Abstract
Many of the proposed physiological functions of α-synuclein, a protein involved in the pathogenesis of Parkinson's disease, are related to its ability to interact with phospholipids. To better understand the conformational changes that occur upon membrane binding of monomeric α-synuclein, we performed EPR analysis of 47 singly labeled α-synuclein derivatives. We show that membrane interaction is mediated by major conformational changes within seven N-terminal 11-aa repeats, which reorganize from a highly dynamic structure into an elongated helical structure devoid of significant tertiary packing. Furthermore, we find that analogous positions from different repeats are in equivalent locations with respect to membrane proximity. These and other findings suggest a curved membrane-dependent α-helical structure, wherein each 11-aa repeat takes up three helical turns. Similar helical structures could also apply to apolipoproteins and other lipid-interacting proteins with related 11-aa repeats.
The protein α-synuclein is the main component of Lewy bodies, a class of intracellular inclusions that is highly characteristic in Parkinson's disease (PD) (1). A causative role of α-synuclein in PD has been supported by genetic studies of familial forms of this disease (2–4) as well as by various animal models (5). In addition to its involvement in PD, α-synuclein may also play important roles in Alzheimer's disease, dementia with Lewy bodies, multiple system atrophy, and Hallervorden-Spatz syndrome (6, 7).
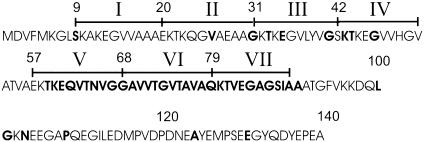
Although not yet fully understood, the physiological function of α-synuclein is likely to involve a role in modulating synaptic plasticity (8), presynaptic vesicle pool size, and neurotransmitter release (9–11), as well as vesicle recycling (12). In agreement with these membrane-related functions, α-synuclein has been shown to interact with liposomes in vitro (13–16). According to circular dichroism analysis, this interaction causes α-synuclein to undergo a conformational change from an unstructured monomer in solution (13, 17, 18) to an α-helical, membrane-bound protein. Based upon sequence analysis, it was recognized early on that the N-terminal portion of α-synuclein was likely to mediate lipid interaction (8, 13). The N terminus of α-synuclein contains seven repeats, each of which is made up of 11 aa (Fig. 1). These repeats are similar to those found in apolipoproteins, and it was proposed that the lipid interaction of α-synuclein could be similar to that of the apolipoproteins (8, 13). The involvement of the N terminus in membrane interaction was subsequently confirmed experimentally by analysis of α-synuclein deletion mutants (15) and NMR studies of liposomebound α-synuclein (18–20). The latter studies revealed an ordering of the N-terminal repeat regions induced upon membrane binding whereas the highly charged C terminus remained unstructured and, therefore, was not involved in membrane interaction. Beyond these data, however, direct structural information, such as the precise location, length, orientation, and number of helices for membrane-bound α-synuclein, has remained unknown.
Fig. 1.
Human α-synuclein sequence. The 140-aa human α-synuclein contains seven N-terminal 11-residue repeats (numbers in roman numerals). Letters in bold denote sites where single substitutions to R1 were made.
More is known about the structure of α-synuclein in the presence of SDS micelles, which also induce a helical structure. NMR studies of SDS-bound α-synuclein (19, 20) have revealed the presence of two extended helices (residues 1–41 and 45–94), with a break at residues 42, 43, and 44. Based on modeling, such extended helical structures were not consistent with a regular helical periodicity of 3.6 aa per turn (20). However, the data could be reconciled by postulating a slight alteration in the periodicity to 3.67 aa per turn, resulting in an α-11/3 helix (11 aa making up 3 turns) (20).
The goals of the present study were to obtain structural information for vesicle-bound α-synuclein, and to determine whether the models proposed for SDS-bound α-synuclein could be applicable to the membrane-bound form. Toward this end, we used site-directed spin labeling (SDSL) combined with EPR spectroscopy (21, 22). The basic strategy of SDSL requires the introduction of a nitroxide-containing side chain to specific sites. The most commonly used side chain, often referred to as R1 (see Fig. 6, which is published as supporting information on the PNAS web site), is generated by labeling specific cysteine residues.
Analysis of the EPR spectral features of R1-labeled proteins can provide structural information with respect to backbone dynamics, secondary and tertiary structure, and inter-residue distances (21, 22). One of the most important features contained in the EPR spectra of R1-labeled proteins is the mobility information. Through analysis of R1 mobility, one can distinguish between at least three different categories: sites introduced in loop or unfolded regions, sites on the surface of ordered structures, or sites that are buried inside the core of a protein. In the case of membrane proteins, additional secondary structure and membrane topography information can be obtained by studying the accessibility of R1 to paramagnetic colliders, such as O2 and chelated nickel [nickel(II)-ethylenediamine-N,N′-diacetic acid (NiEDDA)] (21).
Our SDSL analysis of the membrane interaction of α-synuclein demonstrates that the N-terminal repeat regions form an extended helical structure with few tertiary contacts whereas the C terminus remains unfolded. Equivalent positions within different repeats are located in structurally comparable positions with respect to membrane proximity, suggesting a structural model wherein individual 11-aa repeats take up three helical turns.
Materials and Methods
Generation of α-Synuclein Cysteine Mutants and Protein Purification. Human α-synuclein cloned in a pRK172 plasmid was kindly provided by M. Goedert (Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.). Human α-synuclein does not contain any cysteine residues. The generation of cysteine mutants, as well as purification of wild-type and mutant proteins, has been described (23).
Spin Labeling of Single-Cysteine Proteins. Protein stocks were filtered through 1 × 105 MWCO spin filters (Millipore) to remove oligomeric species. DTT was added to samples to a final concentration of 1 mM. DTT was removed by size exclusion by using PD-10 columns (Pharmacia) in 10 mM Hepes (pH 7.4), 100 mM NaCl buffer. A 5× molar excess of the MTSL spin label [(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate] was incubated with the protein samples for 30 min at room temperature. Excess spin label was removed by size exclusion by using PD-10 columns. Due to the high reactivity of the label and the freely accessible nature of cysteines introduced in poorly folded structures, near quantitative labeling was expected under these conditions. This near quantitative labeling was indeed verified by mass spectrometry of several test cases.
EPR Experiments. Spin-labeled proteins (5 μM) were incubated with small unilamellar vesicles at a 1:250 molar ratio in 10 mM Hepes (pH 7.4), 100 mM NaCl buffer, and concentrated and washed of unbound protein by using YM-100 (Amicon) spin filters. Membrane interaction of spin-labeled α-synuclein was independently confirmed by monitoring the formation of helical structure by using circular dichroism on a Jasco (Easton, MD) J-810 spectrometer. The optically clear unilamellar vesicles contained 30% 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-l-serine)/70% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (wt/wt) (Avanti Polar Lipids) and were generated by repeated sonication of larger vesicles prepared as described (24). Vesicles were further characterized by electron microscopy and found to have an average size of 300–400 Å. EPR spectra were recorded on a Bruker (Billerica, MA) EMX spectrometer fitted with a dielectric resonator at 1.59 mW incident microwave power, by using a field modulation of 1.5 G. The O2 and NiEDDA accessibilities (ΠO2 and ΠNiEDDA) were determined by employing a commonly used power saturation method (25). The final concentration of NiEDDA was 3 mM whereas the final concentration of O2 was that of air in equilibrium with buffer. The stability of our preparation was verified in control experiments, which demonstrated that the EPR spectra and accessibilities did not change over the time course of one week. As described in Results, the spectra of sites that became ordered upon membrane interaction continued to retain residual sharp spectral components. Using software generously provided by C. Altenbach (Department of Chemistry and Biochemistry, University of California, Los Angeles), we were able to baseline correct our spectra by subtracting the sharp spectral components. The amounts subtracted were estimated by double integration of the respective spectra and agreed closely with the previously published estimate (13).
Depth Calibration and Analysis of Φ Parameter. To calibrate the immersion depth of R1 at different sites, we took advantage of the previously established relationship that d[Å] = a·Φ + b, where Φ = ln Π(O2)/Π(NiEDDA). To obtain the parameters for a and b, we used 1-palmitoyl-2-stearoyl-(n-DOXYL)-sn-glycero-3-phosphocholine (Avanti Polar Lipids) with the spin label attached at the 5, 7, and 10 positions on the acyl chains, as published (25). For the spin-labeled phosphatidylcholines in vesicles containing α-synuclein under the conditions described above, we found that a = 5.9 and b = -4.1.
To estimate the error associated with the Φ values obtained from membrane-bound α-synuclein derivatives, triplicate experiments were conducted for several sites, and the error was found to be on the order of 0.1–0.2 units. The periodic oscillation of the Φ parameter as a function of residue number was fit by using the cosine fitting option implemented in the grapher software package (Golden Software, Golden, CO). Variables of the fit were amplitude, offset, phase, and periodicity.
Results
Conformational Changes of α-Synuclein upon Membrane Interaction. To characterize the structural changes induced by membrane interaction, we generated R1-labeled α-synuclein derivatives and recorded their EPR spectra in solution and when bound to membranes. EPR spectra of soluble, monomeric α-synuclein have been published previously (23) but are shown here to illustrate potential spectral changes upon membrane interaction. As shown in Fig. 2 (black), monomeric R1-labeled α-synuclein derivatives gave rise to sharp EPR line shapes that are characteristic for loop or unfolded regions. These results are in good agreement with previous studies, which have suggested that monomeric α-synuclein is largely unfolded (13, 17, 18).
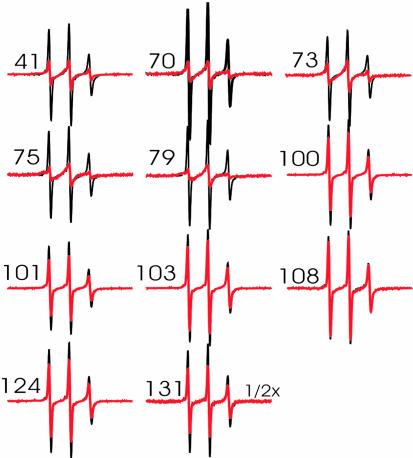
Fig. 2.
Overlay of R1-labeled α-synuclein. Samples were measured in the absence (black) and in the presence (red) of small unilamellar vesicles. Spectra were collected at a scan width of 100 G and normalized to the same number of spins. The numbers denote the sites at which R1 was introduced.
Next, we recorded the EPR spectra of R1-labeled α-synuclein derivatives in the presence of 30% phosphatidylserine/70% phosphatidylcholine-containing small unilamellar vesicles (diameter ≈ 300–400 Å, see Materials and Methods). As shown in Fig. 2 (red), spectral changes were detected when R1 is introduced in the repeat regions (residues 41, 70, 73, 75, and 79). In contrast, little or no changes were observed at positions within the last 40 aa, confirming that conformational changes occur within the repeat regions, but not within the C-terminal portion of the protein (18–20).
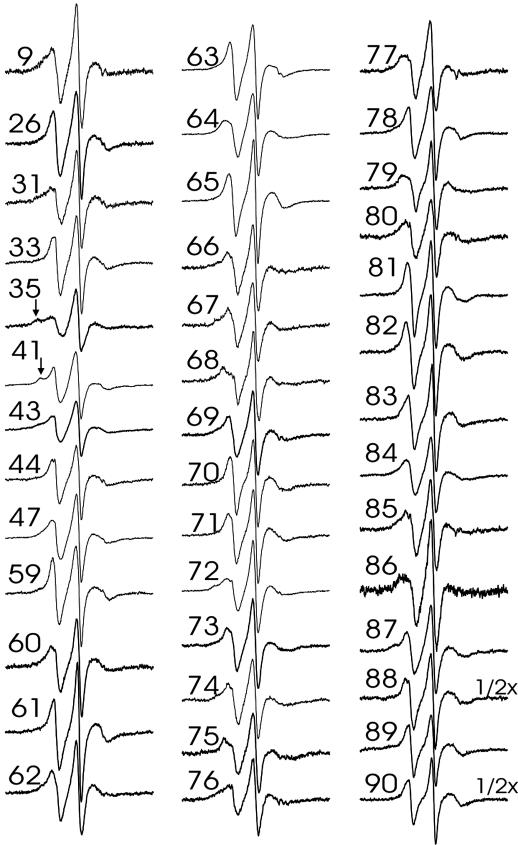
Previous studies have demonstrated that liposome interaction of α-synuclein depends on vesicle dimension, with smaller highly curved vesicles resulting in the strongest interaction (13, 16). However, even for the small unilamellar vesicles used in the present study, membrane interaction is known to be incomplete, with ≈10–15% of the protein remaining unbound (13). Using gel filtration experiments analogous to those described in ref. 13, we obtained similar results for the liposomes used in the present study (data not shown). In agreement with these results, we have observed small amounts of residual sharp components, similar to those of the soluble form, in the EPR spectra of membranebound α-synuclein derivatives. The overall amount of this component is in agreement with 10–15% of unbound protein, although small amounts of nonspecific background labeling could further contribute toward these lines. As described in Materials and Methods, these spectral components could easily be subtracted. All of the resulting spectra of membrane-bound α-synuclein labeled at sites within the repeat regions (Fig. 3) exhibit highly similar line shapes that are characteristic of lipid- or solvent-exposed helix surface sites. In addition, small amounts of immobile components are present in some spectra, for example at positions 35 and 41 (see arrows pointing to additional outer peaks). Although nearest neighbor contacts within the same helix can cause such immobilization (26), occasional collisional encounters between different proteins could further contribute to the observed line shapes. Overall, the degree of immobilization at all sites is, however, still much weaker than what is typically observed at buried sites in globular (27–29) or well packed membrane proteins (30, 31). This finding is further supported by analysis of the central line width of the EPR spectra shown in Fig. 3. The central line width (or its inverse) is a commonly used mobility parameter. Buried sites have highly characteristic spectra, with line width values ranging from 5 to 8 G (27–29). In contrast, all of the spectra shown in Fig. 3 have central line width values between 2.8 and 4.25 G and therefore cannot be considered buried.
Fig. 3.
EPR spectra of membrane-bound α-synuclein. Spectra of R1-labeled α-synuclein bound to small unilamellar vesicles were collected at a scan width of 100 G and normalized to the same number of spins. The residual sharp spectral components were subtracted from the raw spectra (see Materials and Methods). Arrows denote immobilized component.
Secondary Structure and Membrane Topology. In the interest of further refining the secondary structure and topology of membrane-bound α-synuclein, we determined the O2 and NiEDDA accessibilities (ΠO2 and ΠNiEDDA) for residues 59–90, located in repeats V–VII (see Materials and Methods). Nonpolar O2 preferentially partitions into the membrane whereas the more polar NiEDDA preferentially partitions into the solvent (21). Thus, the highly O2-accessible residues are exposed to the membrane whereas the more NiEDDA-exposed residues are exposed to the solvent.
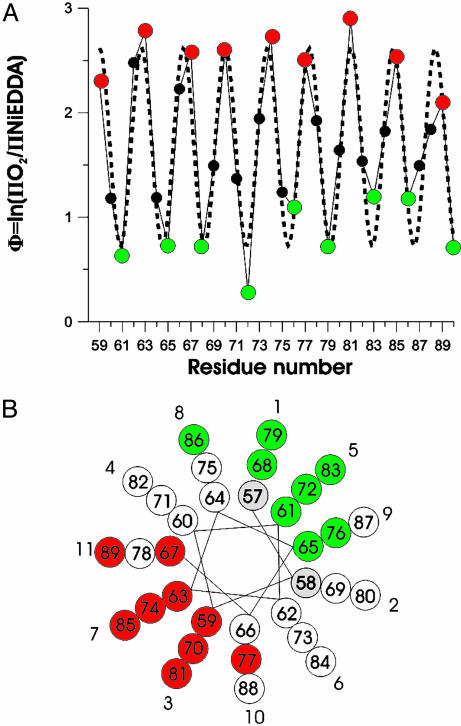
ΠO2 and ΠNiEDDA exhibit out of phase periodicities that are characteristic for an asymmetrically solvated helix, wherein one face is exposed to the phospholipid and the other face is exposed to the solvent (see Fig. 7, which is published as supporting information on the PNAS web site). The membrane topography information contained in the O2 and NiEDDA accessibilities of R1 can conveniently be summarized by the contrast parameter Φ [Φ = ln(ΠO2/ΠNiEDDA)], with larger values indicating deeper membrane insertion. As shown in Fig. 4A, the Φ values reveal a pronounced periodicity in agreement with a helical structure. Residues 59, 63, 67, 70, 74, 77, 81, 85, and 89 have high Φ values and are lipidexposed (Fig. 4A, red) whereas residues 61, 65, 68, 72, 76, 79, 83, 86, and 90 have low Φ values and are solvent-exposed (Fig. 4A, green). The values of the maxima are very similar to one another, suggesting comparable immersion depths of all lipid-exposed sites. This result is in close agreement with the notion of a peripherally bound helix rather than a transmembrane helix; in the latter case, the respective values would first increase linearly as the residues approach the center of the bilayer, then decrease as residues move away from the center (21).
Fig. 4.
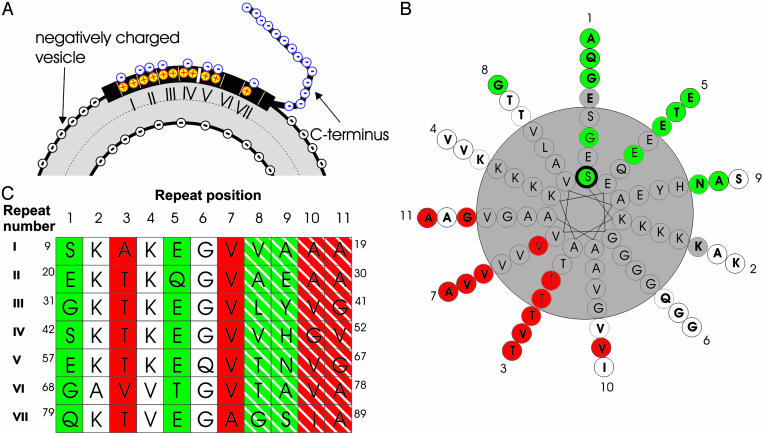
Secondary structure and topology of R1-labeled α-synuclein. (A) The ratios of the O2 and NiEDDA accessibilities are summarized by the contrast parameter Φ. High Φ values indicate lipid-exposed sites (red) whereas low Φ values indicate solvent-exposed sites (green). The dashed line indicates the best fit to a cosine function resulting in a periodicity of 3.67 aa per turn. (B) When residues are plotted on a helical wheel with a periodicity of 3.67 aa per turn, lipid-exposed sites (red) are on one side whereas solvent-exposed sites (green) lie on the opposite side. White circles denote residues with Φ values that are neither maxima nor minima. Residues shown in gray have not been tested. Repeat position (numbered 1–11) are indicated.
Previous SDSL studies have demonstrated that the Φ values are directly proportional to the depth of membrane insertion, and that the depth of insertion can be calibrated through the use of vesicles containing spin-labeled phospholipids (25). Based on the calibration described in Materials and Methods, we estimate the average immersion depth of the lipid-facing R1 side chains (Φ maxima) to be on the order of 11 Å. It should be emphasized that this immersion depth corresponds to the location of the N-O moiety of the R1 side chain. Naturally, in a membrane surface-bound helix, the membrane-facing R1 side chains will point away from the helix and toward the membrane. Thus, the immersion depth of these residues will be deeper than that of the helix backbone. To relate the observed immersion depth to that of the helix backbone, we inspected the crystal structures of proteins wherein R1 was introduced within a helical structure (26). In these structures, the distances between the R1 nitroxide moiety and the center of the helix were in the range of 7–10 Å. Based on these simple geometric considerations, we estimate that the center of the helix is located at an immersion depth of ≈1–4 Å, further supporting the notion that α-synuclein does not penetrate deeply into the membrane (32). This estimate also falls into the range predicted for related apolipoprotein sequences (33) and agrees well with a recent x-ray diffraction study of an 18-aa apolipoprotein A-1-derived peptide, which suggested an immersion depth of ≈3 Å below the phosphates (34).
To further characterize the underlying periodicity in the Φ plot, we fit the data to mathematical functions that also exhibit regular periodic oscillations. Fig. 4A shows the best fit of the data to a cosine curve, in which amplitude, y-offset, phase, and periodicity were variables. As can be seen in the figure, the fit reproduces the periodicity in Φ very well and results in a value of 3.67 aa per turn (R2 = 0.87). The difference between this value and that of an ideal helix (3.6 aa per turn, R2 = 0.48; see Fig. 8, which is published as supporting information on the PNAS web site) suggests the possibility that 11 aa (rather than the ideal 10.8 residues) could make up three turns. In agreement with this idea, we find that a helical wheel based on a periodicity of 3.67 aa per turn represents the data very well. As shown in Fig. 4B, all of the lipid-exposed residues (red circles) fall onto one face of the helix whereas the solvent-exposed sites (green circles) fall onto the opposite face.
According to this model, membrane proximity of a given residue is determined by its position within a repeat. For example, repeat positions 3 and 7 are invariantly lipid-exposed whereas repeat positions 1 and 5 are always solvent-exposed. If the structure of repeats 1–4 were similar, we would expect an analogous pattern. To test this notion, we determined the Φ values for residues at repeat positions 1 and 5 (residues 9, 31, and 35) and for residues at repeat position 3 and 7 (residues 26, 33, and 44) in the earlier repeats. Residues 9, 31, and 35 have Φ values of <1.0 (0.36, 0.56, and 0.63, respectively), which identifies them as solvent-exposed sites, whereas residues 26, 33, and 44 have Φ values of >2.0 (2.20, 2.87, and 2.06, respectively), consistent with lipid exposure of these residues. Thus, the data from these sites are in close agreement with the prediction that the same repeat positions are likely to be in equivalent positions with respect to membrane proximity.
Discussion
The main goal of the present study was to characterize the structural transition that occurs when monomeric α-synuclein binds to phospholipid vesicles. In agreement with earlier studies (15, 18–20), we find that membrane interaction is mediated by the N-terminal region of α-synuclein, which contains seven 11-aa repeats. In contrast, the highly negatively charged C-terminal region of the protein remains unstructured, even in the presence of membranes (Fig. 5A).
Fig. 5.
Structural model of membrane-bound α-synuclein. (A) Schematic illustration of the structural changes that occur upon interaction of α-synuclein with negatively charged membranes. The N-terminal repeat regions of α-synuclein become helical and mediate membrane interaction. In contrast, the highly negatively charged C terminus does not interact with the lipid. Also indicated is the charge distribution. The negative charges (blue circles, minus)of α-synuclein are solvent exposed whereas the Lys residues (red/yellow circles, plus) are on the level of the negative charge of the head groups (black circles, minus). (B) Experimentally determined lipid- and solvent-exposed sites are colored red and green, respectively. Residues colored white are neither maximally lipid-exposed nor solvent-exposed. Gray-colored residues have not been tested. Numbers around the helical wheel indicate repeat positions. (C) Table showing the amino acid sequence for repeats I–VII. Repeat positions are indicated above the table. Green columns denote repeat positions that are likely to be solvent-exposed whereas repeat positions in red are likely to be lipid-exposed. Positions 8 and 9 are striped in green to indicate that both are almost equally solvent-exposed, and there will likely be some variability as to which of the two residues within a given turn is the more solvent-accessible. The same considerations, except with respect to lipid proximity, apply for sites 10 and 11, which are striped in red.
The EPR spectra of membrane-bound α-synuclein from all 41 sites tested within the N-terminal repeat regions are very similar to each other. Each of these sites gives rise to spectra that are indicative of helix surface sites that are in contact with either solvent or phospholipids. The lack of strong R1 immobilization or loop formation at all sites tested strongly argues against a globular or well packed structure in the repeat regions. In agreement with this notion, a consecutive scan of repeats V–VII directly demonstrates the formation of a single, elongated helix for this region.
This helical structure is not only continuous and without significant tertiary contacts, it is also folded in such a way that each of the adjoining 11-aa repeats makes up three turns (Fig. 4). In fact, it might be difficult for consecutive repeats to bind to membranes, if individual repeats were unable to fully complete the helical turns. In that case, ensuing repeats would first have to complete the residual helical turn and thereby continuously start in different positions. This is not the case here because same positions within different repeats fall onto equivalent positions in the helical wheel and have comparable membrane proximities.
Although we have not tested each site in repeats I–IV, analysis of key sites in the first four consecutive repeats strongly suggests that this structure is applicable to all seven repeats, and a comprehensive helical wheel summarizing the accessibility data from all repeats is shown in Fig. 5B.
Inspection of the protein sequence further suggests that this structure would be generally applicable to all repeats (Fig. 5C). For example, all of the negatively charged residues are located exclusively at the solvent-exposed repeat positions 1, 5, 8, and 9 (Fig. 5 A and B). This result is of particular importance because the negatively charged residues would be destabilizing at sites that are exposed to either the hydrophobic membrane interior or the negatively charged head group region.
In contrast to the high density of negatively charged residues at solvent-exposed repeat positions, 11 of the 14 residues at repeat positions 2 and 4 are composed of lysines. Although the positive charge of the lysines would also be destabilizing within the membrane interior, there are several reasons why lysines could be stabilizing at the polar/nonpolar interface at the head group region. First, the positive charge of these residues could interact favorably with the negative charge of the lipid head group (Fig. 5A). In addition, the lysine moiety has a relatively long hydrophobic aliphatic chain, which could interact favorably with the hydrocarbon region of the bilayer. This bipartite interaction of lysine with hydrophobic and hydrophilic regions was proposed initially for apolipoproteins (35), and is commonly referred to as “snorkeling.” The immersion depth estimated in the present study falls into the predicted range for such snorkeling to occur (33).
In agreement with the aforementioned consideration, none of the putative lipid interaction sites contain charged residues. Position 7 is made up almost exclusively of valine residues and one alanine residue (repeat VII). Similarly, positions 10 and 11 lack any charged or polar residues although three glycine residues are present (see below). Repeat position 3, also a main membrane interaction site, consists mainly of threonine residues. Although threonine is a polar amino acid, it has been identified at the lipid-facing surfaces of transmembrane helices and is believed to possess membrane interaction properties similar to those of alanine and valine (36). The direct membrane interaction of residues at repeat position 3 is further supported by site-directed mutagenesis studies, which show that introduction of charged residues at these sites inhibits membrane interaction (15).
Helical curvature is often complementary to alterations in helical periodicities. For example, in the case of left-handed coiled coils, a periodicity of 3.5 aa per turn is modeled by using helical wheels where equivalent positions are spaced 7 aa apart and invariantly fall onto the same positions on the helical wheel. Although helical wheel models are useful for predicting the relative position of the amino acids with respect to one another, in actuality the coiled coils are not straight helices with an altered periodicity. Rather, they are continuously curved α helices. The discrepancy between the helical wheel model and the native structure is caused by the fact that the helical wheels are drawn with respect to a particular reference point. In the case of coiled coils, this reference point is the protein–protein contact face. Due to the winding of helices around each other in the native structure, this reference point is constantly shifting so that analogous positions are placed in positions that are equivalent with respect to the protein–protein interface.
In the case of α-synuclein, the reference point for generating helical wheels is between an α-helix and the membrane surface. Based on these considerations, one can envision the formation of an elongated, continuous α-helical structure that is bent in such a way that analogous repeat positions take up equivalent positions with respect to membrane or lipid proximity. Such a structure might not only be applicable to α-synuclein, but also to other 11-aa-repeat-containing proteins, such as the apolipoproteins (37, 38), which wrap around lipid particles of defined sizes. Whether a general complementarity exists between helix curvature and most favorable vesicle or lipid particle size and what roles amino acid insertions between different repeats could play still need to be determined.
It should be noted that α-synuclein can bind to surfaces that are even more curved than those of small unilamellar vesicles. This ability is evidenced by the formation of an α-helical structure in the presence of SDS micelles, which typically have circumferences on the order of 150 Å (39). However, a key structural difference between the SDS interaction and the one studied here is that SDS interaction results in the formation of two elongated helices interrupted by a loop region containing residues 42–44 (19, 20). Our data on positions 43 and 44 show both sites to be ordered, with residue 44 acting as a membrane interaction site. Inasmuch as a continuous α-helix of 90 or more amino acids would be at least 130 Å long, it would have to almost completely wrap around SDS micelles, but span only ≈20% of the small unilamellar vesicle circumference. The curvature strain of SDS micelles might therefore require breakage of the elongated helix near its midpoint (residues 42–44).
Together with our recent analysis of the structure of amyloid fibrils of α-synuclein (23), the present study illustrates the large range of different structures that can be investigated by SDSL. Besides studying the structural basis for the putative physiological functions of α-synuclein, SDSL could also be used to investigate the conformational changes involved in the transition from unfolded or helical synuclein into toxic oligomeric forms (40). In this respect, it is interesting to note that, at least under some conditions, membrane interaction can facilitate the formation of such oligomers (41–43) and that modulation of the membrane interaction could be responsible for the formation of toxic oligomers in vivo (44, 45). Thus, the molecular understanding of the helical, membrane-bound form discussed here might prove to be an important starting point for our understanding of the misfolding that occurs in disease.
Acknowledgments
We thank L. Te, W. Tse, and M. Kaptein for their technical support. This work was supported by a grant from the Larry L. Hillblom Foundation and the Beckman Foundation (to J.C. and R.L.), the Pew Scholars Program, and National Institutes of Health Grant P50 AG05142 (to R.L.). A.D.-S. is supported by a predoctoral fellowship from the National Institutes of Health and by the National Institute of Dental and Craniofacial Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PD, Parkinson's disease; SDSL, site-directed spin labeling; NiEDDA, nickel(II)-ethylenediamine-N,N′-diacetic acid.
References
- 1.Spillantini, M. G., Schmidt, M. L., Lee, V. M., Trojanowski, J. Q., Jakes, R. & Goedert, M. (1997) Nature 388, 839-840. [DOI] [PubMed] [Google Scholar]
- 2.Polymeropoulos, M. H., Lavedan, C., Leroy, E., Ide, S. E., Dehejia, A., Dutra, A., Pike, B., Root, H., Rubenstein, J., Boyer, R., et al. (1997) Science 276, 2045-2047. [DOI] [PubMed] [Google Scholar]
- 3.Kruger, R., Kuhn, W., Muller, T., Woitalla, D., Graeber, M., Kosel, S., Przuntek, H., Epplen, J. T., Schols, L. & Riess, O. (1998) Nat. Genet. 18, 106-108. [DOI] [PubMed] [Google Scholar]
- 4.Singleton, A. B., Farrer, M., Johnson, J., Singleton, A., Hague, S., Kachergus, J., Hulihan, M., Peuralinna, T., Dutra, A., Nussbaum, R., et al. (2003) Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 5.Maries, E., Dass, B., Collier, T. J., Kordower, J. H. & Steece-Collier, K. (2003) Nat. Rev. Neurosci. 4, 727-738. [DOI] [PubMed] [Google Scholar]
- 6.Goedert, M. (2001) Nat. Rev. Neurosci. 2, 492-501. [DOI] [PubMed] [Google Scholar]
- 7.Trojanowski, J. Q. & Lee, V. M. (2003) Ann. N.Y. Acad. Sci. 991, 107-110. [DOI] [PubMed] [Google Scholar]
- 8.George, J. M., Jin, H., Woods, W. S. & Clayton, D. F. (1995) Neuron 15, 361-372. [DOI] [PubMed] [Google Scholar]
- 9.Abeliovich, A., Schmitz, Y., Farinas, I., Choi-Lundberg, D., Ho, W. H., Castillo, P. E., Shinsky, N., Verdugo, J. M., Armanini, M., Ryan, A., et al. (2000) Neuron 25, 239-252. [DOI] [PubMed] [Google Scholar]
- 10.Murphy, D. D., Rueter, S. M., Trojanowski, J. Q. & Lee, V. M. (2000) J. Neurosci. 20, 3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabin, D. E., Shimazu, K., Murphy, D., Cole, N. B., Gottschalk, W., McIlwain, K. L., Orrison, B., Chen, A., Ellis, C. E., Paylor, R., et al. (2002) J. Neurosci. 22, 8797-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotharius, J. & Brundin, P. (2002) Hum. Mol. Genet. 11, 2395-2407. [DOI] [PubMed] [Google Scholar]
- 13.Davidson, W. S., Jonas, A., Clayton, D. F. & George, J. M. (1998) J. Biol. Chem. 273, 9443-9449. [DOI] [PubMed] [Google Scholar]
- 14.Jo, E., McLaurin, J., Yip, C. M., St. George-Hyslop, P. & Fraser, P. E. (2000) J. Biol. Chem. 275, 34328-34334. [DOI] [PubMed] [Google Scholar]
- 15.Perrin, R. J., Woods, W. S., Clayton, D. F. & George, J. M. (2000) J. Biol. Chem. 275, 34393-34398. [DOI] [PubMed] [Google Scholar]
- 16.Zhu, M. & Fink, A. L. (2003) J. Biol. Chem. 278, 16873-16877. [DOI] [PubMed] [Google Scholar]
- 17.Weinreb, P. H., Zhen, W., Poon, A. W., Conway, K. A. & Lansbury, P. T., Jr. (1996) Biochemistry 35, 13709-13715. [DOI] [PubMed] [Google Scholar]
- 18.Eliezer, D., Kutluay, E., Bussell, R., Jr. & Browne, G. (2001) J. Mol. Biol. 307, 1061-1073. [DOI] [PubMed] [Google Scholar]
- 19.Chandra, S., Chen, X., Rizo, J., Jahn, R. & Sudhof, T. C. (2003) J. Biol. Chem. 278, 15313-15318. [DOI] [PubMed] [Google Scholar]
- 20.Bussell, R., Jr. & Eliezer, D. (2003) J. Mol. Biol. 329, 763-778. [DOI] [PubMed] [Google Scholar]
- 21.Hubbell, W. L., Gross, A., Langen, R. & Lietzow, M. A. (1998) Curr. Opin. Struct. Biol. 8, 649-656. [DOI] [PubMed] [Google Scholar]
- 22.Hubbell, W. L., Cafiso, D. S. & Altenbach, C. (2000) Nat. Struct. Biol. 7, 735-739. [DOI] [PubMed] [Google Scholar]
- 23.Der-Sarkissian, A., Jao, C. C., Chen, J. & Langen, R. (2003) J. Biol. Chem. 278, 37530-37535. [DOI] [PubMed] [Google Scholar]
- 24.Reeves, J. P. & Dowben, R. M. (1969) J. Cell Physiol. 73, 49-60. [DOI] [PubMed] [Google Scholar]
- 25.Altenbach, C., Greenhalgh, D. A., Khorana, H. G. & Hubbell, W. L. (1994) Proc. Natl. Acad. Sci. USA 91, 1667-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langen, R., Oh, K. J., Cascio, D. & Hubbell, W. L. (2000) Biochemistry 39, 8396-8405. [DOI] [PubMed] [Google Scholar]
- 27.McHaourab, H. S., Lietzow, M. A., Hideg, K. & Hubbell, W. L. (1996) Biochemistry 35, 7692-7704. [DOI] [PubMed] [Google Scholar]
- 28.Margittai, M., Fasshauer, D., Pabst, S., Jahn, R. & Langen, R. (2001) J. Biol. Chem. 276, 13169-13177. [DOI] [PubMed] [Google Scholar]
- 29.Isas, J. M., Langen, R., Haigler, H. T. & Hubbell, W. L. (2002) Biochemistry 41, 1464-1473. [DOI] [PubMed] [Google Scholar]
- 30.Perozo, E., Cortes, D. M. & Cuello, L. G. (1998) Nat. Struct. Biol. 5, 459-469. [DOI] [PubMed] [Google Scholar]
- 31.Gross, A., Columbus, L., Hideg, K., Altenbach, C. & Hubbell, W. L. (1999) Biochemistry 38, 10324-10335. [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishnan, M., Jensen, P. H. & Marsh, D. (2003) Biochemistry 42, 12919-12926. [DOI] [PubMed] [Google Scholar]
- 33.Palgunachari, M. N., Mishra, V. K., Lund-Katz, S., Phillips, M. C., Adeyeye, S. O., Alluri, S., Anantharamaiah, G. M. & Segrest, J. P. (1996) Arterioscler. Thromb. Vasc. Biol. 16, 328-338. [DOI] [PubMed] [Google Scholar]
- 34.Hristova, K., Wimley, W. C., Mishra, V. K., Anantharamiah, G. M., Segrest, J. P. & White, S. H. (1999) J. Mol. Biol. 290, 99-117. [DOI] [PubMed] [Google Scholar]
- 35.Segrest, J. P., De Loof, H., Dohlman, J. G., Brouillette, C. G. & Anantharamaiah, G. M. (1990) Proteins 8, 103-117. [DOI] [PubMed] [Google Scholar]
- 36.Wimley, W. C. & White, S. H. (1996) Nat. Struct. Biol. 3, 842-848. [DOI] [PubMed] [Google Scholar]
- 37.Segrest, J. P., Jones, M. K., Klon, A. E., Sheldahl, C. J., Hellinger, M., De Loof, H. & Harvey, S. C. (1999) J. Biol. Chem. 274, 31755-31758. [DOI] [PubMed] [Google Scholar]
- 38.Oda, M. N., Forte, T. M., Ryan, R. O. & Voss, J. C. (2003) Nat. Struct. Biol. 10, 455-460. [DOI] [PubMed] [Google Scholar]
- 39.Gennis, R. B. (1989) Biomembranes: Molecular Structure and Function (Springer, New York).
- 40.Caughey, B. & Lansbury, P. T. (2003) Annu. Rev. Neurosci. 26, 267-298. [DOI] [PubMed] [Google Scholar]
- 41.Perrin, R. J., Woods, W. S., Clayton, D. F. & George, J. M. (2001) J. Biol. Chem. 276, 41958-41962. [DOI] [PubMed] [Google Scholar]
- 42.Necula, M., Chirita, C. N. & Kuret, J. (2003) J. Biol. Chem. 278, 46674-46680. [DOI] [PubMed] [Google Scholar]
- 43.Welch, K. & Yuan, J. (2003) Trends Neurosci. 26, 517-519. [DOI] [PubMed] [Google Scholar]
- 44.Outeiro, T. F. & Lindquist, S. (2003) Science 302, 1772-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharon, R., Bar-Joseph, I., Frosch, M. P., Walsh, D. M., Hamilton, J. A. & Selkoe, D. J. (2003) Neuron 37, 583-595. [DOI] [PubMed] [Google Scholar]