Abstract
Per–Arnt–Sim (PAS) domain-containing kinases are common in prokaryotes, but a mammalian counterpart has only recently been described. Although the PAS domain of the mammalian PAS kinase (PASK) is closely related to the bacterial oxygen sensor FixL, it is unclear whether PASK activity is changed in mammalian cells in response to nutrients and might therefore contribute to signal transduction by these or other stimuli. Here, we show that elevated glucose concentrations rapidly increase PASK activity in pancreatic islet β cells, an event followed by the accumulation of both PASK mRNA and protein. Demonstrating a physiological role for PASK activation, comicroinjection into clonal β cells of cDNA encoding wild-type PASK, or PASK protein itself, mimics the induction of preproinsulin promoter activity by high glucose concentrations. Conversely, anti-PASK antibodies block promoter activation by the sugar, and the silencing of PASK expression by RNA interference suppresses the up-regulation by glucose of preproinsulin and pancreatic duodenum homeobox 1 gene expression, without affecting glucose-induced changes in the levels of mRNAs encoding glucokinase or uncoupling protein 2. We conclude that PASK is an important metabolic sensor in nutrient-sensitive mammalian cells and plays an unexpected role in the regulation of key genes involved in maintaining the differentiated phenotype of pancreatic β cells.
Keywords: PAS, insulin, secretion, pancreatic β cell
Per–Arnt–Sim (PAS) protein domains act as modular sensors of the intracellular environment, able to monitor a variety of parameters such as light, oxygen, or redox state (1). The PAS domain-containing protein kinase, PASK (2) (also termed PASKIN) (3), is a recently identified mammalian relative of the Rhizobia oxygen-sensing protein FixL (4) and the Saccharomyces cerevisiae serine/threonine kinases, PSK1 and PSK2 (5), each of which bears PAS domains essential for function. Whereas PSK1 and PSK2 regulate protein synthesis and glycolytic flux in yeast (6), the role(s) and means of regulation of PASK in mammalian cells are at present unclear (7).
PASK possesses two N-terminal PAS domains and a C-terminal kinase domain regulated by phosphorylation on Thr-1161 in the canonical activation loop. Phosphorylation of Thr-1161 seems to play an analogous role in the activation of PASK to that of Thr-172 in 5′-AMP-activated protein kinase (AMPK), the mammalian ortholog of yeast nutrient-sensing kinase, SNF1 (8), and relative of PASK, by an upstream kinase, LKB1 (9, 10).
Pancreatic islet β cells respond to elevations in blood glucose concentration with an increase in ATP synthesis (11, 12), closure of ATP-sensitive K+ channels (13), and the release of stored insulin (14). We have recently demonstrated that AMPK is involved in controlling the synthesis (15) and secretion (8, 16, 17) of insulin from the pancreatic islet. Thus, increases in glucose concentration lead to a decrease in AMPK activity in clonal β cells (8, 15, 17) and in islets (18), and forced expression of activated AMPK suppresses insulin gene expression (15) and glucose-stimulated insulin secretion (16, 17). Here, we show that PASK is also regulated by glucose in β cells and may play a complementary role in the regulation of gene expression.
Materials and Methods
Materials. The silencer small interfering RNA (siRNA) construction kit was from Ambion (Austin, TX). siRNA oligonucleotides were from Cruachem (Herndon, VA). TransIT-TKO transfection reagent was from Mirus (Madison, WI), human growth hormone (hGH) ELISA kit was from Roche Diagnostics, and rat insulin radioimmunoassay kit was from Linco Research Immunoassay (St. Charles, MO). Tissue culture reagents were from Sigma or GIBCO/BRL. Lipofectamine 2000 was from Invitrogen, collagenase was from Boehringer Mannheim, and human extracellular matrix was from Becton Dickinson. Polyclonal anti-hPASK antibody (U2501) was as described (2). γ-[32P]ATP was from Amersham.
Plasmids. pcDNA3.hGH was a gift from R. Burgoyne (University of Liverpool, Liverpool, U.K.). pINS.LucFF contained nucleotides -260 to -60 bp of the human preproinsulin promoter fused upstream of the minimal herpes simplex thymidine kinase promoter and humanized luciferase (19). Plasmid pCMV.RL encoded Renilla reniformis luciferase under cytomegalovirus promoter control (20). Plasmids based on pcDNA3.1 (Invitrogen) and encoding wild-type human PASK bearing C-terminal c-myc and his epitope tags (pPASK.WT) or inactive PASK mutated at the ATP-binding site (K1028R; pPASK.KD) were as described (2). pPDX1.LucFF, encoding the 5′ flanking region of the mouse pancreatic duodenum homeobox 1 (PDX-1) gene (-2,715 to 0 bp), was generated from MIN6 cell cDNA (Q. Qian and G.A.R., unpublished data). Site-directed mutagenesis was performed using the QuikChange kit (Stratagene) and verified by automated sequencing. Primer sequences are available upon request.
Semiquantitative and Real-Time RT-PCR. Primers against mouse PASK were as follows: PASKIN5.1, 5′-CCACCTTCCCTCTCAGT T TG-3′; and PASKIN3.1, 5′-CAGCTCCAACTGAGCTTCCT-3′ (3). mRNAs encoding hepatic nuclear factor-1α (21), Foxa2 (HNF-3β) (21), MafA (22), NeuroD1/Beta2 (23), and upstream stimulatory factor (USF1/2) (24) were quantified by real-time PCR with Platinum SYBR Green qPCR SuperMix UDG (Invitrogen). Other mRNAs were quantified by using TaqMan (25).
Rat Islet Culture. Rat islets were isolated by intraductal collagenase digestion, purified on a Histopaque (Sigma) gradient (11), and cultured for 16–24 h in DMEM (GIBCO) containing 11 mM glucose and 30% FCS in a humidified atmosphere at 37°C with 5% CO2. Islets were plated on glass coverslips precoated with human extracellular matrix (26) and cultured in DMEM containing 10% FCS for 10 days. The glucose concentration was lowered to 3 mM 16 h before microinjection.
MIN6 Cell Culture. MIN6 β cells (27) (passages 19–30) were grown in DMEM containing 15% FCS, 25 mM glucose, 5.4 mM KCl, 2 mM glutamine, 100 mM 2-mercaptoethanol, 100 units/ml-1 penicillin, and 100 μg/ml-1 streptomycin in a humidified atmosphere at 37°C with 5% CO2, and seeded onto poly(L)-lysinecoated coverslips for microinjection. Cells were transfected with Lipofectamine 2000 or TransIT-TKO (for siRNA). Culture was continued for 24 h as above then at 3 mM glucose for 16 h.
siRNA Construction. siRNA was generated by using the Ambion Silencer siRNA construction kit (25). Target sequences were derived from the cDNA sequence for mouse PASK (GenBank accession no. NM_080850). Target and control sequences, each with the sequence 5′-CCTGTCTC-3′ at the 3′ end, were as follows: PASK target, 5′-AATTTATGGAGTCAACCACAGCTT-3′; and scrambled 5′-AAGTCAACGCTTCACTTTATGGAA-3′.
Western (Immuno-) Blotting. Cells were washed twice in ice-cold PBS, scraped in ice-cold lysis buffer (PBS/1% Triton X-100/5 μg/ml-1 pepstatin/5 μg/ml-1 antipain/5 μg/ml-1 leupeptin/2 mM benzamidine/0.5 mM DTT), and vortex-mixed. Protein was assayed with a BCA kit (Pierce) against BSA type V (Sigma) standards. Total protein extracts (50 μg) were resolved by SDS/PAGE (10% wt/vol acrylamide) and transferred to nitro-cellulose membranes, followed by immunoblotting with anti-PASK antibody (1:1,000). Secondary antibodies were revealed by using BM chemiluminescence blotting substrate (Roche Diagnostics).
Single-Cell Reporter Gene Assay. Intranuclear microinjection of plasmids, antibody, and protein was performed by using an Eppendorf 5121/5246 micromanipulator (20) at plasmid concentrations of 0.1 (pINS.LucFF) and 0.05 (pCMV.RL, pPASK.WT, pPASK.KD) mg/ml-1. Antibody against the kinase domain of PASK, purified wild-type PASK, or BSA (Sigma) as control was microinjected at 1.0 mg/ml-1. Individual experiments involved the injection of 100–200 separate cells (nucleus plus cytosol) per condition, with an efficiency of 5–20% productive injections as assessed by the expression of R. reniformis luciferase activity. Cells were imaged 6 h after microinjection. Photon-counting imaging was performed by using an Olympus IX-70 inverted microscope (×10 air objective, 0.4 numerical aperture) and an intensified charge-coupled device camera (Photek, East Sussex, U.K.) (20).
hGH Secretion. MIN6 cells, seeded in six-well plates (Falcon), were grown to 70% confluency and transfected with 1 μg of pXhGH and 1 μg of pPASK.WT or pPASK.KD with ≈30% cotransfection efficiency. Culture was continued for 24 h in DMEM containing 25 mM glucose and then at 3 mM glucose for a further 16 h. Cells were washed in PBS and incubated in modified Krebs–Ringer medium containing either 3 or 30 mM glucose. Incubations were performed for 20 min at 37°C in a shaking water bath. Secreted and total hGH was measured by ELISA.
Measurement of Intramitochondrial-Free ATP Concentration. MIN6 cells were microinjected with a plasmid encoding mitochondrially targeted firef ly luciferase (11) and empty pcDNA3, pPASK.WT, or pPASK.KD, as indicated in legends to Figs. 1, 2, 3, 4, 5. Cells were cultured for 24 h in DMEM-based medium containing 25 mM glucose, then in medium containing 3 mM glucose for 16 h before photon counting (11).
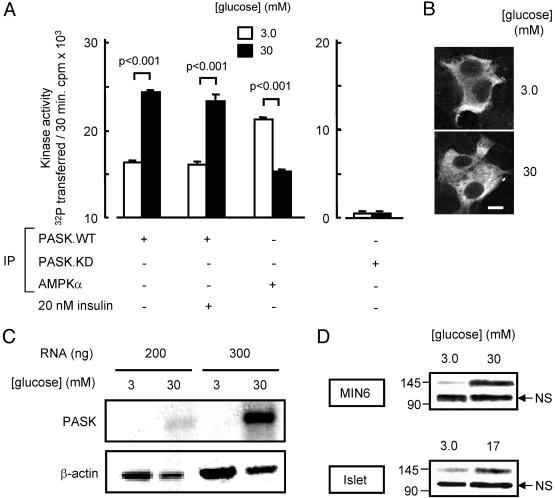
Fig. 1.
Glucose regulates PASK activity and gene expression but not its intracellular localization. (A) MIN6 cells were transfected with pcDNA3 or plasmids pPASK.WT or pPASK.KD, cultured for 16 h in 3 mM glucose (Materials and Methods), then incubated in modified Krebs–Ringer medium for 1 h at the glucose and insulin concentrations indicated. The activities of immunoprecipitated PASK or AMPK were assessed by SAMS peptide assay and are given as means ± SEM of three separate experiments. (B) MIN6 cells were transfected with pPASK.WT, then incubated for 1 h as in A, before immunocytochemical analysis (see Materials and Methods). Scale bar, 10 μm. (C) Untransfected MIN6 cells were cultured for 16 h at 3 mM glucose and then for 6 h at 3 or 30 mM glucose as indicated, before RNA extraction and RT-PCR analysis (30 cycles; see Materials and Methods). The fold increase in PASK mRNA was 14.8 ± 0.4 (mean of three separate experiments involving 30 PCR cycles and 200 ng of template RNA). (D) Immunoblot analysis of endogenous PASK protein. Cells or islets were cultured for 24 h at the indicated glucose concentrations before protein extraction and analysis. PASK migrates as a 142-kDa band. NS, nonspecific. Results in B–D are representative of four independent experiments.
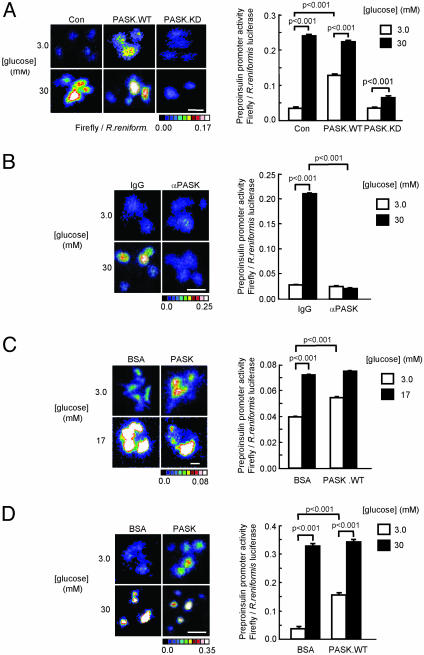
Fig. 2.
Regulation of preproinsulin promoter activity by glucose and PASK. MIN6 (A, B, and D) or cultured islet cells (C) were microinjected with plasmid pINS.LucFF and pCMV.RL (see Materials and Methods) plus (A) pcDNA3 (Con) pcDNA3PASK.WT or pcDNA3.PASK.KD, (B) control IgG or anti-PASK antibody (1.0 mg/ml-1), or (C and D) BSA or wild-type PASK protein (1.0 mg/ml-1) before incubation for 6 h followed by imaging of firefly and R. reniformis luciferases (see Materials and Methods). Data are from three experiments on a total of 30–120 single cells per condition. (Bars = 20 μm.)
Fig. 3.
Regulation of wild-type and mutated preproinsulin and PDX-1 promoters by PASK. MIN6 cells were injected with unmodified (A) preproinsulin or (B) PDX-1 promoter constructs, with purified wild-type PASK protein or BSA (-), as indicated. Cells were incubated in the presence or absence of diazoxide (DAO, 10 μM), as given. C is as A, but with wild-type or mutant preproinsulin promoters (34): mA3, -220CTCTCCTGACC; mA2, -138CGGACCTTGCAC; mE1, -114GCCCGCTG. D as in C but with wild-type PDX-1 promoter or mutants: Foxa2 binding (area I, region II, a1, -2630GTTTTTGGGGTATTA or a2, -2630GTTTTTGTTGGGGTATCC) (47), PDX-1-binding (area 1, region 1, -2648TATCCTTGG) (48) (mutated residues underlined). See Fig. 2 for other details.
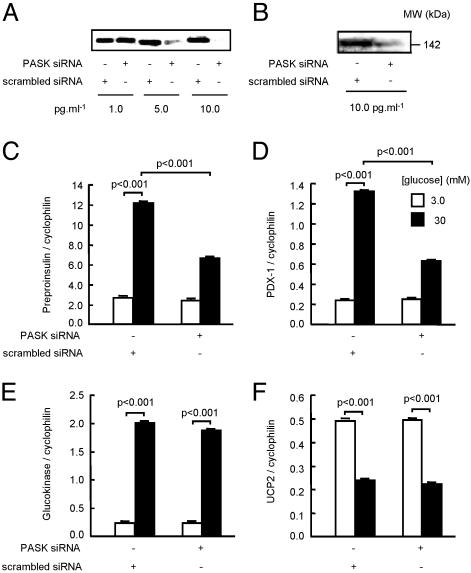
Fig. 4.
Involvement of PASK activation in the regulation of endogenous MIN6 β cell genes by glucose. (A and B) Cells were transfected for 72 h with the indicated concentrations of scrambled or PASK siRNA duplexes before (A) measurement of PASK mRNA by RT-PCR or (B) PASK protein after SDS gel electrophoresis and immunoblotting. Quantification by RT-PCR of preproinsulin (C), PDX-1 (D), glucokinase (E), and UCP2 mRNA (F) in MIN6 cells transfected with control (scrambled) siRNA or anti-PASK siRNA (10 pg/ml-1) as indicated. In C–F, cells were cultured at 3 mM glucose for 16 h before culture for 6 h at 3 or 30 mM glucose as indicated. Results are representative of (A and B) or are the means of (C–F) data from three experiments.
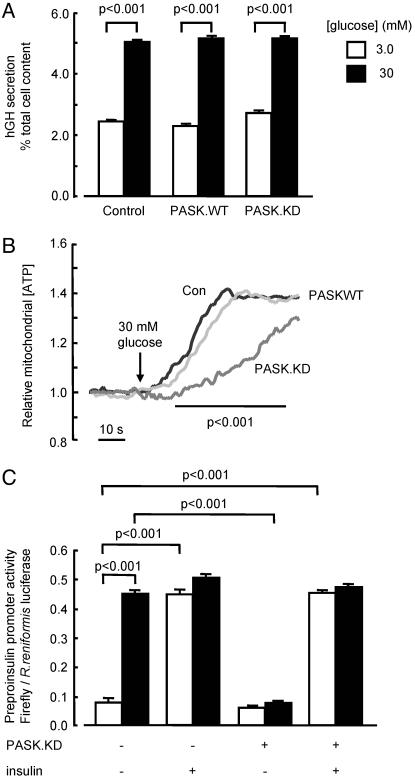
Fig. 5.
Changes in PASK activity have no effect on the acute stimulation of peptide secretion by glucose. (A) MIN6 cells were transfected with plasmids encoding hGH (1.0 μg/ml-1) and empty vector, pPASK.WT, or pPASK.KD. Transfected cells were cultured for 16 h at 3 mM glucose, before incubation for 20 min in modified modified Krebs–Ringer medium and measurement of released and total cellular hGH by ELISA (see Materials and Methods). (B) MIN6 cells were microinjected with plasmids encoding mitochondrially targeted luciferase and control (black trace) or PASK.WT (light gray) or PASK.KD (dark gray). Injected cells were cultured for 16 h in normal medium, then for 16 h at 3 mM glucose before transfer to modified Krebs–Ringer medium initially containing 3 mM glucose, and imaging; 30 mM glucose was added as indicated. Data are combined from three separate experiments. (C) Preproinsulin promoter activity was assessed in 30–120 single cells (three separate experiments) after microinjection of reporter constructs (Fig. 2) plus either empty vector (50 μg/ml-1) or pPASK.KD, before incubation for 6 h at the indicated concentrations of glucose and in the absence or presence of 20 nM insulin.
PASK Activity Assay. MIN6 cells were cultured in six-well plates to 70% confluency and transfected with 1 μg of pPASK.WT or pPASK.KD. Twenty-four hours posttransfection, cells were transferred to medium containing 3 mM glucose and cultured for 16 h before further culture at 3 or 30 mM glucose, in the absence or presence of 20 nM insulin, for 1 h (Fig. 1). Cells were washed once with PBS, and the plates were quick-frozen in liquid nitrogen. The cells were resuspended in 1 ml of ice-cold homogenization buffer (50 mM Tris, pH 7.5/1 mM EDTA/100 mM NaCl/100 mM NaF/1 mg/ml-1 each pepstatin, antipain, and leupeptin/1 mM NaVO4) and then disrupted by using a syringe. Cell debris was removed by centrifugation for 10 min at 25,000 × g. Two microliters (0.8 μg) of anti-c-myc antibody (Roche) and 100 μl of 1:1 suspension of protein G agarose in homogenization buffer were added to the supernatant. This was mixed at 4°C for 30 min and then centrifuged at 20,000 × g for 10 s. The resultant pellet was washed four times with 1 ml of homogenization buffer and resuspended in 200 μl of SAMS kinase assay buffer before assay (15).
Statistical Analysis. Data are given as means ± SEM for the number of experiments indicated. Comparisons were performed by one-tailed Student's t test by using Microsoft excel.
Results
Glucose Stimulates PASK Activity in Clonal MIN6 β Cells. We sought first to examine the effect of glucose on PASK activity in MIN6 cells, a glucose-responsive insulinoma-derived cell line (20, 27). Because the endogenous enzyme could not be efficiently immunoprecipitated (results not shown), we overexpressed c-myc epitope-tagged PASK and immunoprecipitated the expressed protein before assay by phosphotransfer (Fig. 1 A). Incubation of cells for 1 h at 30 mM (versus 3 mM) glucose before cell extraction, and immunoprecipitation increased the activity of the expressed wild-type kinase by ≈50%. This effect seemed to be unrelated to the activated release of insulin, because addition of a high concentration (20 nM) of the exogenous hormone had no effect on PASK activity at either low or high glucose concentrations (Fig. 1 A). By contrast, the activity of endogenous AMPK, immunoprecipitated with a pan AMPKα subunit antibody that recognizes complexes containing either isoform of the AMPK catalytic subunit (α1 and α2), was reduced by >30% at 30 mM glucose (Fig. 1 A), in line with previous measurements in this cell type (15, 17). The observed changes in PASK activity were not associated with detectable alterations in either the protein level at 1 or 6 h (results not shown) or in subcellular distribution (Fig. 1B), which remained essentially extranuclear at both concentrations of the sugar.
Measured after 6 h by semiquantitative RT-PCR, PASK mRNA levels were significantly increased in cells exposed to 30 mM glucose (Fig. 1C). Correspondingly, the higher concentration of glucose led to a marked accumulation of PASK protein after a 24-h incubation of either MIN6 cell or whole islet homogenates (Fig. 1D).
Wild-Type PASK Mimics, Whereas Inactive PASK Blocks, the Effects of Glucose on Insulin Promoter Activity. Preproinsulin promoter activity, assessed in single MIN6 cells microinjected with a luciferase reporter (see Materials and Methods and ref. 28), was strongly induced by incubation at 30 mM (versus 3 mM) glucose (Fig. 2A). This effect of 30 mM glucose was partially mimicked by the comicroinjection of an expression plasmid encoding wild-type PASK but reversed by the introduction of cDNA encoding an inactive form of PASK, mutated at the ATP-binding site (K1028R, Fig. 2 A) and likely to act as a dominant-negative toward endogenous PASK. Consistent with the requirement for PASK activity in the stimulation of preproinsulin promoter activity, microinjection of an anti-PASK antibody into MIN6 cells also completely abrogated the transcriptional response to 30 mM glucose (Fig. 2B).
Confirming that PASK was able to regulate preproinsulin transcription in the context of primary β cells, microinjection of purified wild-type PASK into cultured rat islet cells significantly activated the preproinsulin promoter at basal, but not elevated (17 mM), glucose concentrations (Fig. 2C), in line with the stimulatory effects of the wild-type protein in MIN6 cells (Fig. 2D).
Indicating that the effects of PASK overexpression were unlikely to involve changes in ATP-sensitive K+ channel activity, activation by wild-type PASK of the preproinsulin (Fig. 3A) and pancreatic duodenum homeobox-1 (PDX-1, Fig. 3B) promoters was fully preserved in the presence of the ATP-sensitive K+ channel opener, diazoxide.
To further dissect the possible sites of action of PASK, we monitored the activity of promoter constructs bearing mutations at key transcription factor-binding sites. Mutation of the PDX-1 (A3) (29) and MafA (A2) (22) sites of the preproinsulin promoter (30) blocked the activation by either glucose or coinjected PASK, whereas mutation of NeuroD1/Beta2 (E1)-binding site had much smaller effects (Fig. 3C). Similarly, deletion of the PDX-1-binding site of the PDX-1 promoter blocked activation of the latter by either agent, whereas PDX-1 promoter activity was unaffected by mutation of Foxa2 (HNF3β)-binding sites (Fig. 3D).
Silencing of PASK Expression Selectivity Inhibits Glucose-Stimulated Gene Expression. To determine whether changes in the levels of PASK may affect the expression of the endogenous preproinsulin and other glucose-regulated genes, MIN6 cells were incubated with increasing concentrations of an siRNA duplex targeting the kinase. siRNA treatment progressively diminished PASK mRNA levels (Fig. 4A), whereas transfection with a scrambled siRNA was without effect (Fig. 4A), consistent with previous studies (25, 31, 32). At the optimal siRNA concentration (10 pg/ml-1), levels of immunoreactive PASK were reduced by 93.7 ± 0.03% (mean ± SEM of three separate experiments, Fig. 4B). Loss of PASK expression had no effect on the basal levels of preproinsulin mRNA (at 3 mM glucose) but reduced by >60% the induction of the message observed after a 6-h culture at 30 mM glucose in the presence of scrambled siRNA (Fig, 4C). Similarly, levels of mRNA encoding PDX-1 were markedly reduced in PASK siRNA-treated cells (Fig. 4D), whereas MafA mRNA levels were slightly but significantly reduced at 30 mM glucose (MafA/cyclophilin mRNA ratios: 0.030 ± 0.0003 vs. 0.028 ± 0.0002, P < 0.01, scrambled or anti-PASK siRNAs, respectively). Levels of mRNAs encoding glucokinase (Fig. 4E) uncoupling protein 2 (UCP-2, Fig. 4F), USF1/2, foxa2 (HNF3β), HNF1α, and NeuroD1/Beta2 (data not shown) were unaffected by PASK silencing.
Changes in PASK Activity Have No Effect on Glucose-Stimulated Insulin Release. We (25, 33) and others (34, 35) have previously proposed that activation of insulin gene expression occurs at least in part as a downstream consequence of the release of insulin and the activation of β cell insulin receptors. Suggesting that suppression of insulin release was unlikely to be responsible for the effects of PASK inhibition (Fig. 2) or depletion (Fig. 4) of insulin gene expression, the release of cotransfected human growth hormone, used as surrogate for insulin (36), was unaffected by the expression of either wild-type or kinase-inactive PASK (Fig. 5A). Correspondingly, neither form of the kinase exerted any significant effect on the steady-state mitochondrial-free ATP concentration, measured by using a recombinant firefly luciferase (11), although expression of inactive PASK decreased the rate of increase in free ATP concentration in response to glucose (Fig. 5B). Finally, the addition of exogenous insulin completely reversed the effects of dominant-negative PASK on preproinsulin promoter activity, consistent with the action of the kinase on intracellular glucose signaling (Fig. 5C).
Discussion
Regulation of PASK Activity by Glucose. We demonstrate here that PASK activity is regulated in islet β cells by exogenous glucose. This provides the demonstration in the mammalian cell type that PASK activity is altered in response to a physiological stimulus, consistent with the proposed behavior of the yeast orthologs (1, 6). The present measurements of the acute (1-h) response to elevated glucose concentrations were performed by using the overexpressed epitope-tagged kinase, because the endogenous enzyme could not be immunoprecipitated with an anti-PASK kinase domain antibody (the only anti-mouse PASK antibody currently available). Because the increase in kinase activity at this time point did not involve any change in the level of the exogenous protein, this likely reflects a covalent modification of the overexpressed enzyme. These observations seem most consistent with the increased phosphorylation of both overexpressed and endogenous PASK at the regulatory site, Thr-1161 (2).
We have previously speculated that the PAS domains repress the activity of PASK under basal conditions, but that removal of these domains (by mutation or physiological stimulus) permits full activation of PASK by autophosphorylation or conceivably through transphosphorylation by an as-yet-unknown upstream kinase. Whether glucose leads to the activation of the putative upstream PASK kinase or changes the susceptibility of PASK to auto or transphosphorylation (as seems to be the case for AMPK) (9) is presently unclear.
An intriguing aspect of the present findings is that, whereas AMPK and PASK possess homologous kinase domains, the activities of the two enzymes in β cells change in opposite directions by glucose, i.e., high glucose concentrations diminish AMPK activity (15) while increasing PASK activity. In the case of AMPK, the inhibition of activity is likely to involve a decrease in intracellular [AMP]/[ATP] ratio and a loss of binding of AMP to the γ subunit (37). Exposure of β cells to glucose also leads to increased concentrations of a number of molecules, which may conceivably interact with PAS domains (38) and affect a change in PASK tertiary structure and hence phosphorylation state. These include reduced pyridine nucleotides (39, 40), ATP and ADP (11, 41), and Ca2+ (42), in addition to a range of glycolytic intermediates, amino acids (43), and fatty acids (44), as well as a decrease in O2 tension (45) and membrane potential (46). Identification of those parameters that play a role in regulating PASK activity requires further investigation.
Role of PASK in Glucose Sensing by Pancreatic β Cells. mRNA encoding PASK is expressed in most mouse and human tissues (3), with particularly high levels in the thymus and testis (7). Although mice inactivated for PASK expression are normally fertile and display no apparent defects in growth and development (7), the impact of PASK deletion on glucose homeostasis or adaptation to metabolic stress is unknown. In the present studies on pancreatic β cells, overexpression of either a wild-type or a likely dominant-negative form of PASK had only subtle effects on glucose metabolism and had no impact on the release of stored insulin. By contrast, overexpression or direct microinjection of wild-type PASK was sufficient to cause marked increases in preproinsulin promoter activity at low glucose concentrations. Conversely, silencing of PASK expression markedly decreased the accumulation of preproinsulin and PDX-1 mRNAs in response to high glucose. The latter effects were at least partly selective, because the responses of two other glucose-regulated genes (glucokinase and UCP-2), as well as a variety of transcription factors, were unaltered by PASK depletion. Interestingly, regulation by PASK or glucose of both the preproinsulin and PDX-1 promoters was completely abrogated by deletion of PDX-1-binding sites, implicating PDX-1 and possibly MafA (see Results) as targets of PASK action.
The effects of AMPK on preproinsulin gene expression are mediated in part through changes in insulin release and rebinding to β cell insulin receptors (17) and in part by an insulin-independent intracellular signaling mechanism. The former pathway is unlikely to explain the effects of PASK, given that (i) overexpression or inactivation of PASK had no effect on glucose-stimulated exocytosis, and (ii) overexpressed PASK enhanced preproinsulin and PDX-1 promoter activities when insulin secretion was blocked. These observations are thus consistent with the view that glucose and insulin can regulate preproinsulin gene expression via distinct, but complementary, pathways (25).
Conclusion
We show that (i) PASK activity is regulated at multiple levels in pancreatic β cells and (ii) appears to be involved in mediating the direct (intracellular) effects of glucose on gene transcription. Decreases in PASK activity in β cells may thus contribute to some forms of type 2 diabetes, whereas activation of the enzyme may provide a new therapeutic strategy for this disease.
Acknowledgments
This work was supported by a Wellcome Trust Program grant and Research Leave Fellowship (to G.A.R.) and a Sara and Frank McKnight Foundation Fellowship (to J.R.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AMPK, 5′-AMP-activated protein kinase; hGH, human growth hormone; PASK, mammalian Per–Arnt–Sim domain protein kinase; PDX-1, pancreatic duodenum homeobox 1; siRNA, small interfering RNA.
References
- 1.Wilson, W. A. & Roach, P. J. (2002) Cell 111, 155-158. [DOI] [PubMed] [Google Scholar]
- 2.Rutter, J., Michnoff, C. H., Harper, S. M., Gardner, K. H. & McKnight, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 8991-8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofer, T., Spielmann, P., Stengel, P., Stier, B., Katschinski, D. M., Desbaillets, I., Gassmann, M. & Wenger, R. H. (2001) Biochem. Biophys. Res. Commun. 288, 757-764. [DOI] [PubMed] [Google Scholar]
- 4.David, M., Daveran, M. L., Batut, J., Dedieu, A., Domergue, O., Ghai, J., Hertig, C., Boistard, P. & Kahn, D. (1988) Cell 54, 671-683. [DOI] [PubMed] [Google Scholar]
- 5.Gu, Y. Z., Hogenesch, J. B. & Bradfield, C. A. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 519-561. [DOI] [PubMed] [Google Scholar]
- 6.Rutter, J., Probst, B. L. & McKnight, S. L. (2002) Cell 111, 17-28. [DOI] [PubMed] [Google Scholar]
- 7.Katschinski, D. M., Marti, H. H., Wagner, K. F., Shibata, J., Eckhardt, K., Martin, F., Depping, R., Paasch, U., Gassmann, M., Ledermann, B., et al. (2003) Mol. Cell. Biol. 23, 6780-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salt, I. P., Johnson, G., Ashcroft, S. J. & Hardie, D. G. (1998) Biochem. J. 335, 533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawley, S. A., Boudeau, J., Reid, J. L., Mustard, K. J., Udd, L., Makela, T. P., Alessi, D. R. & Hardie, D. G. (2003) J. Biol. 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong, S. P., Leiper, F. C., Woods, A., Carling, D. & Carlson, M. (2003) Proc. Natl. Acad. Sci. USA 100, 8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy, H. J., Pouli, A. E., Jouaville, L. S., Rizzuto, R. & Rutter, G. A. (1999) J. Biol. Chem. 274, 13281-13291. [DOI] [PubMed] [Google Scholar]
- 12.Ainscow, E. K., Zhao, C. & Rutter, G. A. (2000) Diabetes 49, 1149-1155. [DOI] [PubMed] [Google Scholar]
- 13.AguilarBryan, L. & Bryan, J. (1999) Endocr. Rev. 20, 101-135. [DOI] [PubMed] [Google Scholar]
- 14.Rutter, G. A. (2001) Mol. Aspects Med. 22, 247-284. [DOI] [PubMed] [Google Scholar]
- 15.da Silva Xavier, G., Leclerc, I., Salt, I. P., Doiron, B., Hardie, D. G., Kahn, A. & Rutter, G. A. (2000) Proc. Natl. Acad. Sci. USA 97, 4023-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eto, K., Yamashita, T., Matsui, J., Terauchi, Y., Noda, M. & Kadowaki, T. (2002) Diabetes 51, S414-S420. [DOI] [PubMed] [Google Scholar]
- 17.da Silva Xavier, G., Leclerc, I., Varadi, A., Tsuboi, T., Moule, S. K. & Rutter, G. A. (2003) Biochem. J. 371, 761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leclerc, I., Woltersdorf, W. W., da Silva Xavier, G., Rowe, R. L., Cross, S. E., Korbutt, G. S., Rajotte, R. V., Smith, R. & Rutter, G. A. (February 10, 2004) Am. J. Physiol., 10.1152/ajpendo.00532.2003. [DOI] [PubMed]
- 19.Rafiq, I., da Silva Xavier, G., Hooper, S. & Rutter, G. A. (2000) J. Biol. Chem. 275, 15977-15984. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy, H. J., Viollet, B., Rafiq, I., Kahn, A. & Rutter, G. A. (1997) J. Biol. Chem. 272, 20636-20640. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz, R. E., Reyes, M., Koodie, L., Jiang, Y., Blackstad, M., Lund, T., Lenvik, T., Johnson, S., Hu, W. S. & Verfaillie, C. M. (2002) J. Clin. Invest. 109, 1291-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajihara, M., Sone, H., Amemiya, M., Katoh, Y., Isogai, M., Shimano, H., Yamada, N. & Takahashi, S. (2003) Biochem. Biophys. Res. Commun. 312, 831-842. [DOI] [PubMed] [Google Scholar]
- 23.Laybutt, D. R., Weir, G. C., Kaneto, H., Lebet, J., Palmiter, R. D., Sharma, A. & Bonner-Weir, S. (2002) Diabetes 51, 1793-1804. [DOI] [PubMed] [Google Scholar]
- 24.MacFarlane, W. M., Cragg, H., Docherty, H. M., Read, M. L., James, R. F., Aynsley-Green, A. & Docherty, K. (1997) FEBS Lett. 413, 304-308. [DOI] [PubMed] [Google Scholar]
- 25.da Silva Xavier, G., Qian, Q., Cullen, P. J. & Rutter, G. A. (2004) Biochem. J. 377, 149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson, G. H., Knobel, S. M., Arkhammar, P., Thastrup, O. & Piston, D. W. (2000) Proc. Natl. Acad. Sci. USA 97, 5203-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyazaki, J., Araki, K., Yamato, E., Ikegami, H., Asano, T., Shibasaki, Y., Oka, Y. & Yamamura, K. (1990) Endocrinology 127, 126-132. [DOI] [PubMed] [Google Scholar]
- 28.Rafiq, I., Kennedy, H. & Rutter, G. A. (1998) J. Biol. Chem. 273, 241-247. [DOI] [PubMed] [Google Scholar]
- 29.McKinnon, C. M. & Docherty, K. (2001) Diabetologia 44, 1203-1214. [DOI] [PubMed] [Google Scholar]
- 30.Melloul, D., Marshak, S. & Cerasi, E. (2002) Diabetologia 45, 309-326. [DOI] [PubMed] [Google Scholar]
- 31.Varadi, A., Tsuboi, T., Johnson-Cadwell, L. I., Allan, V. J. & Rutter, G. A. (2003) Biochem. Biophys. Res. Commun. 311, 272-282. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell, K. J., Tsuboi, T. & Rutter, G. A. (2004) Diabetes 53, 393-400. [DOI] [PubMed] [Google Scholar]
- 33.da Silva Xavier, G., Varadi, A., Ainscow, E. & Rutter, G. A. (2000) J. Biol. Chem. 275, 36269-36277. [DOI] [PubMed] [Google Scholar]
- 34.Leibiger, I. B., Leibiger, B., Moede, T. & Berggren, P. O. (1998) Mol. Cell 1, 933-938. [DOI] [PubMed] [Google Scholar]
- 35.Kulkarni, R. N., Bruning, J. C., Winnay, J. N., Postic, C., Magnuson, M. A. & Kahn, C. R. (1999) Cell 96, 329-339. [DOI] [PubMed] [Google Scholar]
- 36.Emmanouilidou, E., Teschemacher, A., Pouli, A. E., Nicholls, L. I., Seward, E. P. & Rutter, G. A. (1999) Curr. Biol. 9, 915-918. [DOI] [PubMed] [Google Scholar]
- 37.Rutter, G. A., da Silva Xavier, G. & Leclerc, I. (2003) Biochem. J. 375, 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, B. L. & Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panten, U., Christians, J., Kriegstein, E. V., Poser, W. & Hasselblatt, A. (1973) Diabetologia 9, 477-482. [DOI] [PubMed] [Google Scholar]
- 40.Pralong, W. F., Bartley, C. & Wollheim, C. B. (1990) EMBO J. 9, 53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Detimary, P., Gilon, P. & Henquin, J. C. (1998) Biochem. J. 333, 269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grapengiesser, E., Gylfe, E. & Hellman, B. (1988) Biochem. Biophys. Res. Commun. 150, 419-425. [DOI] [PubMed] [Google Scholar]
- 43.Maechler, P. & Wollheim, C. B. (1999) Nature 402, 685-689. [DOI] [PubMed] [Google Scholar]
- 44.Prentki, M., Vischer, S., Glennon, M. C., Regazzi, R., Deeney, J. T. & Corkey, B. E. (1992) J. Biol. Chem. 267, 5802-5810. [PubMed] [Google Scholar]
- 45.Aspinwall, C. A., Lakey, J. R. & Kennedy, R. T. (1999) J. Biol. Chem. 274, 6360-6365. [DOI] [PubMed] [Google Scholar]
- 46.Ashcroft, F. M. & Rorsman, P. (1990) Prog. Biophys. Mol. Biol. 54, 87-143. [DOI] [PubMed] [Google Scholar]
- 47.Marshak, S., BenShushan, E., Shoshkes, M., Havin, L., Cerasi, E. & Melloul, D. (2000) Mol. Cell. Biol. 20, 7583-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerrish, K., Grannon, M., Shih, D., Henderson, E., Stoffel, M., Wright, C. V. E. & Stein, R. (2000) J. Biol. Chem. 275, 3485-3492. [DOI] [PubMed] [Google Scholar]