Grötsch et al. find that the AP-1 transcription factor Fra-1 limits the generation of antibody-producing plasma cells. Absence of Fra1 in B cells results in abnormally high numbers of plasma cells and increased antibody responses after vaccination.
Abstract
The cornerstone of humoral immunity is the differentiation of B cells into antibody-secreting plasma cells. This process is tightly controlled by a regulatory gene network centered on the transcriptional repressor B lymphocyte–induced maturation protein 1 (Blimp1). Proliferation of activated B cells is required to foster Blimp1 expression but needs to be terminated to avoid overshooting immune reactions. Activator protein 1 (AP-1) transcription factors become quickly up-regulated upon B cell activation. We demonstrate that Fra1, a Fos member of AP-1, enhances activation-induced cell death upon induction in activated B cells. Moreover, mice with B cell–specific deletion of Fra1 show enhanced plasma cell differentiation and exacerbated antibody responses. In contrast, transgenic overexpression of Fra1 blocks plasma cell differentiation and immunoglobulin production, which cannot be rescued by Bcl2. On the molecular level, Fra1 represses Blimp1 expression and interferes with binding of the activating AP-1 member c-Fos to the Blimp1 promoter. Conversely, overexpression of c-Fos in Fra1 transgenic B cells releases Blimp1 repression. As Fra1 lacks transcriptional transactivation domains, we propose that Fra1 inhibits Blimp1 expression and negatively controls plasma cell differentiation through binding to the Blimp1 promoter. In summary, we demonstrate that Fra1 negatively controls plasma cell differentiation by repressing Blimp1 expression.
The terminal differentiation of B cells into antibody-secreting cells (ASCs) is the basis of humoral immunity. After birth, B cell development begins in the BM from where selected immature B cells migrate to the spleen. There, immature B cells progress into T2 B cells and subsequently into the B2 B cell lineage, namely into marginal zone (MZ) B cells, or follicular (FO) B cells that recirculate through the lymphoid follicles of spleen and lymph nodes (Loder et al., 1999). Another B cell subtype, called B1 B cells, is found predominantly in the pleural and intraperitoneal cavities either as B1a B cells (CD11b, CD5 double positive) or B1b B cells (CD11b positive, CD5 negative; Martin et al., 2001). Upon activation, B cells divide several times and can differentiate into plasmablasts, plasma cells, or memory B cells (Manz et al., 2005).
Depending on the activating signal, distinct B cell subsets preferentially contribute to the humoral immune response. MZ and B1 B cells have the unique capacity to quickly respond to specific bacterial side products like LPS, and differentiate into plasmablasts and short-lived plasma cells producing large amounts of IgM as well as isotype-switched antibodies (Lopes-Carvalho and Kearney, 2004; Kallies et al., 2007). In the case of protein antigens, FO B cells can produce long-lived plasma cells after provision of survival and differentiation signals by T helper cells, and formation of germinal centers (GCs; Klein and Dalla-Favera, 2008; Victora and Nussenzweig, 2012). In GCs, activated FO B cells undergo hypermutation of Ig genes and class switch recombination (CSR). The GCs also support affinity maturation of the B cell response through the selection of B cells expressing the B cell receptor (BCR) variants of highest affinity for a given antigen (Rajewsky, 1996; Klein and Dalla-Favera, 2008). Thereby, memory B cells or plasma cells secreting high affinity class-switched antibodies are generated. Collectively, GC plasma cells usually home back into the BM where they can reside as long-lived plasma cells (Moser et al., 2006). Several differentiation pathways can therefore lead from a naive B cell to an ASC.
Two principles determine the propensity of activated B cells to develop into plasma cells. The first one is a regulatory gene network centered on the transcriptional repressor B lymphocyte–induced maturation protein 1 (Blimp1), encoded by the Prdm1 gene. The second is that the proportion of B cells that undergo CSR or differentiation into ASC is proportionally linked to consecutive cell divisions (Nutt et al., 2011). Contrastingly, B cell proliferation needs to be stopped to allow plasma cell differentiation driven by Blimp1. Thus, the proper balance between proliferation and differentiation of activated B cells to plasma cells is of key importance to humoral immunity.
Although differentiation of activated B cells into short-lived, cycling, and antibody-secreting pre-plasmablasts can occur in the absence of Blimp1, it is absolutely required for the generation of mature and terminally differentiated plasma cells (Kallies et al., 2007). Blimp1 expression increases concomitantly with the terminal differentiation of B cells into long-lived plasma cells (Kallies et al., 2004). In fact, all plasma cells express Blimp1 at high levels, and Blimp1 ablation in differentiated BM ASC results in their rapid loss (Shapiro-Shelef et al., 2005). It is of considerable interest to decipher the molecular mechanisms controlling the expression of Blimp1 and the formation of highly effective ASC.
Blimp1 expression is tightly controlled by an interdependent complex network of transcriptional repressors and activators (Nutt et al., 2011). For instance, Pax5, which specifies B cell identity by repressing non–B cell lineage genes (Nutt et al., 1999), also represses genes required for ASC differentiation including Blimp1 (Reimold et al., 1996; Rinkenberger et al., 1996; Delogu et al., 2006; Nera et al., 2006). Similarly, Bcl6 and Bach2 also repress Blimp1 and inhibit ASC development (Tunyaplin et al., 2004; Ochiai et al., 2006). In contrast, IFN regulatory factor 4 (Irf4), which is expressed at low levels in resting B cells and is up-regulated during B cell activation and plasma cell differentiation, induces Blimp1 expression (Sciammas et al., 2006; Kwon et al., 2009). Strikingly, the transcriptional networks characterizing B cells and plasma cells are mutually antagonistic. Indeed, while being repressed in nonactivated B cells, Blimp1 is a transcriptional repressor itself, which, upon induction, erases the mature B cell gene expression signature, for instance genes involved in proliferation and survival of activated B cells such as Pax5, c-Myc, or Bcl6 (Lin et al., 2002; Shaffer et al., 2002; Vasanwala et al., 2002). Apart from Pax5 and Irf4, there may be other transcriptional pathways regulating Blimp1 expression. Among them are transcription factors of the activator protein 1 (AP-1) family that, based on their induction and their capacity to regulate cytokines, have long been proposed to control lymphoid cell proliferation and differentiation (Foletta et al., 1998; Wagner and Eferl, 2005).
AP-1s are heterodimeric transcription factors formed by dimerization of one member of the Fos protein (c-Fos, FosB, Fra1, or Fra2) with one of the members of the Jun family (c-Jun, JunB, or JunD). They display pro- or anti-proliferative and pro- or anti-apoptotic functions, depending on the identity of the components associated in the dimer as well as on the cellular context (Shaulian and Karin, 2002). Thus, AP-1 can either stimulate or repress transcription (Hess et al., 2004; Luther et al., 2011). Of note, both the human and the mouse Blimp1 promoter contain AP-1 binding sites (AP1BSs; Vasanwala et al., 2002; Yu et al., 2012) to which c-Fos can bind to trans-activate Blimp1 transcription and subsequently promote the generation of ASCs (Ohkubo et al., 2005). Interestingly, Bcl6, which is a potent inhibitor of plasma cell differentiation, represses Blimp1 transcription by hijacking the activating AP-1 subunit Jun (Vasanwala et al., 2002). Collectively, these data suggested that AP-1 members may play important roles in the regulation of Blimp1 expression in B cells and thereby in the generation of ASC.
These observations prompted us to analyze the role of the Fos-related antigen 1 (Fra1) which was originally cloned as a Fos member induced after B cell activation in vitro (Huo and Rothstein, 1996). Using loss or gain of function mouse strains, we demonstrate that Fra1 suppresses plasma cell generation from activated FO B cells. The inhibition of plasma cell development by Fra1 is mediated by direct binding to the Prdm1/Blimp1 promoter, which prevents binding of c-Fos. Collectively, our data identify the AP-1 member Fra1 as a novel key regulator of plasma cell differentiation.
RESULTS
Increased humoral immune responses in Fra1ΔBcell mice
We first analyzed Fra1 mRNA expression in unstimulated B cells and in B cells activated with LPS, or via the BCR and CD40 in presence of IL-4. Resting B cells expressed low levels of Fra1 that was strongly up-regulated upon stimulation, both in FO (IgMlow/CD23+/CD21+) and MZ (IgM+/CD23−/CD21+) B cells (unpublished data). These data confirmed previous results (Huo and Rothstein, 1996) and suggested that Fra1 might contribute to humoral immune responses. Based on this hypothesis, we sought to examine the function of Fra1 in B cells in vivo and generated mice specifically lacking Fra1 in B cells by crossing mice carrying the floxed allele of Fra1 (Fra1fl/fl; Eferl et al., 2004) with the B cell–specific deleter mice (mb1-Cre; Hobeika et al., 2006), thereafter called Fra1ΔBcell mice.
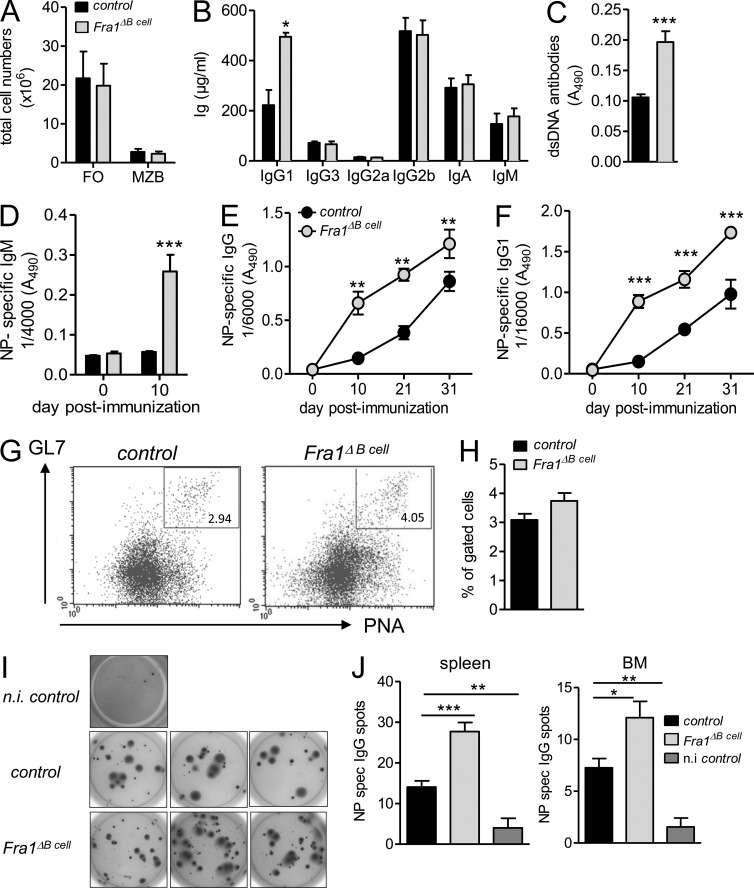
B cell development was essentially normal in Fra1ΔBcell mice compared with Fra1fl/fl littermate controls (Fig. 1 A). However, adult Fra1ΔBcell mice exhibited increased levels of serum IgG1, whereas other Ig isotypes were not affected (Fig. 1 B). Furthermore, Fra1ΔBcell mice had higher titers of anti–double-stranded DNA (dsDNA) IgG antibodies than control mice, suggesting a role for Fra1 in the control of overshooting and unspecific B cell responses (Fig. 1 C). Immunization with a T cell–independent type I antigen, nitrophenol (NP)-Ficoll, did not reveal any difference in antigen-specific IgM production between Fra1ΔBcell and control mice (unpublished data). However, immunization with the alum-precipitated T cell–dependent (TD) antigen NP-KLH resulted in a significantly stronger increase in NP-specific IgM (Fig. 1 D), IgG (Fig. 1 E), and IgG1 (Fig. 1 F) in Fra1ΔBcell mice. Because TD immune reactions involve GC responses (Rajewsky, 1996), our data suggested that either enhanced GC responses or enhanced TD plasma cell differentiation might occur in the absence of Fra1 in B cells. To determine whether loss of Fra1 in B cells would enhance the GC reaction, we immunized mice with sheep RBCs (SRBCs), which is known to induce strong GC responses. SRBC immunization elicited a slight but not significant increase in GC formation in Fra1ΔBcell mice at day 7 (Fig. 1, G and H). To confirm this result, we also immunized mice with NP-KLH and performed kinetic analyses of GC formation. Compatible with SRBC immunization, we observed a significantly increased GC compartment in Fra1ΔBcell mice at day 7, a small increase at day 10, and no difference at later time points (unpublished data). Next, we quantified ASCs in BM and spleen by antigen-specific ELISPOT. This experiment revealed a significant increase in cells secreting antigen-specific antibodies in NP-KLH immunized Fra1ΔBcell mice (Fig. 1, I and J). Collectively, our data suggest that Fra1 is involved in TD B cell responses and might be a negative regulator of TD plasma cell generation in vivo.
Figure 1.
Increased humoral immune responses in Fra1ΔB cell mice. (A) Splenic MZ and FO B cells in Fra1ΔBcell and Fra1fl/f littermate control mice (n = 7 mice) were enumerated by flow cytometry. Data are shown as total cell numbers. (B and C) Basal levels of immunoglobulin subclasses (B) and of anti-dsDNA antibodies (C) in sera of 11-wk-old Fra1ΔBcell and Fra1fl/fl littermate control mice (n ≥ 4 mice) were assessed by ELISA. (D–F) Fra1ΔBcell and Fra1fl/fl littermate control mice were immunized with alum-precipitated NP-KLH, and levels of NP-specific serum IgM (D; n = 4 mice), NP-specific total IgG (E; n ≥ 3 mice), and NP-specific IgG1 (F; n ≥ 3 mice) were measured by ELISA. (G and H) Fra1ΔBcell and Fra1fl/fl littermate control mice were immunized with SRBC, and GC B cells (B220+GL7+PNA+; G) were quantified at day 7 by flow cytometry (n ≥ 5). (I and J) Mice were immunized with NP-KLH, and NP-specific IgG-secreting cells in spleen and BM were visualized (I) and quantified by ELISPOT assay (J). Control mice were injected with alum/PBS (n = 4 mice). Mean ± SEM is shown for all statistical analyses. Data (A–F, H, and J) are pooled from at least 2 independent experiments, whereas data in G and I show one representative experiment out of 2 independent performed. *, P < 0.05; **, P < 0.005; ***, P < 0.001, unpaired Student’s t test (A–D and J) or two-way-ANOVA (E and F).
Decreased humoral immune responses in Fra1tg mice
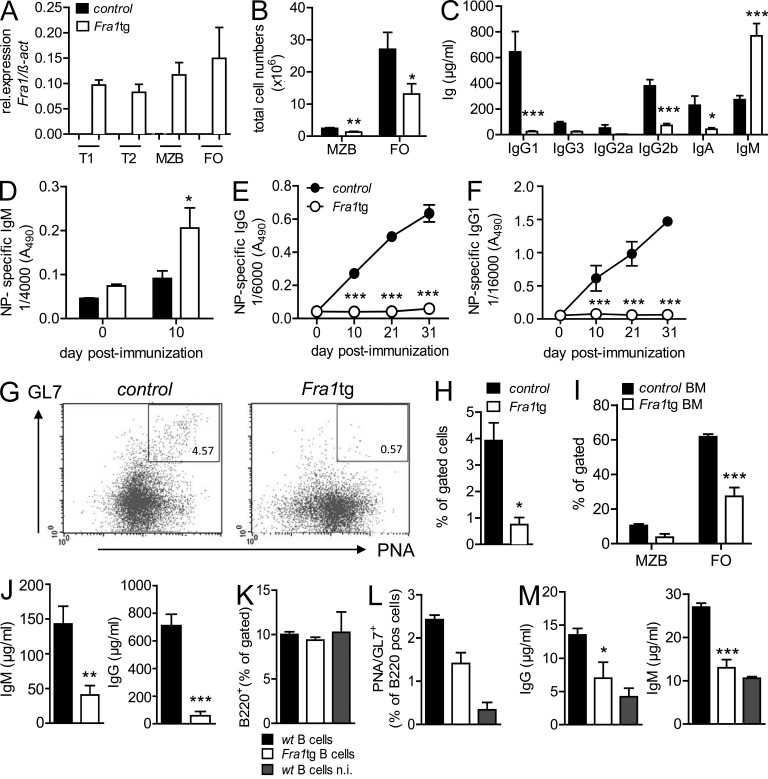
If Fra1 was a negative regulator of plasma cell differentiation, enhanced Fra1 expression in B cells should impair the generation of antibody-secreting plasma cells in response to TD immunization. To test this possibility, we analyzed the humoral immune response of transgenic mice ubiquitously overexpressing Fra1 (Fra1 transgenic mice, thereafter called Fra1tg mice; Jochum et al., 2000). The increased Fra1 expression in all splenic B cell lineages of the Fra1tg mice was confirmed by qPCR (Fig. 2 A) and by immunohistochemistry of the spleen (not depicted). FO and MZ B cells were present in the spleen of Fra1tg mice, albeit both of them in reduced numbers compared with control mice (Fig. 2 B). Fra1tg mice also had decreased numbers of B2 cells in the peritoneal cavity, yet the number and proportion of CD11b-expressing B1b B cells were significantly increased (unpublished data). Fra1tg mice had normal numbers of CD8 T cells and increased numbers of CD4 T cells in the spleen (unpublished data). In concordance with the data obtained from Fra1ΔBcell mice, constitutive overexpression of Fra1 led to strongly decreased serum levels of all IgG subclasses compared with control mice (Fig. 2 C). Similar data were obtained in Fra1tg mice on a mixed C57/Bl6; Sv129 background demonstrating that the phenotype was independent of the background (unpublished data). Surprisingly, Fra1tg mice displayed increased levels of IgM (Fig. 2 C). Nevertheless, even upon immunization with NP-KLH, no antigen-specific IgG or IgG1 could be detected in the Fra1tg mice, whereas antigen-specific IgM was increased (Fig. 2, D–F). The increased basal and induced IgM in Fra1tg mice was unexpected and was possibly due to the increased numbers of B1 B cells that secrete polyreactive IgM. The latter could be due to nonhematopoietic factors. However, in full accordance with missing IgG production, GC numbers were significantly reduced in Fra1tg mice after SRBC immunization (Fig. 2, G and H). Identical results were obtained after NP-KLH immunization (unpublished data).
Figure 2.
Decreased humoral immune responses in Fra1tg mice. (A) Fra1 expression in sorted splenic B cell subsets from WT and Fra1tg mice was assessed by QPCR. (B) Total numbers of splenic MZ and FO B cells in Fra1tg and wt littermate control mice were assessed by flow cytometry. (C) Basal levels of immunoglobulin subclasses in sera of 11-wk-old Fra1tg and wt control mice were quantified by ELISA. (D–F) Fra1tg and wt control mice were immunized with alum-precipitated NP-KLH at days 0 and 21. Serum levels of NP-specific IgM (D), NP-specific total IgG (E), and NP-specific IgG1 (F) were measured by ELISA. (G and H) Fra1tg and control mice were immunized with SRBC, and GC B cells were identified (G) and quantified (H) by flow cytometry. (I–M) Rag1−/− mice were transplanted with BM from Fra1tg or control mice. 8 wk after the transfer, splenic MZB and FO B cells were identified by flow cytometry using the same markers as in G and quantified (I) as percentages of gated cells. (J) 6 wk after the BM transfer, levels of total IgM and total IgG were analyzed by ELISA. (K–M) Rag1−/− mice were reconstituted with wt T cells and wt or Fra1tg B cells and immunized with SRBC or left unimmunized (n.i.). B cells (K) and GC B cells (L) were analyzed by flow cytometry 7 d after immunization. (M) Serum IgG and IgM was quantified by ELISA 7 d after immunization. n ≥ 3 mice for all experimental setups in at least 2 independent experiments (A–F and H–M). G shows one representative experiment out of 2 independent experiments. Mean ± SEM for all statistical analyses (A–M). *, P < 0.05; **, P < 0.005; ***, P < 0.001, unpaired Student’s t test (A–D, H–J, and M) or two-way ANOVA (E and F).
To test the possibility that the increased IgM levels observed in Fra1tg mice are due to nonhematopoietic factors and to determine whether the lack of IgG production could be cell autonomous, we reconstituted Rag1−/− mice with Fra1tg BM. Decreased FO and MZ B cell subpopulations were still observed in Rag1−/− mice reconstituted with Fra1tg BM (Fig. 2 I), indicating that this decrease was not a consequence of the osteosclerosis that leads to an almost complete obliteration of the BM cavity in Fra1tg mice (Jochum et al., 2000). Moreover, in full agreement with the data from Fra1ΔBcell mice, basal IgM and IgG titers were strongly reduced in Rag1−/− mice reconstituted with Fra1tg BM (Fig. 2 J), revealing that Fra1 overexpression acts in an opposite manner to Fra1 deletion in a non-osteosclerotic environment.
To determine whether the lack of B cell response could be a consequence of Fra1 overexpression in the T cell lineage, we further reconstituted Rag1−/− mice with WT T cells and WT B cells or Fra1tg B cells (Fig. 2 K), immunized these mice with SRBC, and assessed GC formation and antibody production. Fra1tg B cells generated less GC B cells (Fig. 2 L) than WT B cells and produced only minimal amounts of IgG and IgM that were hardly above the levels of nonimmunized mice (Fig. 2 M). In summary, these observations generally support the notion that Fra1 overexpression hampers GC formation and leads to impaired TD humoral immunity, i.e., a cell-autonomous impaired antibody production by B cells.
Fra1 is a negative regulator of plasma cell differentiation in vitro
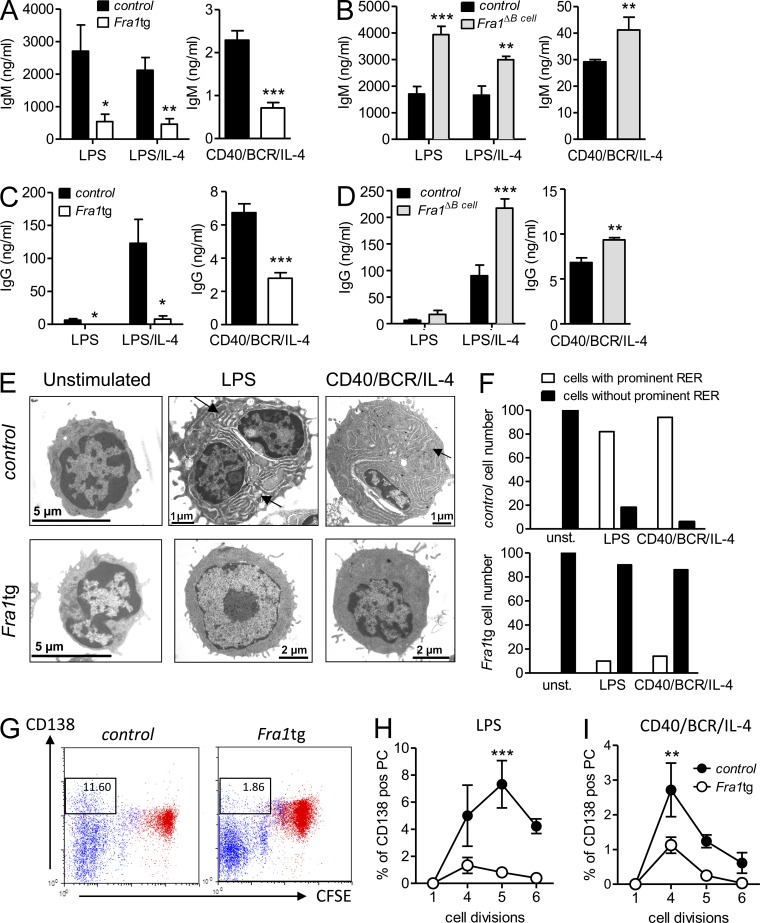
Because our data indicated that Fra1 might limit humoral immunity, we addressed whether it could act as a cell-autonomous inhibitor of B cell differentiation into ASC. To this end, we compared the antibody production of sorted FO B cells from WT, Fra1ΔBcell, and Fra1tg mice stimulated with LPS or LPS/IL-4, or, via CD40, the BCR and IL-4. Significantly reduced IgM was found in the supernatant of Fra1tg B cells, whereas increased IgM secretion was detected in supernatants of Fra1ΔBcell cells (Fig. 3, A and B). Remarkably, an almost complete block in IgG secretion was observed in supernatants of LPS and LPS/IL-4–stimulated Fra1tg B cells, and IgG was also significantly reduced in cells stimulated via CD40, the BCR, and IL-4 (Fig. 3 C). In contrast, IgG secretion by Fra1-deficient B cells was significantly enhanced compared with littermate controls in response to all three stimulations (Fig. 3 D). In support of this notion, electron microscopy revealed the absence of expanded endoplasmic reticulum, which characterizes actively immunoglobulin-secreting WT plasma cells, in Fra1tg FO B cells stimulated with LPS or via the BCR/CD40/IL-4 (representative pictures and quantification in Fig. 3, E and F). These observations indicated that Fra1 indeed could possibly act as an inhibitor of plasma cell differentiation. In contrast, the reduced GC reactions of Fra1tg B cells in reconstituted Rag1−/− mice (Fig. 2, G and H) suggested a cell-autonomous involvement of Fra1 in B cell proliferation or apoptosis. To address this issue, we sought to dissect a possibly dual function of Fra1 during B cell proliferation and differentiation into plasma cells. In fact, FACS analysis of Fra1-overexpressing FO B cells stimulated for 3 d with LPS showed a strong reduction in the proportion of CD138-positive cells that had undergone several rounds of proliferation (Fig. 3 G). The proportion of B cells that undergo CSR or differentiation into ASC is proportionally linked to consecutive cell divisions (Nutt et al., 2011), and it appeared indeed that Fra1tg B cells proliferated less. To dissect between the respective effect of Fra1 in B cell proliferation and plasma cell differentiation, we calculated the frequencies of CD138-positive B cells per cell division (Fig. 3, H and I). Importantly, the frequency of CD138-positive cells was lower in the portion of Fra1tg B cells that did undergo the number of cell divisions required for efficient plasma cell differentiation in the case of WT cells. From these data, we conclude that Fra1 is a transcription factor that inhibits plasma cell differentiation of FO B cells in a cell-autonomous manner. This could be the consequence of the effect of Fra1 on proliferation and apoptosis of B cells as well as of a direct inhibition of plasma cell differentiation.
Figure 3.
Fra1 is a negative regulator of plasma cell differentiation in vitro. (A) IgM secretion after 7 d stimulation with LPS or LPS/IL-4 or via the BCR/CD40/IL-4 of splenic IgD+/IgMlow FO B cells sorted from Fra1tg and wt littermate control were measured by ELISA (n = 5). (B) FO B cells sorted from Fra1ΔBcell and Fra1fl/fl littermate control mice were analyzed as in A (LPS and LPS/IL-4, n = 7; BCR/CD40/IL-4, n = 5). (C and D) IgG secretion of Fra1tg and wt control (C; LPS and LPS/IL-4, n = 4; BCR/CD40/IL-4, n = 6) and Fra1ΔBcell and Fra1fl/fl littermate control mice (D; LPS/IL-4, n = 7; BCR/CD40/IL-4, n = 6). (E) Electron microscopy of IgD+/IgMlow FO B cells sorted from WT control and Fra1tg mice unstimulated or stimulated with LPS or via the BCR/CD40/IL-4 for 3 d. Arrows indicate the extended endoplasmic reticulum in stimulated WT cells. (F) Quantification of cells with expanded rough endoplasmic reticulum (RER). 100 cells were counted for each condition directly at the electron microscope. (G) CD138 expression of CFSE-labeled control and Fra1tg FO B cells after 3 d stimulation with LPS (black) or unstimulated cells (red) was assessed by flow cytometry. (H and I) Quantification of the proportion of CD138-positive cells per cell division of LPS (H) or BCR/CD40/IL-4–stimulated WT and Fra1tg B cells (I; n = 4). Data are shown as mean ± SEM (A–I). Experiments were performed in ≥3 (A–D) and 2 (H and I) independent experiments, whereas G shows 1 representative out of 2 independent experiments performed. *, P < 0.05; **, P < 0.005; ***, P < 0.001, unpaired Student’s t test (A–D) or two-way ANOVA (H and I).
Fra1 is a negative regulator of proliferation and a positive regulator of apoptosis in B cells
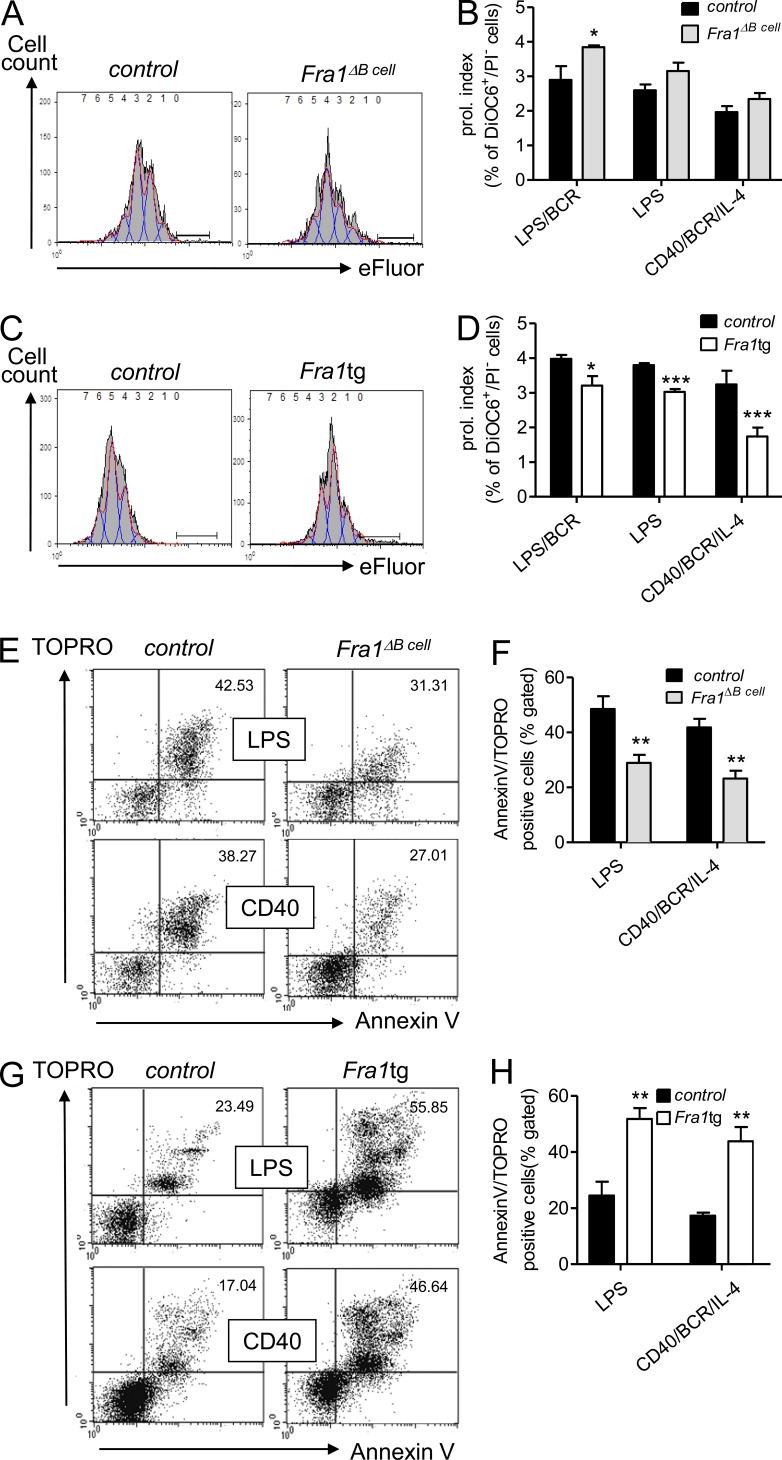
We therefore compared the proliferation and apoptosis of IgMlowIgDhigh FO B cells sorted from Fra1tg and Fra1ΔBcell mice to their corresponding littermate controls. The decline in fluorescence of eFluor647-marked FO B cells was used to evaluate proliferation. To exclude dilution of the proliferation marker through apoptosis, we only analyzed live cells that excluded PI and had a high mitochondrial membrane potential (DiOC6high). This revealed an increased proliferation in B cells lacking Fra1 and a decreased proliferation in B cells overexpressing Fra1, and, although not statistically significant, an increased proliferation in B cells lacking Fra1 after stimulation via LPS or CD40/BCR/IL-4 (Fig. 4, A–D). To ensure that we only addressed proliferation without interference by terminal differentiation, we also included BCR stimulation to LPS cultures to prevent plasma cell differentiation (Knödel et al., 2001) with similar results. From this experiment, we conclude that Fra1 negatively regulates B cell proliferation. Similarly, we addressed whether Fra1 was involved in the control of B cell apoptosis. Although decreased B cell apoptosis was observed in Fra1ΔBcell cells, increased apoptosis rates were observed in Fra1tg B cells compared with controls (Fig. 4, E–H). Notably, although loss of Fra1 was not associated with a significant difference in LPS-induced proliferation, it clearly resulted in a decreased sensitivity to apoptosis. These data suggested that the inhibition of plasma cell differentiation by Fra1 might be a consequence of increased apoptosis.
Figure 4.
Fra1 is a negative regulator of proliferation and a positive regulator of apoptosis in B cells. (A) Proliferation analysis assessed by dilution of eFluor647 labeling of FO (IgD+/IgMlow) Fra1ΔBcell and control FO B cells stimulated by BCR/CD40/IL-4 for 72 h. Red, draw model; blue, draw model components; gray, total division profile; the bar marks undivided cells. (B) Quantification of the proliferation index of purified FO B cells stimulated via LPS/BCR LPS and CD40/IL-4/BCR (n = 4, WT; n = 5, Fra1ΔBcell). (C and D) Fra1tg and wt control B cells were stimulated via LPS/BCR LPS and CD40/IL-4/BCR and analyzed as in A (n = 4). (E and F) Activation-induced cell death of BCR/CD40/IL-4 or LPS-stimulated Fra1ΔBcell and control FO B cells by flow cytometry (E) and quantification of the proportion of apoptotic cells as Annexin V/TOPRO double-positive cells calculated as percentage of all lymphocyte gated cells (F; n ≥ 3 mice). (G and H) Fra1tg sorted FO B cells and control B cells were analyzed as in E and F. Data are shown as mean ± SEM (B, D, F, and H). Experiments were performed in 2 (A–D) and ≥3 (E–H) independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.001, two-way-ANOVA (B and D) or unpaired Student’s t test (F and H).
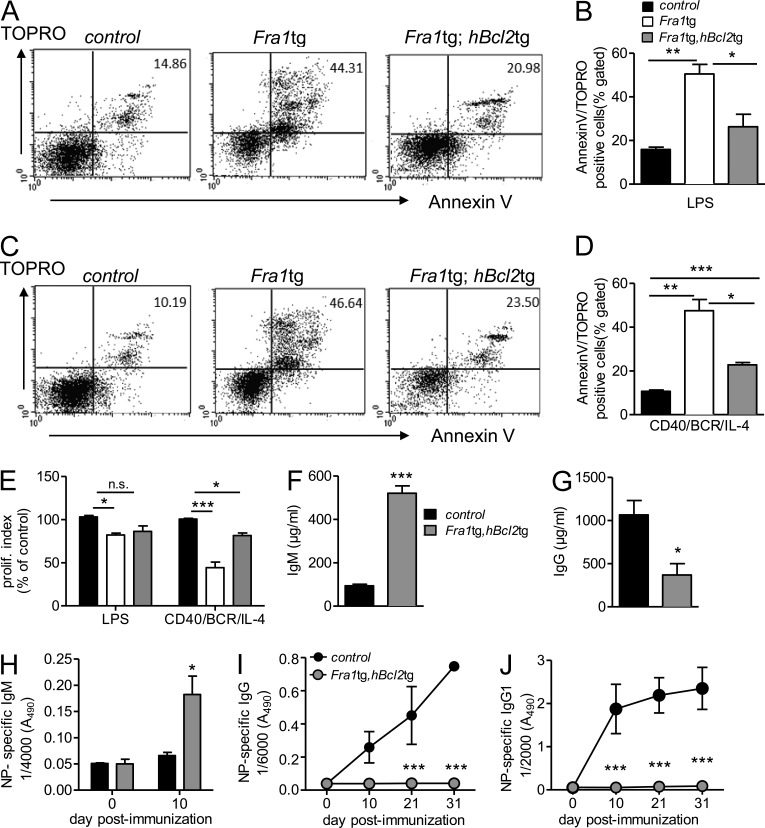
To assess the role of apoptosis in the inhibition of plasma cell differentiation by Fra1, we crossed Fra1tg mice with mice overexpressing the anti-apoptotic factor Bcl2 in B cells (Eμ bcl2-22; Strasser et al., 1991). As expected, an almost complete rescue of the Fra1-dependent apoptotic phenotype was observed in FO B cells overexpressing both Fra1 and Bcl2 (Fig. 5, A–D). We next asked whether the rescue of apoptosis by Bcl2 expression was also able to rescue proliferation. Indeed, Bcl2 was at least partially able to rescue proliferation in response to LPS or BCR/CD40/IL-4 stimulation (Fig. 5 E). As in the Fra1tg mouse, increased IgM serum levels and strongly decreased basal IgG were still observed in the double transgenic mice (Fig. 5, F and G). Most important, however, although Bcl2 overexpression rescued Fra1tg B cells from apoptosis and enhanced their proliferative capacity, it was not at all able to correct the defect in immunoglobulin production in immunized mice (Fig. 5, H–J). These data demonstrate that the lack of IgG-secreting cells is not caused by Fra1-induced apoptosis, and most likely not by defects in proliferation, but rather by an Fra1-mediated inhibition of plasma cell differentiation.
Figure 5.
Characterization of Fra1tg;hBcl2tg mice. (A) Activation-induced cell death of LPS-stimulated Fra1tg and Fra1tg;hBcl2tg and WT control sorted FO B cells assessed by combined Annexin V/Topro staining by flow cytometry. (B) Quantification of the proportion of Annexin V/Topro double-positive cells (n ≥ 3 mice in 2 independent experiments). (C and D) BCR/CD40/IL-4–stimulated Fra1tg and Fra1tg;hBcl2tg and WT control sorted FO B cells were analyzed as in A and B. (E) Proliferation index of purified WT, Fra1tg, and Fra1tg,hBcl2tg FO B cells stimulated by LPS or CD40/IL-4/BCR (n = 4 from 2 independent experiments). (F and G) Levels of circulating IgM (F) and IgG (G) in 8-wk-old control, Fra1tg, and Fra1tg;hBcl2tg mice. (H–J) NP-specific serum IgM (H), NP-specific total IgG (I), and NP-specific IgG1 (J) in sera of control and Fra1tg;hBcl2tg mice immunized with alum-precipitated NP-KLH (n ≥ 3 mice in 3 independent experiments). Data are shown as mean ± SEM for all statistical analyses (B, D, and E–J). *, P < 0.05; **, P < 0.005; ***, P < 0.001, unpaired Student’s t test (B, D, F, and G) or two-way-ANOVA (E, I, and J).
Fra1 inhibits Blimp1 expression
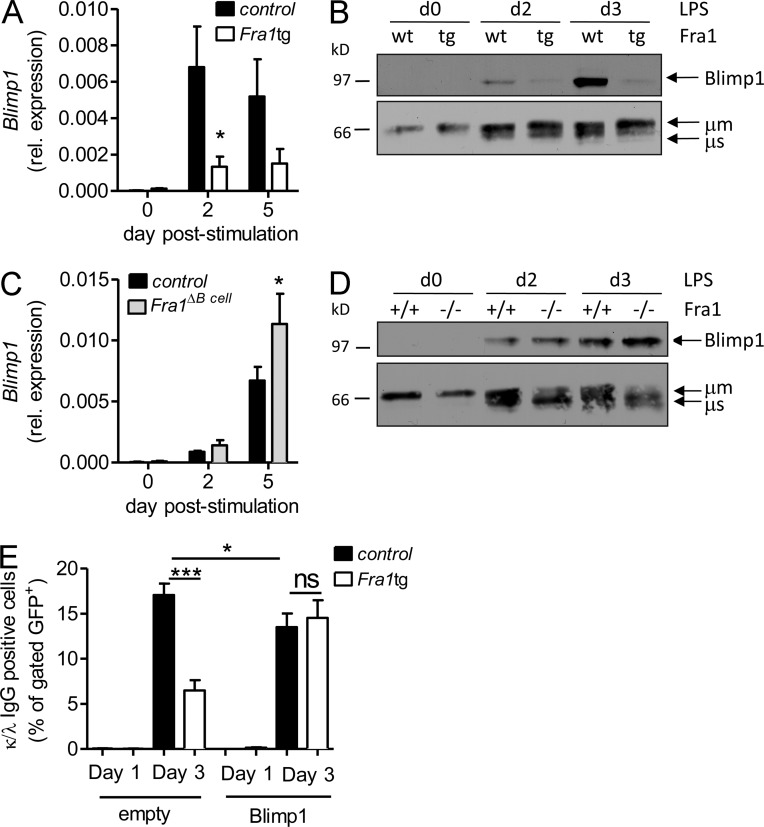
To uncover the molecular mechanism underlying the inhibition of plasma cell differentiation by Fra1, we analyzed the effects of Fra1 on the expression of Blimp1, a key transcriptional mediator of plasma cell maturation, using in vitro–stimulated sorted FO B cells. The expected up-regulation of Blimp1 in response to LPS was clearly inhibited in FO B cells overexpressing Fra1 both on RNA (Fig. 6 A) and protein level (Fig. 6 B). The inhibitory function of Fra1 in regulating Blimp1 was further confirmed by the increased Blimp1 expression observed in LPS-treated Fra1-deficient B cells on RNA and protein level (Fig. 6, C and D). The specificity of this regulation was confirmed by unaltered expression of IRF4 protein (unpublished data), as well as by clear up-regulation of the μ heavy chain in LPS-activated Fra1tg B cells (Fig. 6 B). These data strongly indicated that Fra1 acts as a negative regulator of Blimp1 transcription, thereby inhibiting plasma cell differentiation.
Figure 6.
Fra1 inhibits Blimp1 expression. (A and B) QPCR analysis (A) and Western Blot analysis (B) of Blimp1 expression in LPS-stimulated IgD+/IgMlow FO B cells sorted from WT and Fra1tg mice (*, P < 0.05, two way ANOVA, n ≥ 3 mice). The Western blot is representative of one of two independent experiments. (C and D) LPS-stimulated FO B cells sorted from Fra1fl/fl littermate control and Fra1ΔBcell mice were analyzed as in A and B. (E) Rescue of plasma cell differentiation by ectopic Blimp1 expression. CD43-depleted B cells isolated from Fra1tg or control WT mice were stimulated with LPS/IL-4 for 24 h, infected with an IRES-GFP (empty) or a Blimp1-IRES-GFP (Blimp1)–expressing retrovirus, and stimulated for another 48 h with LPS/IL-4. GFP-expressing plasma cells were quantified as IgG1 and intracellular κ/λhigh double-positive cells (n ≥ 3 from 2 independent experiments). Data are shown as mean ± SEM. *, P < 0.05; ***, P < 0.001, two-way-ANOVA (A and C) or unpaired Student’s t test (E).
We postulated that if Blimp1 repression by Fra1 was responsible for the block in plasma cell differentiation, this phenotype should be rescued by ectopically expressing Blimp1 in Fra1 transgenic B cells under the control of a distinct promoter. We therefore transduced WT or Fra1tg FO B cells with retroviral particles encoding for GFP alone or Blimp1 in combination with GFP (Knödel et al., 1999) and induced their differentiation by LPS/IL-4 stimulation. Plasma cell differentiation was evaluated by intracellular IgL (κ/λ) and IgG1 staining of GFP gated cells. As expected, a pronounced defect in IgG1 production was found in activated Fra1tg B cells infected with the GFP-expressing virus. Importantly, a complete recovery in high κ/λ and IgG1 expression was obtained by reexpressing Blimp1 in the Fra1tg B cells (Fig. 6 E), confirming that Fra1 acts as negative regulator of plasma differentiation by repressing Blimp1.
Fra1 binds to the Prdm1/Blimp1 promoter and displaces c-Fos
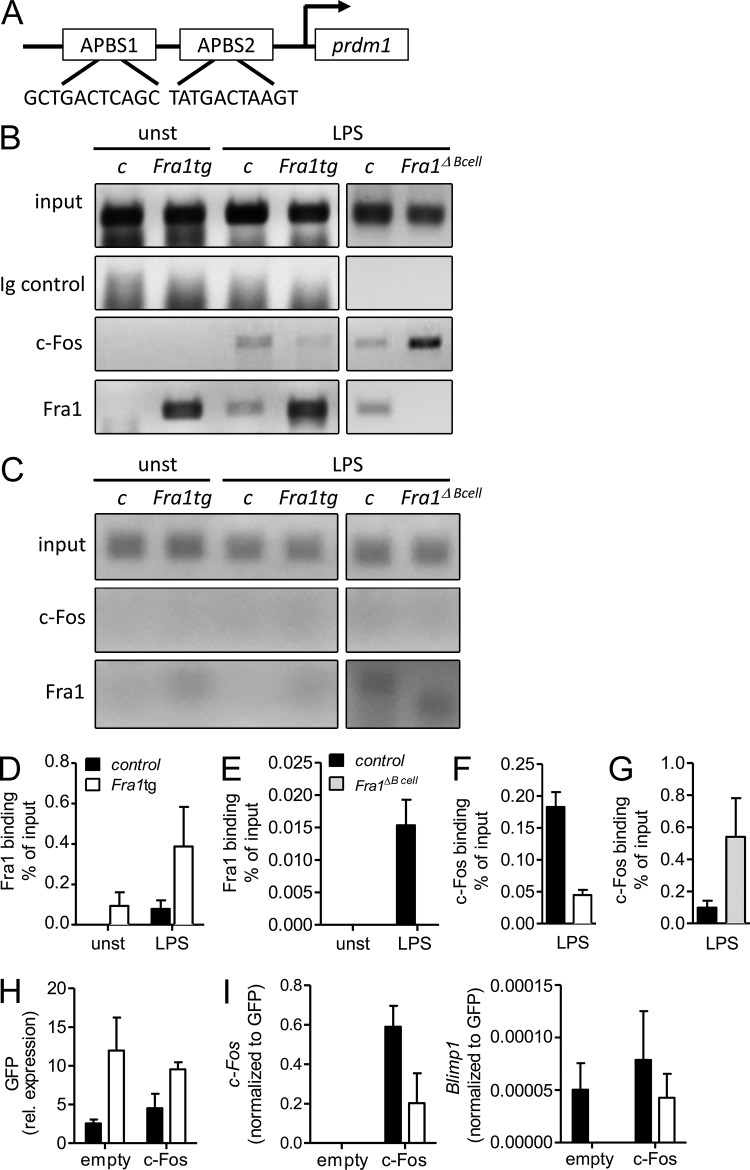
AP1BSs have been described in the human and mouse promoters of Prdm1, the gene encoding for Blimp1 (Ohkubo et al., 2005; Yu et al., 2012; Fig. 7 A). Of note, these AP1BS were proposed to mediate c-Fos/AP-1–dependent up-regulation of Blimp1, as suggested by the direct binding of c-Fos to the AP1BS of the Prdm1/Blimp1 promoter in primary B cells and other cell types (Ohkubo et al., 2005; Yu et al., 2012). These observations therefore suggested that Fra1, acting putatively as a transcriptional repressor of Prdm1/Blimp1, might exert its effect via interfering with binding of other AP-1 factors to the Prdm1/Blimp1 promoter. To analyze whether Fra1 indeed binds to the Prdm1/Blimp1 promoter in primary B cells, we isolated nuclear extracts from FO B cells stimulated with LPS and performed ChIP that we quantified by quantitative PCR using primers flanking the AP1BS2 of the Prdm1/Blimp1 promoter. We demonstrated a clear binding of Fra1 in LPS–stimulated WT cells. Importantly, this binding activity was absent in the nuclear extract of the cells lacking Fra1 (Fig. 7, B and E). In addition, a constitutive binding of Fra1 to AP1BS2 was already observed in the unstimulated cells overexpressing Fra1 and this binding was further increased by LPS stimulation (Fig. 7, A, B, and D). Binding of c-Fos to the AP1BS2 of the prdm1/Blimp1 promoter was also observed in LPS-stimulated WT B cells (Fig. 7, B and F). However, this binding was strongly reduced in cells overexpressing Fra1, whereas it was markedly increased in Fra1-deficient B cells (Fig. 7, B, F, and G). The specificity of c-Fos and Fra1 binding to AP1BS2 was further demonstrated by the absence of any detectable amplification of a nonrelevant fragment of the Prdm1/Blimp1 promoter after ChIP with the anti–c-Fos or anti-Fra1 antibodies (Fig. 7 C).
Figure 7.
Fra1 binds to the Prdm1/Blimp1 promoter. (A) Schematic drawing and sequence of the AP1BSs (AP1BS1 and AP1BS2) in the mouse Prdm1/Blimp1 promoter. (B) c-Fos and Fra1 binding to the AP1BS2 determined by ChIP of unstimulated (unst) or LPS-stimulated FO B cells isolated from Fra1tg, Fra1ΔBcell, and their corresponding wt and Fra1fl/fl littermate control mice. 10% of the input was loaded as control (input) and chromatin was immunoprecipitated with rabbit IgG as nonspecific isotype control, anti-Fra1, or anti–c-Fos antibodies. PCR was performed with primers flanking the AP1BS2 in the Prdm1/Blimp1 promoter (one representative experiment out of three is shown). (C) PCR with primers from a control region in the Prdm1/Blimp1 promoter showing the specificity of the binding of c-Fos and Fra1 to the AP1BS2 in one representative experiment. (D and E) QPCR quantification of Fra1 binding to AP1BS2 in 72h LPS treated Fra1tg (n = 6) or Fra1ΔBcell (n = 6). (F and G) Quantification of c-Fos binding to AP1BS2 in 72 h LPS-treated Fra1tg (n = 6) or Fra1ΔBcell (n = 6). (H and I) Rescue of plasma cell differentiation by ectopic c-Fos expression. CD43-depleted B cells isolated from Fra1tg or control mice were stimulated with LPS/IL-4 for 24 h, infected with an IRES-GFP (empty) or a c-Fos-IRES-GFP (c-Fos)–expressing retrovirus, and stimulated for another 24 h with LPS/IL-4. Relative GFP (H), and c-Fos and Blimp1 (I) expression normalized to GFP were measured by QPCR. Data represent 2 (C) or 3 (B and D–G) independent experiments. Data are shown as mean ± SEM.
We next asked whether the Fra1-mediated repression of Blimp1 transcription could be a consequence of a competition with c-Fos–mediated transcription of Blimp1. Therefore, we infected WT and Fra1tg FO B cells with a retrovirus encoding GFP alone or in combination with c-Fos and analyzed Blimp1 expression by quantitative PCR. In agreement with our hypothesis, a recovery of Blimp1 expression was observed in Fra1tg B cells overexpressing c-Fos (Fig. 7, H and I). Collectively, these observations document that Fra1 and c-Fos bind to the AP1BS2 site in an inversely proportional manner upon B cell activation, suggesting that Fra1, which interferes with c-Fos binding to the AP1BS2, opposes c-Fos–dependent activation of Prdm1/Blimp1 transcription.
DISCUSSION
Herein, we demonstrate that the AP-1 transcription factor Fra1 is a potent negative regulator of B cell function that inhibits plasma cell differentiation and therefore acts as a limiting factor during the humoral immune response. The decision of an activated B cell to become a plasma cell is a stochastic event, requiring several cell divisions, adequate survival signals, and a specific transcriptional program, resulting in expression of Blimp1 (Nutt et al., 2011; Peperzak et al., 2012). If insufficient numbers of cell divisions are achieved, the plasma cell differentiation program cannot work appropriately. We demonstrate here that Fra1 exerts two effects on plasma cell generation: (1) an anti-proliferative and pro-apoptotic function; and (2) Blimp1 repression, most likely through interfering with the activation of the Prdm1/Blimp1 promoter by the other AP-1 member c-Fos (Ohkubo et al., 2005; Yu et al., 2012).
Fra1 overexpression can block proliferative and survival signals provided through BCR/CD40/IL-4, LPS, and LPS/BCR stimulation of B cells. Conversely, enhanced proliferation was observed in B cells lacking Fra1 after LPS/BCR stimulation, reflecting a specific involvement of Fra1 in proliferation induced through LPS/BCR signaling, as BCR triggering of LPS-stimulated B cells prevents their differentiation but maintains proliferation (Knödel et al., 2001). In contrast, apoptosis was negatively regulated by Fra1 in LPS and BCR/CD40/IL-4–stimulated B cells. As recently suggested, Fra1 opposes NF-κB signals during bone development through Jun N-terminal kinase activation (Krum et al., 2010). Thus, the absence of Fra1 could equip late GC B cells with increased anti-apoptotic NF-κB activity, thereby facilitating plasma cell differentiation (De Silva et al., 2012). These notions are in agreement with our concept of Fra1 being a negative regulator of plasma cell differentiation.
Although Fra1 overexpression reduced proliferation in response to LPS, LPS/BCR, and BCR/CD40/IL-4 stimulation, a proportion of cells was still able to complete the number of cell division required for plasma cell differentiation but still did not express CD138. In addition, whereas the apoptotic phenotype was reversed by overexpressing Bcl2 and also resulted in a significant rescue of the proliferative capacity of Fra1-overexpressing B cells, there was no recovery of the TD immune response in vivo. Hence, our data strongly suggest that Fra1 has an effect on plasma cell differentiation that is uncoupled from apoptosis. In contrast, we cannot absolutely exclude that the inhibitory effect of Fra1 on plasma cell differentiation is totally independent of the decreased proliferation. This could indeed limit the final rounds of B cell proliferation required for plasma cell differentiation (Nutt et al., 2011). However, GC B cell numbers were not significantly increased in immunized Fra1ΔBcell mice but antigen-specific BM plasma cell numbers and antigen-specific antibody titers were. This suggests that apart from a possible involvement of Fra1 in B cell proliferation in vivo, Fra1 also limits plasma cell development directly. Thus, the combined reduced proliferation and inhibition of differentiation are likely responsible for repression of plasma cell generation by Fra1.
Our conclusion is also supported by key observations. First, electron microscopy provided direct in vitro evidence for a role of Fra1 in controlling plasma cell differentiation. Indeed, we could correlate the increase in Fra1 expression to an absence of large numbers of ER cisternae that morphologically characterizes actively secreting plasma cells. In agreement with these data, a correlation between Blimp1 expression and the number of ER cisternae was also observed by others (D. Tarlinton, personal communication). Second, increased immunoglobulin amounts were detected in the supernatants of in vitro stimulated Fra1-deficient B cells, whereas secretion was decreased in the Fra1-overexpressing cells. Finally, decreased numbers of CD138-positive cells were found even in the proliferated fraction of LPS-stimulated FO B cells overexpressing Fra1. Thus, Fra1 appears as a negative regulator of plasma cell differentiation. The increase in IgM observed in Fra1tg mice does not contradict this conclusion because this phenotype was not seen in Rag1−/− mice transplanted with Fra1tg BM. Thus, the high frequency of B1 B cells in combination with the altered environment in Fra1tg mice (Jochum et al., 2000) would account for the abundance of IgM in these mice. In accordance, our in vitro data revealed that Fra1tg B cells are still able to produce residual IgM and still can up-regulate IgM synthesis in response to LPS stimulation. We therefore propose that Fra1tg B cells are still capable of differentiating into antibody secreting preplasmablasts as observed in Blimp1-deficient mice (Kallies et al., 2007).
Although increased apoptosis and decreased proliferation occurred in B cells overexpressing Fra1, the inhibition of plasma cell differentiation was likely not a consequence of apoptosis because it could not be corrected by Bcl2 overexpression. Our data rather strongly support the hypothesis that the repression of Blimp1 expression is at least one molecular mechanism by which Fra1 inhibits plasma cell differentiation. Blimp1 is known to be essential for plasma cell differentiation (Kallies et al., 2007). Importantly, IgG1 production by FO B cells overexpressing Fra1 could be rescued through ectopic Blimp1 expression. We believe that the Blimp1-mediated rescue of IgG1 expression in LPS/IL-4–stimulated Fra1-overexpressing B cells is due to effects on plasma cell differentiation rather than due to positive regulation of CSR. In agreement with this concept, Blimp1 is a known transcriptional repressor rather than promoter of CSR. In addition, Blimp1 has been shown to repress Bcl6, which is required for CSR (Nutt et al., 2011).
Blimp1 expression is inversely correlated to the gain-of-function and loss-of-function mutants of Fra1, clearly indicating that Fra1 is a regulator of Blimp1 expression. Interestingly, JunD, a potential Jun partner of Fra1, is a major enhancer of Bcl6 expression (Arguni et al., 2006) and Bcl6 can also interact with JunD to repress Blimp1 (Vasanwala et al., 2002). These data could point to a putative positive-feedback loop where B cell activation, leading to the up-regulation of JunD, enforces Bcl6 expression and establishment of GC (Ohkubo et al., 2005). However, GCs were not significantly enhanced in Fra1tg mice. Thus it is unlikely that Fra1-mediated activation of Bcl6 represses Blimp1 and this would also not be relevant for differentiation of B cells in vitro.
However, Fra1 can act as a potent transcriptional repressor (Hess et al., 2004; Luther et al., 2011) and could therefore repress Blimp1 transcription. This notion is supported by our ChIP experiments that show the binding of Fra1 to the AP1BS of the Prdm1/Blimp1 gene, AP1BS2. Consistent with this hypothesis, Fra1 binds to AP1BS2 of the Prdm1/Blimp1 gene in an inversely correlated manner to that of c-Fos, which has been shown to promote Blimp1 expression and the formation of ASCs (Körholz et al., 1992; Ohkubo et al., 2005). A similar mechanism was proposed to explain the repression of Blimp1 by Bcl6 that reinforced the potential stimulatory effect of c-Fos on Blimp1 expression. In that case, Bcl6 was shown to dimerize with Jun to uncouple the association of Jun with c-Fos, thereby blocking the AP-1–dependent transactivation of the Prdm1 promoter (Vasanwala et al., 2002). Finally, c-Jun was shown to directly activate Blimp1 in transfection experiments of cell lines. Importantly, this activation could be reduced by Fra1 co-transfection, and up-regulation of Fra1 upon serum induction coincided with down-regulation of serum-induced Blimp1 expression in A549 lung cancer cells, whereas Fra2 or c-Fos did not (Yu et al., 2012). Finally, we demonstrated that overexpressing c-Fos in Fra1tg FO B cells releases the repression of Blimp1. Whether this effect is a result of a direct or indirect competition between c-Fos and Fra1 for the binding to AP1BS2 remains to be elucidated. In summary, our previously published data clearly indicate that different AP-1 subunits differentially modulate Blimp1 expression and formation of ASCs.
The Fos components of AP-1 are generally sequentially induced in response to cell stimulation. This is also the case in stimulated B cells where the induction of c-Fos precedes Fra1 expression in response to CD40 ligand (Huo and Rothstein, 1995, 1996). This finding is in agreement with Fra1 being a direct target gene of c-Fos (Matsuo et al., 2000). c-Fos and Fra1 are structurally similar but not identical. Indeed, in contrast to c-Fos, which acts as a strong activator of transcription due to the presence of transactivator domains (Sutherland et al., 1992), these domains are absent in Fra1, which therefore cannot effectively activate transcription. This difference has functional consequences as shown by the knock-in of Fra1 in the c-Fos locus that overcomes some but not all of the phenotypes caused by the absence of c-Fos (Fleischmann et al., 2000; Wenzel et al., 2002; Gass et al., 2004). In addition, Fra1 was not able to rescue the expression of typical c-Fos target genes in isolated fibroblasts (Fleischmann et al., 2000). Remarkably, Fra1 could rescue the phenotype of c-Fos deletion wherever the transactivation domain of c-Fos is not needed (Matsuo et al., 2000; Gass et al., 2004). Our data therefore suggest that c-Fos and Fra1 play opposing roles in plasma cell differentiation via the regulation of Blimp1. Although c-Fos is a known activator of Blimp1 that contributes to plasma cell differentiation (Ohkubo et al., 2005; Yu et al., 2012), it is possible that a negative auto-regulatory loop is induced by c-Fos–driven Fra1 up-regulation, which might block Blimp1 expression by interfering with c-Fos, thereby limiting humoral immune responses.
In summary, we demonstrate that Fra1 acts as a key suppressor of B cell differentiation into plasma cells. Fra1 orchestrates the major steps of B cell activation—proliferation and apoptosis—and specifically binds to the Blimp1 promoter and silences Blimp1 expression, plasma cell differentiation, and IgG production. These insights can help to build an overreaching concept of molecular control of plasma cell differentiation based on modulation of the Blimp1–Bcl6 axis by AP-1. In addition, they provide a molecular explanation for the physiological control of plasma cell responses during immune activation and also suggest a role for Fra1/AP-1 in the break of humoral immune tolerance and increased autoantibody production as seen in human autoimmune disease.
MATERIALS AND METHODS
Animals.
Fra1 (H2-Fra1-LTR) transgenic mice (Jochum et al., 2000) were backcrossed into C57BL/6 background by more than nine successive crossings. To generate B cell–specific Fra1-deficient mice, Fra1flox/flox mice of a mixed background (C57BL/6 crossed with SV129; Eferl et al., 2004) were crossed with Mb1-cre mice (129S-C57BL/6 mixed background; Hobeika et al., 2006). To generate Fra1/Bcl-2 double transgenic mice, Fra1 transgenic mice were crossed with Eμ bcl2-22 transgenic mice (D. Vöhringer, University of Erlangen-Nuremberg, Erlangen, Germany; Strasser et al., 1991) of a mixed background (BALB/c crossed with C57BL/6). All animals were maintained in an SPF facility. Animal experiments were approved by the local ethic committee, license Az:54-2532.1-36/12, “Fra1 in B-Zellen” delivered by the Regierung von Mittelfranken, Germany.
BM chimeras.
Recipient Rag1−/− mice were irradiated at 7 Gray using orthovoltage irradiation. The next day, mice were reconstituted by intravenous injection of 5 × 106 BM cells in Medium 199 (Sigma-Aldrich) containing Hepes (Gibco), DNase (Sigma-Aldrich), and Gentamycin (Sigma-Aldrich) and analyzed 6 wk after transplantation.
B cell reconstitution.
Splenic wt and Fra1tg CD43− B cells, as well as wt CD4+ T cells, were isolated by Macs separation. Subsequently, isolated wt T cells were mixed 1:1 with either wt or Fra1tg CD43− B cells and ∼1.5 × 107 cells of the B–T cell mixture were injected retro-orbitally into Rag1−/− mice. On day 2 after injection, mice were immunized with ∼109 SRBCs (Fiebig Nährstofftechnik, Idstein-Niederauroff/Ts.) i.p. in PBS and analyzed 7 d after immunization.
Immunizations and antigen-specific ELISA.
Mice were injected with 50 µg NP(36)-KLH (Biosearch Technologies) in PBS + 50 µl Adjuvans Imject Alum (Thermo Fisher Scientific) in a 1:1 dilution i.p. for primary immunization and with 10 µg NP(36)-KLH i.p. for boost immunization. Alternatively, mice were challenged with 50 µg NP-Ficoll (Biosearch Technologies) or with ∼109 SRBCs (Fiebig Nährstofftechnik, Idstein-Niederauroff/Ts.) i.p. in PBS.
For detection of NP-specific antibodies, microtiter plates were coated overnight with NP-BSA IgG1 at 4°C and blocked with PBS/2% FCS. Sera were diluted in PBS/2% FCS, starting with a 1/3,000 dilution for IgG or 1/1,000 for IgM and IgG1, plated in eightfold serial dilutions and incubated for 1 h at room temperature. HRP-conjugated IgM/IgG and IgG1 detection antibodies were obtained from SouthernBiotech. For basal Ig, Elisa microtiter plates were coated with 1 µg/ml Ig purified unlabeled antibodies (SouthernBiotech).
For quantification of anti-dsDNA antibody titers, microtiter plates were precoated with 50 µl poly-l-lysine in Tris EDTA (TE) buffer, pH 8, overnight at 4°C, washed three times with TE buffer, and coated with calf thymus dsDNA in TE buffer over night with the following detection procedure as described previously. All microtiter plates were analyzed at 490 nm on a SpectraMax 190 Microplate Reader (Molecular Devices).
ELISPOT assay.
To assess cells secreting NP-specific IgG, 96-well multiscreen plates (Millipore) were coated with 2 mg/ml NP-BSA in PBS. Plates were incubated with BM and splenic cell suspensions (5 × 106, 2.5 × 106, or 1.25 × 106 cells) of NP-KLH immunized mice (day 31) in triplicates overnight in a humidified incubator containing 5% CO2. After incubation, plates were washed and incubated with HRP-conjugated goat anti–mouse IgG (Southern Biotech) for 1 h at room temperature. Membrane-bound IgG was stained by addition of tetramethylbenzidine one-component membrane peroxidase (Kirkegaard & Perry Laboratories). The numbers of spots were counted with a video-based automatic ELISPOT reader (AID Diagnostics).
Isolation and in vitro cell stimulation of splenic B cells.
Spleens were digested in 5 ml Hank’s buffer supplemented with 1 mg/ml collagenase D (Roche) and 200 U/ml DNase I (Roche) for 30 min at 37°C. The reaction was stopped with 150 µl 0.5 M EDTA and the tissue was squashed through a 70-µm cell strainer before single cell suspension.
For peritoneal cell isolation, the outer skin was cut and gently pulled back to expose the inner skin lining the peritoneal cavity, and 1 ml PBS was injected into the peritoneum. After injection, the peritoneum was gently massaged to dislodge any attached cells into the PBS solution and PBS was collected out of the peritoneum.
B cells were purified by negative selection on CD43 MACS beads (Miltenyi Biotec) or sorted based on their surface markers using a MoFlo cell sorter (DAKO). Purified cells were cultured in RPMI1640 (10% FCS, 1 mM pyruvate, 2 mM l-glutamine, 100 U/ml penicillin/streptomycin, and 50 µM β-mercaptoethanol), and stimulated with 10 µg/ml LPS, 10 µg/ml LPS+100 U/ml recombinant mouse IL-4 (Miltenyi Biotec), 10 µg/ml LPS+anti-BCR antibody b.7.6, anti-CD40 mAb (clone FGK), or a mixture of 10 µg/ml anti-BCR antibody b.7.6 anti-CD40 mAb (clone FGK) and IL-4.
Flow cytometry and cell sorting.
The following antibodies were used (from BD if not indicated otherwise): FITC-conjugated-conjugated anti-CD5 (53–7.3), PE-conjugated anti-CD5 (53–7.3), Alexa Fluor 647 (AF647)–conjugated CD19 (eBio1D3), PE-conjugated anti-CD23 (FceRII; B3B4), FITC-conjugated anti-CD45R (B220; RA3-6B2), peridinin chlorophyll protein (PerCP)–conjugated anti-CD45R (B220; Miltenyi Biotec), APC-conjugated anti-CD138 (281–2), bio-conjugated anti-CD138 (281–2), AF647-GL7 (GL-7), fluorescein peanut agglutinin (PNA; Vector Laboratories), bio-conjugated anti-IgD (SouthernBiotech), APC-conjugated anti-IgG1 (X56), PE-conjugated anti-IgG1 (H143.225.8; SouthernBiotech), FITC-conjugated anti-IgM (SouthernBiotech), Cyanine (Cy) 5–conjugated anti-IgM (SouthernBiotech), PE-conjugated streptavidin, PE-indotricarbocyanine (Cy7)–conjugated streptavidin, and PerCP-conjugated streptavidin.
Analyses of the expression of cell surface molecules on a single cell level were performed by flow cytometry with a FACSCalibur (BD) flow cytometer. 6 × 105 or 2 × 106 cells/staining were washed with 1 ml FACS buffer (2% FCS, 0.02% Sodium azide, and PBS), resuspended in 100 µl FACS buffer, and incubated with sufficient amounts of antibody for 20 min at 4°C in the dark. Afterward, cells were washed in FACS buffer, analyzed in 300 µl FACS buffer, or stained with a secondary antibody. For analysis of intracellular proteins, cells were fixed and permeabilized with the Fix&Perm kit (An der Grub) according to the manufacturer’s instructions and afterward stained and analyzed as mentioned before.
For cell sorting, splenocytes were incubated in 1 ml antibody solution in 2% fetal calf serum and 2 mM EDTA in PBS buffer (MACS buffer) for 30 min at 4°C and then sorted as follows: FO B cells, IgMlow/IgD+ or IgMlow/CD23+/CD21+; MZ B cells, CD23−/IgM+/CD21+; T1 B cells, CD23−/IgM+/CD21−; T2 B cells, CD23+/CD21+/IgM+. Cells were sorted using a MoFlo cell sorter (DAKO instrumentations). Purities of 98–99% were generally achieved.
Proliferation analysis and cell death.
For eFluor staining, 5 × 106 sorted FO cells were mixed 1:1 with a 10 µM eFluor-solution in PBS (cell proliferation dye eFluor 670; eBioscience). Cell solution was incubated for 10 min at 37°C, stopped with 4–5 volumes R10+ and incubated for 5 min at 4°C. The stained cells were resuspended in R10+ and either measured at day 0 by flow cytometry or stimulated for 4 d with LPS and CD40/BCR/IL-4 and measured every day. To analyze only viable cells before each measurement, 3′3-dihexyloxacarbocyanine iodide (80 nM DiOC6) was supplemented to the media and incubated for 30 min at 37°C. Afterward, cells were washed with PBS, resuspended in 200 µl PI solution, and immediately measured by flow cytometry.
For quantification of the proportion of CD138-positive cells per cell division, sorted FO B cells were labeled with CSFE, allowed to proliferate upon stimulation, and stained with anti-CD138 antibody. The proliferation-induced dilution of CSFE was quantified and numbers of achieved cell divisions were calculated using the proliferation analysis option in FlowJo. For cell death analysis, stimulated cells were stained with Cy3–Annexin V for 20 min in Annexin V binding buffer (140 mM NaCl, 20 mM Hepes, pH 7.2, and 2.5 mM CaCl2), washed, and resuspended in Annexin V binding buffer in the presence of TO-PRO-3 Iodide (Invitrogen) and measured by flow cytometry.
Electron microscopy.
For electron microscopy, sorted FO B cells were stimulated with LPS for 72 h. Viable cells were collected by a Ficoll density gradient centrifugation and were fixed overnight in 2.5% glutaraldehyde. Cells were further processed and analyzed as described previously (Meister et al., 2010).
ChIP.
ChIP experiments were performed with ChIP-IT Express kit (Active Motif) according to the manufacturer’s protocol. 14 µl Fra1 and c-Fos antibodies (Santa Cruz Biotechnology, Inc.), as well as a rabbit control serum, were used for the immunoprecipitation. Primer sequence for APBS2 binding site: forward, 5′-AAGGAAAGCAGGGTAAACCGTG-3′; reverse, 5′-GTTAGCTTGCTCTTGTGCCAGG-3′. Primer sequence for negative control: forward, 5′-CAGACTGGGGCTGAAGGAAA-3′; reverse, 5′-TCTAAGTCCTCCGGATCGCT-3′.
Transient transfection of adherent cells using calcium phosphate.
1 d before transfection, 4 × 106 Phoenix-eco cells were seeded in a 10-cm-well plate and incubated overnight at 37°C and 5% CO2 in D10 medium. 20 µg DNA was mixed with 125 µl 2 M CaCl2 and 50 µl 10 mM chloroquine to a final volume of 1 ml with dH2O. Under bubbling conditions, the mixture was added to 1 ml 2× HBS (50 mM Hepes, 10 mM potassium chloride, 12 mM Dextrose, 280 mM sodium chloride, and 1.5 mM Na2HPO4, pH 7.4). Precipitates were trickled onto cells, and after 6–8 h of incubation at 37°C and 5% CO2, cells were provided with 6 ml of fresh D10 medium. Viral supernatant was removed after 24, 48, and 72 h, filtered, and stored at −80°C for infection.
Infection of primary stimulated B lymphoid cells.
0.5 × 106 primary CD43− B cells, LPS stimulated for 24 h, were infected with retroviral supernatants supplemented with 2 µl transfection reagent (QIAGEN). Infection mixture was centrifuged for 3.5 h at 33,000 rpm at 33°C. Infected cells were stimulated for 3 d with LPS+IL-4 and analyzed by flow cytometry.
RNA extraction and quantitative real-time PCR.
Sorted stimulated and unstimulated cells were isolated in 500 µl TRIzol reagent (Invitrogen). RNA isolation was performed according to the manufacturer’s instructions and reverse transcribed into cDNA using oligo d(T) primers. QPCRs were performed using SYBR Green I-dTTP (Eurogentec). Samples were analyzed in duplicate and normalized to the level of β-actin mRNA. The following primer sequences were used for real time analysis: β-actin forward, 5′-TGTCCACCTTCCAGCAGATGT-3′; β-actin reverse, 5′-AGTCAGTAACAGTCCGCCTAGA-3′; Blimp1 forward, 5′-GACAGAGGCCGAGTTTGAAG-3′; Blimp1 reverse, 5′-GGCATTCTTGGGAACTGTGT-3′; Fra1 forward, 5′-GAGACGCGAGCGGAACAAG-3′; Fra1 reverse, 5′-CTTCCAGCACCAGCTCAAGG-3′; c-fos forward, 5′-CGGGTTTCAACGCCGACTAC 3′; and c-fos reverse, 5′-CAGGTCTGGGCTGGTGGAGA-3′.
Western blot.
Proteins were electrophoresed on 10% SDS polyacrylamide gels and transferred to a nitrocellulose membrane (1.5 h, 110 mA). Transfer efficiency was determined by Ponceau S. Membranes were blocked in 5% milk powder or 5% BSA in TBS-T and probed with anti–Blimp-1, and anti-µ (all antibodies were obtained from Santa Cruz Biotechnology, Inc.) at 4°C overnight. Membranes were washed in TBS-T (3 × 15 min) and incubated with the secondary antibody anti–mouse/rabbit IgG HRP conjugate (1:25,000; Promega) for 1 h at room temperature. Bands were detected by ECL (Thermo Fisher Scientific). For re-blotting, membranes are stripped using ReBlot Plus Stripping Solution (Millipore).
Statistical analysis.
All statistical analyses were performed using Prism (GraphPad Software). Statistical significances were calculated by Student’s t test or one-way ANOVA using Prism 6 software.
Acknowledgments
We thank Dr. David Tarlinton for sharing unpublished data, Dr. David Vöhringer for Eμ bcl2-22 mice, and Dr. Wolfgang Baum for performing the retro-orbital injections.
This work was supported by grants from the German Science Foundation (Deutsche Forschungsgemeinschaft; SPP1468 to J.-P. David and D. Mielenz [DA1067/7-2], Mi932/2-2 and TRR130 to G. Schett, and GK1660 to J.-P. David and G. Schett; an Emmy Noether Stipend to A. Bozec; and CRG643 to G. Schett) and the Interdisciplinary Research Center Erlangen (IZKF; A7, E8 to D. Mielenz). E. Hobeika and M. Reth were supported by the Excellence Initiative of the German Federal and State Governments (EXC 294), by ERC grant 322972, and by the Deutsche Forschungsgemeinschaft through SFB746 and TRR130.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ASC
- antibody-secreting cell
- AP1BS
- AP-1 binding site
- BCR
- B cell receptor
- Blimp1
- B lymphocyte–induced maturation protein 1
- ChIP
- chromatin immunoprecipitation
- CSR
- class switch recombination
- dsDNA
- double-stranded DNA
- FO
- follicular
- GC
- germinal center
- Irf4
- IFN regulatory factor 4
- MZ
- marginal zone
- NP
- nitrophenol
- SRBC
- sheep RBC
- TD
- T cell dependent
References
- Arguni, E., Arima M., Tsuruoka N., Sakamoto A., Hatano M., and Tokuhisa T.. 2006. JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. Int. Immunol. 18:1079–1089. 10.1093/intimm/dxl041 [DOI] [PubMed] [Google Scholar]
- De Silva, N.S., Simonetti G., Heise N., and Klein U.. 2012. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol. Rev. 247:73–92. 10.1111/j.1600-065X.2012.01113.x [DOI] [PubMed] [Google Scholar]
- Delogu, A., Schebesta A., Sun Q., Aschenbrenner K., Perlot T., and Busslinger M.. 2006. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 24:269–281. 10.1016/j.immuni.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Eferl, R., Hoebertz A., Schilling A.F., Rath M., Karreth F., Kenner L., Amling M., and Wagner E.F.. 2004. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 23:2789–2799. 10.1038/sj.emboj.7600282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann, A., Hafezi F., Elliott C., Remé C.E., Rüther U., and Wagner E.F.. 2000. Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev. 14:2695–2700. 10.1101/gad.187900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletta, V.C., Segal D.H., and Cohen D.R.. 1998. Transcriptional regulation in the immune system: all roads lead to AP-1. J. Leukoc. Biol. 63:139–152 [DOI] [PubMed] [Google Scholar]
- Gass, P., Fleischmann A., Hvalby O., Jensen V., Zacher C., Strekalova T., Kvello A., Wagner E.F., and Sprengel R.. 2004. Mice with a fra-1 knock-in into the c-fos locus show impaired spatial but regular contextual learning and normal LTP. Brain Res. Mol. Brain Res. 130:16–22. 10.1016/j.molbrainres.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Hess, J., Angel P., and Schorpp-Kistner M.. 2004. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 117:5965–5973. 10.1242/jcs.01589 [DOI] [PubMed] [Google Scholar]
- Hobeika, E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, L., and Rothstein T.L.. 1995. Receptor-specific induction of individual AP-1 components in B lymphocytes. J. Immunol. 154:3300–3309 [PubMed] [Google Scholar]
- Huo, L., and Rothstein T.L.. 1996. Isolation and characterization of murine fra-1: induction mediated by CD40 and surface Ig is protein kinase C dependent. J. Immunol. 157:3812–3818 [PubMed] [Google Scholar]
- Jochum, W., David J.P., Elliott C., Wutz A., Plenk H. Jr, Matsuo K., and Wagner E.F.. 2000. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nat. Med. 6:980–984. 10.1038/79676 [DOI] [PubMed] [Google Scholar]
- Kallies, A., Hasbold J., Tarlinton D.M., Dietrich W., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200:967–977. 10.1084/jem.20040973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies, A., Hasbold J., Fairfax K., Pridans C., Emslie D., McKenzie B.S., Lew A.M., Corcoran L.M., Hodgkin P.D., Tarlinton D.M., and Nutt S.L.. 2007. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 26:555–566. 10.1016/j.immuni.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Klein, U., and Dalla-Favera R.. 2008. Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol. 8:22–33. 10.1038/nri2217 [DOI] [PubMed] [Google Scholar]
- Knödel, M., Kuss A.W., Lindemann D., Berberich I., and Schimpl A.. 1999. Reversal of Blimp-1-mediated apoptosis by A1, a member of the Bcl-2 family. Eur. J. Immunol. 29:2988–2998. [DOI] [PubMed] [Google Scholar]
- Knödel, M., Kuss A.W., Berberich I., and Schimpl A.. 2001. Blimp-1 over-expression abrogates IL-4- and CD40-mediated suppression of terminal B cell differentiation but arrests isotype switching. Eur. J. Immunol. 31:1972–1980. [DOI] [PubMed] [Google Scholar]
- Körholz, D., Gerdau S., Enczmann J., Zessack N., and Burdach S.. 1992. Interleukin 6-induced differentiation of a human B cell line into IgM-secreting plasma cells is mediated by c-fos. Eur. J. Immunol. 22:607–610. 10.1002/eji.1830220248 [DOI] [PubMed] [Google Scholar]
- Krum, S.A., Chang J., Miranda-Carboni G., and Wang C.Y.. 2010. Novel functions for NFκB: inhibition of bone formation. Nat Rev Rheumatol. 6:607–611. 10.1038/nrrheum.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, H., Thierry-Mieg D., Thierry-Mieg J., Kim H.P., Oh J., Tunyaplin C., Carotta S., Donovan C.E., Goldman M.L., Tailor P., et al. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 31:941–952. 10.1016/j.immuni.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K.I., Angelin-Duclos C., Kuo T.C., and Calame K.. 2002. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol. Cell. Biol. 22:4771–4780. 10.1128/MCB.22.13.4771-4780.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loder, F., Mutschler B., Ray R.J., Paige C.J., Sideras P., Torres R., Lamers M.C., and Carsetti R.. 1999. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J. Exp. Med. 190:75–89. 10.1084/jem.190.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Carvalho, T., and Kearney J.F.. 2004. Development and selection of marginal zone B cells. Immunol. Rev. 197:192–205. 10.1111/j.0105-2896.2004.0112.x [DOI] [PubMed] [Google Scholar]
- Luther, J., Driessler F., Megges M., Hess A., Herbort B., Mandic V., Zaiss M.M., Reichardt A., Zech C., Tuckermann J.P., et al. 2011. Elevated Fra-1 expression causes severe lipodystrophy. J. Cell Sci. 124:1465–1476. 10.1242/jcs.079855 [DOI] [PubMed] [Google Scholar]
- Manz, R.A., Hauser A.E., Hiepe F., and Radbruch A.. 2005. Maintenance of serum antibody levels. Annu. Rev. Immunol. 23:367–386. 10.1146/annurev.immunol.23.021704.115723 [DOI] [PubMed] [Google Scholar]
- Martin, F., Oliver A.M., and Kearney J.F.. 2001. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity. 14:617–629. 10.1016/S1074-7613(01)00129-7 [DOI] [PubMed] [Google Scholar]
- Matsuo, K., Owens J.M., Tonko M., Elliott C., Chambers T.J., and Wagner E.F.. 2000. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat. Genet. 24:184–187. 10.1038/72855 [DOI] [PubMed] [Google Scholar]
- Meister, S., Frey B., Lang V.R., Gaipl U.S., Schett G., Schlötzer-Schrehardt U., and Voll R.E.. 2010. Calcium channel blocker verapamil enhances endoplasmic reticulum stress and cell death induced by proteasome inhibition in myeloma cells. Neoplasia. 12:550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, K., Tokoyoda K., Radbruch A., MacLennan I., and Manz R.A.. 2006. Stromal niches, plasma cell differentiation and survival. Curr. Opin. Immunol. 18:265–270. 10.1016/j.coi.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Nera, K.P., Kohonen P., Narvi E., Peippo A., Mustonen L., Terho P., Koskela K., Buerstedde J.M., and Lassila O.. 2006. Loss of Pax5 promotes plasma cell differentiation. Immunity. 24:283–293. 10.1016/j.immuni.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Nutt, S.L., Heavey B., Rolink A.G., and Busslinger M.. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562. 10.1038/44076 [DOI] [PubMed] [Google Scholar]
- Nutt, S.L., Taubenheim N., Hasbold J., Corcoran L.M., and Hodgkin P.D.. 2011. The genetic network controlling plasma cell differentiation. Semin. Immunol. 23:341–349. 10.1016/j.smim.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Ochiai, K., Katoh Y., Ikura T., Hoshikawa Y., Noda T., Karasuyama H., Tashiro S., Muto A., and Igarashi K.. 2006. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J. Biol. Chem. 281:38226–38234. 10.1074/jbc.M607592200 [DOI] [PubMed] [Google Scholar]
- Ohkubo, Y., Arima M., Arguni E., Okada S., Yamashita K., Asari S., Obata S., Sakamoto A., Hatano M., O-Wang J., et al. 2005. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J. Immunol. 174:7703–7710. 10.4049/jimmunol.174.12.7703 [DOI] [PubMed] [Google Scholar]
- Peperzak, V., Vikstrom I.B., and Tarlinton D.M.. 2012. Through a glass less darkly: apoptosis and the germinal center response to antigen. Immunol. Rev. 247:93–106. 10.1111/j.1600-065X.2012.01123.x [DOI] [PubMed] [Google Scholar]
- Rajewsky, K.1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. 10.1038/381751a0 [DOI] [PubMed] [Google Scholar]
- Reimold, A.M., Ponath P.D., Li Y.S., Hardy R.R., David C.S., Strominger J.L., and Glimcher L.H.. 1996. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J. Exp. Med. 183:393–401. 10.1084/jem.183.2.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkenberger, J.L., Wallin J.J., Johnson K.W., and Koshland M.E.. 1996. An interleukin-2 signal relieves BSAP (Pax5)-mediated repression of the immunoglobulin J chain gene. Immunity. 5:377–386. 10.1016/S1074-7613(00)80263-0 [DOI] [PubMed] [Google Scholar]
- Sciammas, R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., and Singh H.. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236. 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., and Staudt L.M.. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 17:51–62. 10.1016/S1074-7613(02)00335-7 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef, M., Lin K.I., Savitsky D., Liao J., and Calame K.. 2005. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J. Exp. Med. 202:1471–1476. 10.1084/jem.20051611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian, E., and Karin M.. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131–E136. 10.1038/ncb0502-e131 [DOI] [PubMed] [Google Scholar]
- Strasser, A., Whittingham S., Vaux D.L., Bath M.L., Adams J.M., Cory S., and Harris A.W.. 1991. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc. Natl. Acad. Sci. USA. 88:8661–8665. 10.1073/pnas.88.19.8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, J.A., Cook A., Bannister A.J., and Kouzarides T.. 1992. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 6:1810–1819. 10.1101/gad.6.9.1810 [DOI] [PubMed] [Google Scholar]
- Tunyaplin, C., Shaffer A.L., Angelin-Duclos C.D., Yu X., Staudt L.M., and Calame K.L.. 2004. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 173:1158–1165. 10.4049/jimmunol.173.2.1158 [DOI] [PubMed] [Google Scholar]
- Vasanwala, F.H., Kusam S., Toney L.M., and Dent A.L.. 2002. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J. Immunol. 169:1922–1929. 10.4049/jimmunol.169.4.1922 [DOI] [PubMed] [Google Scholar]
- Victora, G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Wagner, E.F., and Eferl R.. 2005. Fos/AP-1 proteins in bone and the immune system. Immunol. Rev. 208:126–140. 10.1111/j.0105-2896.2005.00332.x [DOI] [PubMed] [Google Scholar]
- Wenzel, A., Iseli H.P., Fleischmann A., Hafezi F., Grimm C., Wagner E.F., and Remé C.E.. 2002. Fra-1 substitutes for c-Fos in AP-1-mediated signal transduction in retinal apoptosis. J. Neurochem. 80:1089–1094. 10.1046/j.0022-3042.2002.00807.x [DOI] [PubMed] [Google Scholar]
- Yu, Z., Sato S., Trackman P.C., Kirsch K.H., and Sonenshein G.E.. 2012. Blimp1 activation by AP-1 in human lung cancer cells promotes a migratory phenotype and is inhibited by the lysyl oxidase propeptide. PLoS ONE. 7:e33287. 10.1371/journal.pone.0033287 [DOI] [PMC free article] [PubMed] [Google Scholar]