stabilized by interactions of its C terminus with helix α3, which is unique among structures of dsRBDs determined to date. Deletion of helix α3 results in expression of insoluble protein (not shown), consistent with a stabilizing role for this helix.
Fig. 5.
Comparison of positions of dsRBDs on RNA. Rnt1p dsRBD bound to snR47h (A and B) and modeled dsRNA (C and D). Side (A and C) and top (B and D) views are shown. (E) Side view of Xlrbpa dsRBD bound to dsRNA. (F) Comparison of Rnt1p (yellow) and Xlrpba (blue) α1 and α2 orientations on snR47h. The cylinder representations of the α-helices of Rnt1p (yellow) and Xlrpba (blue) dsRBDs were superimposed in F on their β-sheets, N termini of α2, and the β1–β2 loops. In A–E, the RNA is shown as solvent-accessible surface.
Comparison to Xlrbpa and Staufen Complexes with RNA. The structure of the Rnt1p dsRBD–snR47h complex shows that the dsRBD-binding site is equivalent in length to 15 bp including the tetraloop. In the Xlrbpa complex, the binding site is 16 bp. However, the protein is interacting with two 10-bp duplexes that are stacked in a way that results in a widened major groove at their junction (Fig. 5E). It is not clear whether this widening of the major groove is required for binding of Xlrpba, although in the structure it appears to be important for the α1 contacts to the minor groove. In any case, significant widening of the major groove does not appear to be a general requirement of dsRBD–RNA interactions, because it is not observed in the Rnt1p dsRBD–snR47h complex. In both complexes, the protein sits on top of the major groove and contacts the sugar-phosphate backbone on either side and the side chains do not insert deeply into the groove, resulting in a small gap between the protein and the RNA major groove bases (Figs. 2B and 5 A and E).
In the solution structure of the Staufen dsRBD3-RNA complex, the dsRBD interacts with a 13-bp duplex capped by a nonphysiologically relevant UUCG tetraloop. The β1–β2 loop and helix α1 interact with the minor groove at the bottom of the helix and in the region of the tetraloop, respectively. There was no experimental data for contacts with the major groove. In light of the specific interaction of Rnt1p dsRBD with the AGNN tetraloop fold, the observed interaction of Staufen dsRBD with the minor groove side of the UUCG tetraloop raises the question of whether this fortuitous interaction is biologically relevant. Because the RNA duplex in the complex was 13 bp long (including the terminal base pair), shorter than the Xlrpba and Rnt1p dsRBD-binding sites, it is unlikely that Staufen could form a complex with the RNA without contacting the UUCG tetraloop. In the model structure of the complex the RNA is severely bent, leaving a large gap between the protein and the major groove, to accommodate observed NOE contacts at both ends of the hairpin. Both the limited number of observed NOEs and resonance line broadening provided substantial evidence for conformational dynamics at the protein–RNA interface. This dynamic behavior was not observed in the Rnt1p dsRBD–snR47h complex.
Relevance of dsRBD Binding to Function of Rnt1p in Vivo. In addition to the structural evidence for the importance of the recognition of the tetraloop fold by helix α1 for dsRBD binding, modification interference data on the RNA and mutagenesis of helix α1 provide support for the importance of this interaction in the context of the full-length protein, both for binding in vitro and correct processing in vivo, respectively. Sites of interaction of helix α1 with the tetraloop and the upper part of the stem correspond to positions where base modifications have been shown to interfere with Rnt1p binding (Fig. 1C) (15). These base modifications likely disrupt the conserved fold of the tetraloop and top of the stem, which is required for structure specific binding of the dsRBD.
Helix α1 residues are highly variable in dsRBDs (3), but they show interesting conservation among fungal RNase III dsRBDs (Fig. 4A). In particular, R372 and S376 are conserved in Hemiascomycetes species that recognize AGNN tetraloops, but not in Yarrowia lipolytica, which does not rely on AGNN tetraloop recognition for cleavage (13). This phylogenetic conservation correlates well with the contacts observed between R372 and S376 and the two nonconserved nucleotides of the AGNN tetraloop. Mutation of these residues affects RNA processing in vivo, showing that the helix α1–RNA contacts observed in the Rnt1p dsRBD–snR47h complex are important for Rnt1p function. These mutations affect RNA processing to a different extent, depending on the nature of the substitution. The S376E mutation showed a strong phenotype (Fig. 4B), likely because the negatively charged residue is electrostatically unfavorable. R372A and R372T mutations showed only subtle phenotypes, likely because the Arg was replaced with smaller side chains. Replacement of R372 with a Pro, which probably puts a kink in the middle of the helix, resulted in a temperature sensitive Rnt1p phenotype. This suggests that the integrity of helix α1, which may also be interpreted as the spacing between D367 and S376, is important in tetraloop recognition.
Mutation of K371, which binds in the minor groove at the top of the stem, strongly perturbed RNA processing of all substrates in vivo. This residue is conserved not only in tetraloop-dependent RNases III, but also in Y. lipolytica, E. coli, and Schizosaccharomyces pombe RNases III, which do not rely on tetraloop binding for substrate recognition. This suggests that K371 probably contributes strongly to the binding of the substrate, but that in contrast to residues R372 and S376 has little effect on substrate specificity. This residue is not conserved in dsRBDs outside the RNase III family, and its position is N-terminal to residues that contact the RNA in the Xlrpba and Staufen complexes, further suggesting that K371 is important for substrate recognition only in the RNase III family of dsRBD proteins.
Overall, these results indicate that the observed interactions between helix α1 in the dsRBD–RNA complex in vitro are required for substrate recognition in the context of the entire protein in vivo. The endonuclease domain of Rnt1p is almost immediately N-terminal to the helix α1. This helix includes four additional residues at its N terminus compared to the dsRBD consensus. Two of these contribute to the extensive contacts of helix α1 with the RNA. We speculate that this longer helix serves the additional role of correctly positioning the endonuclease domain on the RNA for cleavage 14–16 bp away from the tetraloop.
Acknowledgments
This work was supported by National Science Foundation Grant MCB0111060 (to J.F.), National Institutes of Health Grants GM37254 (to J.F.) and GM61518 (to G.C.), and a Human Frontier Science Program Organization long-term fellowship (to A.H.).
Abbreviations: dsRNA, double-stranded RNA; sdRBD, dsRNA-binding domain; Xlrbpa, Xenopus laevis RNA-binding protein A; snoRNA, small nucleolar RNA; RDC, residual dipolar coupling.
Data deposition: The atomic coordinates of the Rnt1p dsRBD–snR47h RNA complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID 1T4L).
References
- 1.St. Johnston, D., Brown, N. H., Gall, J. G. & Jantsch, M. (1992) Proc. Natl. Acad. Sci. USA 89, 10979-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyle, M. & Jantsch, M. F. (2002) J. Struct. Biol. 140, 147-153. [DOI] [PubMed] [Google Scholar]
- 3.Fierro-Monti, I. & Mathews, M. B. (2000) Trends Biochem. Sci. 25, 241-246. [DOI] [PubMed] [Google Scholar]
- 4.Saunders, L. R. & Barber, G. N. (2003) FASEB J. 17, 961-983. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- 6.Knight, S. W. & Bass, B. L. (2001) Science 293, 2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutvagner, G., McLachlan, J., Pasquinelli, A. E., Balint, E., Tuschl, T. & Zamore, P. D. (2001) Science 293, 834-838. [DOI] [PubMed] [Google Scholar]
- 8.Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J. & Plasterk, R. H. (2001) Genes Dev. 15, 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. (2003) Nature 425, 415-419. [DOI] [PubMed] [Google Scholar]
- 10.Ryter, J. M. & Schultz, S. C. (1998) EMBO J. 17, 7505-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos, A., Grunert, S., Adams, J., Micklem, D. R., Proctor, M. R., Freund, S., Bycroft, M., St. Johnston, D. & Varani, G. (2000) EMBO J. 19, 997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle, M. & Jantsch, M. F. (2003) J. Cell Biol. 161, 309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanfreau, G. (2003) Eukaryot. Cell 2, 901-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danin-Kreiselman, M., Lee, C. Y. & Chanfreau, G. (2003) Mol. Cell 11, 1279-1289. [DOI] [PubMed] [Google Scholar]
- 15.Chanfreau, C., Buckle, M. & Jacquier, A. (2000) Proc. Natl. Acad. Sci. USA 97, 3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagel, R. & Ares, M. (2000) RNA 6, 1142-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu, H., Pok, K., Butcher, S. E., Kang, S., Chanfreau, G. & Feigon, J. (2001) EMBO J. 20, 7240-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebars, I., Lamontagne, B., Yoshizawa, S., Aboul Elela, S. & Fourmy, D. (2001) EMBO J. 20, 7250-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavanagh, J., Fairbrother, W. J., Palmer, A. G., III, & Skelton, N. J. (1996) Protein NMR Spectroscopy: Principles and Practice (Academic, San Diego).
- 20.Dieckmann, T. & Feigon, J. (1997) J. Biomol. NMR 9, 259-272. [DOI] [PubMed] [Google Scholar]
- 21.Marion, D., Kay, L. E., Sparks, S. W., Torchia, D. A. & Bax, A. (1989) J. Am. Chem. Soc. 111, 1515-1517. [Google Scholar]
- 22.Peterson, R. D., Theimer, C. A., Wu, H. & Feigon, J. (2004) J. Biomol. NMR 28, 59-67. [DOI] [PubMed] [Google Scholar]
- 23.Ruckert, M. & Otting, G. (2000) J. Am. Chem. Soc. 122, 7793-7797. [Google Scholar]
- 24.Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. (2003) J. Magn. Reson. 160, 65-73. [DOI] [PubMed] [Google Scholar]
- 25.Clore, G. M., Gronenborn, A. M. & Tjandra, N. (1998) J. Magn. Reson. 131, 159-162. [DOI] [PubMed] [Google Scholar]
- 26.Chanfreau, G., Rotondo, G., Legrain, P. & Jacquier, A. (1998) EMBO J. 17, 3726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storici, F., Lewis, L. K. & Resnick, M. A. (2001) Nat. Biotechnol. 19, 773-776. [DOI] [PubMed] [Google Scholar]
- 28.Kharrat, A., Macias, M. J., Gibson, T. J., Nilges, M. & Pastore, A. (1995) EMBO J. 14, 3572-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bycroft, M., Grunert, S., Murzin, A. G., Proctor, M. & Stjohnston, D. (1995) EMBO J. 14, 3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chanfreau, G., AbouElela, S., Ares, M. & Guthrie, C. (1997) Genes Dev. 11, 2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abou Elela, S., Igel, H. & Ares, M. (1996) Cell 85, 115-124. [DOI] [PubMed] [Google Scholar]
- 32.Kufel, J., Dichtl, B. & Tollervey, D. (1999) RNA 5, 909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]