Molawi et al. examine the origin and cellular dynamics of macrophages in the heart during postnatal development. Cardiac macrophages derived from CX3CR1+ embryonic progenitors persist into adulthood, but the contribution of these cells to resident macrophages declines after birth with diminished self-renewal as the mice age. Over time, the heart is progressively reconstituted with bone marrow–derived macrophages, even in the absence of inflammation.
Abstract
Cardiac macrophages (cMΦ) are critical for early postnatal heart regeneration and fibrotic repair in the adult heart, but their origins and cellular dynamics during postnatal development have not been well characterized. Tissue macrophages can be derived from embryonic progenitors or from monocytes during inflammation. We report that within the first weeks after birth, the embryo-derived population of resident CX3CR1+ cMΦ diversifies into MHCII+ and MHCII− cells. Genetic fate mapping demonstrated that cMΦ derived from CX3CR1+ embryonic progenitors persisted into adulthood but the initially high contribution to resident cMΦ declined after birth. Consistent with this, the early significant proliferation rate of resident cMΦ decreased with age upon diversification into subpopulations. Bone marrow (BM) reconstitution experiments showed monocyte-dependent quantitative replacement of all cMΦ populations. Furthermore, parabiotic mice and BM chimeras of nonirradiated recipient mice revealed a slow but significant donor contribution to cMΦ. Together, our observations indicate that in the heart, embryo-derived cMΦ show declining self-renewal with age and are progressively substituted by monocyte-derived macrophages, even in the absence of inflammation.
Cardiovascular disease represents a leading cause of death in the developed world, and many pathologies result from insufficient repair of cardiac injury. Mammalian wound healing mechanisms in the adult heart involve scar formation, but for a short time window after birth the neonatal heart maintains full regeneration capacity of cardiac tissue (Porrello et al., 2011), a process which requires macrophages (Aurora et al., 2014). Studies of cardiac repair after myocardial infarction in the adult have highlighted the critical role of infiltrating monocytes and monocyte-derived macrophages for the healing process (Nahrendorf et al., 2007; Swirski et al., 2009). The focus of these studies has been on monocyte-derived cells, consistent with the traditional view that macrophages are part of the mononuclear phagocyte system and monocyte-derived (van Furth and Cohn, 1968; Geissmann et al., 2010). Recent evidence, however, suggests that monocyte contribution to macrophages only represents an emergency pathway, as many tissue macrophage populations get seeded in the developing embryo and can self-maintain without major monocyte contribution (Chorro et al., 2009; Ginhoux et al., 2010; Hoeffel et al., 2012; Schulz et al., 2012; Hashimoto et al., 2013; Yona et al., 2013). Macrophages can massively expand in the tissue by local proliferation in response to challenge (Jenkins et al., 2011; Hashimoto et al., 2013; Sieweke and Allen, 2013) and can extensively self-renew without loss of differentiated function in culture upon inactivation of MafB and cMaf transcription factors (Aziz et al., 2009). This has led to the proposition that tissue macrophages may have a long-term self-renewal capacity akin to that of stem cells (Sieweke and Allen, 2013).
Examples of tissue macrophages that are independent from monocytes are microglia in the brain and epidermal Langerhans cells (Ajami et al., 2007; Chorro et al., 2009; Ginhoux et al., 2010; Hoeffel et al., 2012). In contrast, intestinal or dermal macrophages have a high turnover rate and are constantly replaced from Ly6C+ blood monocytes (Zigmond et al., 2012; Tamoutounour et al., 2013). Thus, origin and turnover of tissue macrophages are highly tissue specific and need to be assessed individually for different organ systems (Sieweke and Allen, 2013).
Resident macrophages can be found in all tissues, where they fulfill a variety of tissue-specific functions contributing to homeostasis, development, and regeneration (Davies et al., 2013), making them prominent candidates for therapeutic intervention. This holds in particular for the heart, where tissue-resident cardiac macrophages (cMΦ) have been described as CX3CR1+ cells that can be found throughout the myocardium (Pinto et al., 2012). A recent study identified several cMΦ populations, including short-lived monocyte-derived cells that resemble monocyte-derived DCs as well as tissue-resident cMΦ, which were suggested to be primarily of embryonic origin and to self-maintain under homeostatic conditions (Epelman et al., 2014).
Here, we examined the origin and turnover of tissue-resident cMΦ during postnatal development. Using genetic lineage tracing, parabiotic mice, and unconditioned BM chimeras, we demonstrate that embryo-derived cMΦ are gradually replaced by monocyte-derived macrophages with age and in the absence of inflammation or injury. This replacement is mirrored by dynamic changes in the resident cMΦ population composition and decreasing cMΦ self-renewal over time.
RESULTS AND DISCUSSION
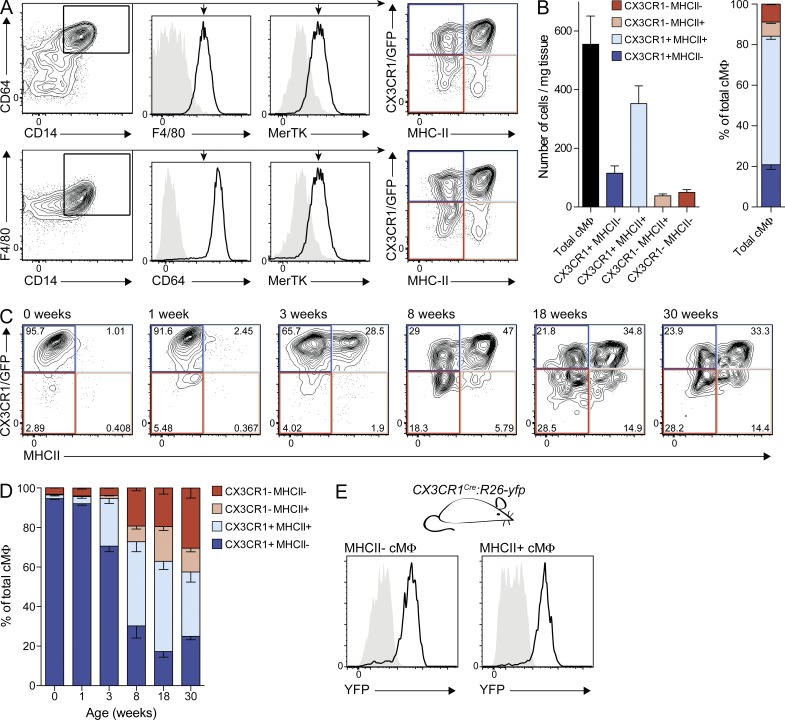
We focused on the resident cMΦ population and applied a rigorous flow cytometry gating strategy to exclude potential infiltrating leukocytes (Fig. S1, A–C). Tissue-resident macrophages were identified as positive for the core macrophage signature markers CD14, CD64, and MerTK (Gautier et al., 2012; Tamoutounour et al., 2013) and the classical macrophage marker F4/80 (Fig. 1 A). The chemokine receptor CX3CR1 is widely expressed in the mononuclear phagocyte system (Jung et al., 2000) and cMΦ have been reported to be CX3CR1-positive (Pinto et al., 2012). Therefore, we refined our analysis of the global cMΦ population by testing CX3CR1 expression in CX3CR1GFP/+ knock-in mice (Jung et al., 2000). Additionally, we included MHCII in our analysis, which can be differentially expressed on tissue macrophages (Tamoutounour et al., 2013). We found that in 8-wk-old mice, the majority of cMΦ was CX3CR1+ (∼80%) and MHCII+ (∼70%), allowing the delineation of four distinct cMΦ subpopulations (Fig. 1, A and B). Additional populations of CD11c+MHCII+ or Ly6C+ cells, referred to as cMΦ by others (Epelman et al., 2014), were excluded from our analysis (Fig. S1 B), as data from other tissues suggest that these are monocytes and monocyte-derived dendritic cells (Tamoutounour et al., 2013). Consistent with this, these cells have been shown to be short-lived and monocyte-derived (Epelman et al., 2014).
Figure 1.
cMΦ develop into 4 subpopulations after birth. (A and B) Cytometry analysis of cMΦ from adult CX3CR1GFP/+ mice. (A) FACS profiles of cMΦ populations, pregated on living single CD11b+ cells with low CD11c and Ly6C expression. (B) Mean of total and subpopulations of cMΦ in absolute numbers (left) or as percentage of total (right) as determined by bead-normalized flow cytometry. Error bars represent SEM (n = 4). (C and D) Cytometry analysis (C) and mean percentage (D) of cMΦ subpopulations from CX3CR1GFP/+ mice of indicated age. Error bars represent SEM (n = 3–4). (E) MHCII− and MHCII+ cMΦ from adult CX3CR1cre:R26-yfp mice (black line) were analyzed for YFP expression and compared with Cre− littermate controls (gray area; n = 3–4). Data in all panels are representative of at least two independent experiments.
To investigate the development of the four cMΦ populations, we analyzed CX3CR1 and MHCII expression in cMΦ from newborn to 30-wk-old mice (Fig. 1, C and D). We observed that almost all embryo-derived cMΦ present at birth were CX3CR1+MHCII−. This homogenous cMΦ compartment diversified with age into four subpopulations with a progressive increase of MHCII+ cMΦ and a decrease of CX3CR1+ cMΦ. Importantly, genetic fate mapping analysis using CX3CR1cre mice crossed to R26-yfp reporter mice (CX3CR1cre:R26-yfp; Yona et al., 2013) revealed that all adult cMΦ subpopulations must have developed from a CX3CR1+ stage (Fig. 1 E). These results suggested two not mutually exclusive possibilities for cMΦ subpopulation development: persistence and diversification of embryo-derived cMΦ as suggested elsewhere (Epelman et al., 2014), or replacement of embryo-derived cMΦ by adult monocyte-derived macrophages.
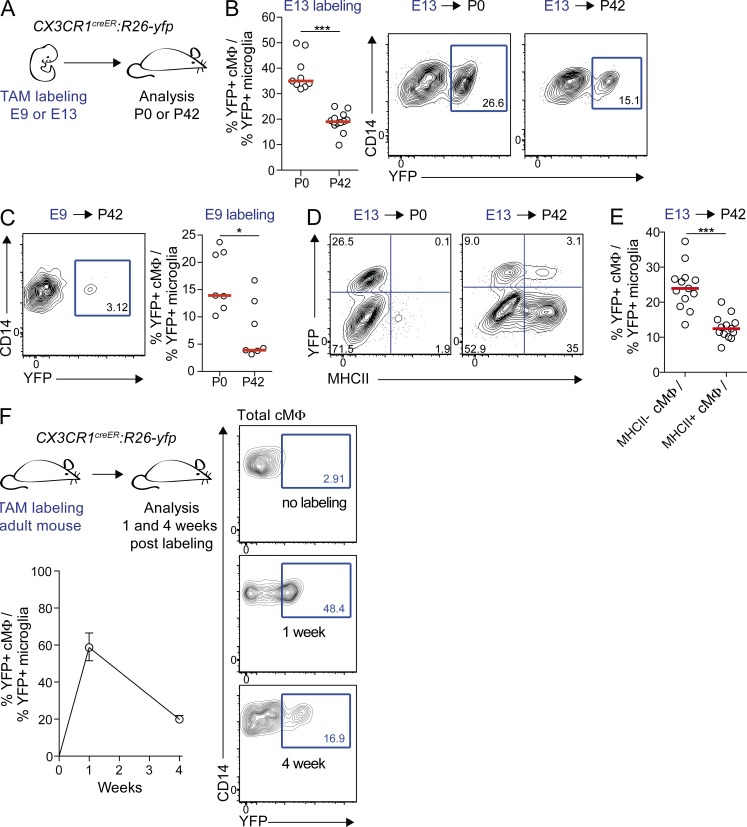
To address these alternatives, we first analyzed the persistence of embryo-derived cMΦ by genetic lineage tracing using CX3CR1-driven tamoxifen (TAM)-inducible Cre recombinase (CreERT2) and R26-yfp reporter mice (CX3CR1creER:R26-yfp; Fig. 2 A; Yona et al., 2013). Tamoxifen-induced pulse labeling at E9 permits us to identify cells derived from yolk sac (YS) CX3CR1+ macrophages before the onset of definitive hematopoiesis. Labeling at E13 cannot distinguish YS- and hematopoietic stem cell (HSC)–derived macrophages, but should increase cMΦ labeling because the heart is colonized by CX3CR1+ macrophages at this time (Epelman et al., 2014). cMΦ labeling was normalized to YFP+ microglia, which are YS-derived, remain CX3CR1+ throughout development, self-maintain without contribution of HSC-derived cells (Ginhoux et al., 2010; Kierdorf et al., 2013), and therefore represent an internal control for maximal CX3CR1 labeling efficiency. E13 labeling revealed that relative cMΦ labeling declined from ∼35% in newborn mice to ∼18% in 6-wk-old mice (Fig. 2 B), indicating a loss of embryo-derived cMΦ with age. E9 labeling showed that embryo-derived cMΦ were at least partially of YS origin and similarly declined from ∼14% in neonates to ∼4% in 6-wk-old mice (Fig. 2 C). Interestingly, all embryo-derived YFP+ cMΦ were MHCII− at birth but had partially differentiated into MHCII+ cMΦ after 6 wk (Fig. 2 D). Although this demonstrated that both populations can develop from cells of embryonic origin, the relative contribution of embryo-derived YFP+ cMΦ to MHCII+ cMΦ was significantly lower than to MHCII− cMΦ, (Fig. 2 E).
Figure 2.
Decreased contribution of embryo-derived macrophages to cMΦ with age. (A) CX3CR1creER:R26-yfp embryos were treated with TAM on E9 or E13 and analyzed on the day of delivery (P0) or 6 wk after delivery (P42). (B) Percentage of microglia-normalized YFP+ cMΦ and representative cytometry plots pregated on total cMΦ at P0 and P42 after E13 labeling. Bars show median (n = 9–13). ***, P ≤ 0.005, Mann-Whitney test. Data were pooled from two independent experiments. (C) Representative cytometry plot showing YFP+ within total cMΦ at P42 and percentage of microglia-normalized YFP+ cMΦ at P0 and P42 after E9 labeling. Bars show median (n = 7) *, P ≤ 0.05. Data were pooled from three independent experiments. (D) Representative cytometry plots showing YFP+ cells within MHCII− and MHCII+ cMΦ at P0 and P42 after E13 labeling (n = 9–13). (E) Quantification of YFP+ cells within MHCII− and MHCII+ cMΦ at P42 after E13 labeling. Bars show the median (n = 9–13). ***, P ≤ 0.005, Mann-Whitney test. Data were pooled from two independent experiments. (F) Adult TAM-treated CX3CR1creER:R26-yfp mice were analyzed for YFP+ cMΦ and microglia at 1 and 4 wk after treatment. Percentage of microglia-normalized YFP+ cMΦ is shown as median with extreme samples as error bars (n = 3). Representative cytometry plots show YFP+ within total cMΦ. Unless otherwise indicated data in all panels are representative of two independent experiments.
We further assessed the turnover of CX3CR1+ cMΦ in the adult heart by pulse labeling adult CX3CR1creER:R26-yfp (Fig. 2 F). 1 wk after treatment, labeling in cMΦ corresponded to ∼60% of microglia labeling but drastically decreased to ∼20% after 4 wk. By comparison, microglia labeling was nearly complete after 1 wk and remained unchanged thereafter (Goldmann et al., 2013). Collectively, our lineage tracing experiments argue that a significant proportion of macrophages established in early embryonic development is still present at birth but is gradually lost with age.
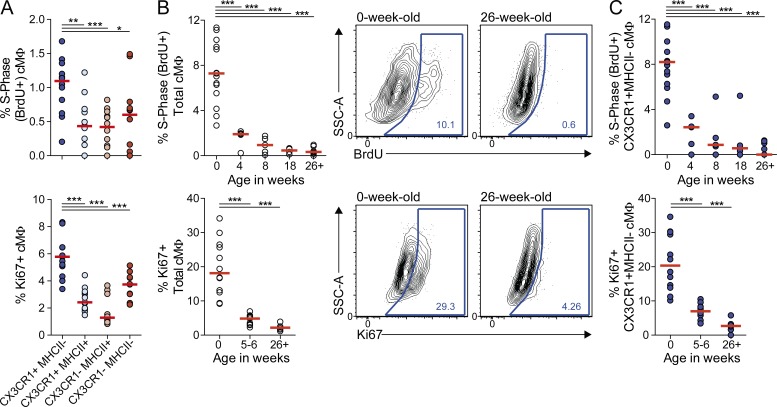
The relatively fast turnover of labeled tissue-resident cMΦ suggested that local self-renewal was insufficient to maintain the resident cMΦ pool. We therefore analyzed the proliferative activity of cMΦ subpopulations in adult mice by BrdU incorporation after a 4-h pulse labeling period and by Ki67 staining (Fig. 3 A). The overall number of proliferating macrophages in adult hearts was low, particularly in MHCII+ populations, but the proliferative rate of CX3CR1+MHCII− cMΦ significantly exceeded that of all other subpopulations. Interestingly, this population was also the only cMΦ subset present at birth but declined with age at the expense of the other cMΦ populations (Fig. 1, C and D). We therefore analyzed the evolution of cMΦ proliferation with age. Both BrdU incorporation and Ki67 staining showed a high proliferative rate in newborn CX3CR1+MHCII− cMΦ, which gradually decreased by 8–10-fold both in total cMΦ (Fig. 3 B) and in the CX3CR1+MHCII− cMΦ population (Fig. 3 C). Together, these data suggest that both a successive loss of the cell population with the highest proliferative rate (CX3CR1+MHCII− cMΦ) and a strong decrease of the proliferation rate collectively result in progressively decreasing self-renewal of the tissue-resident cMΦ pool.
Figure 3.
Decreased proliferation rate of embryo-derived cMΦ with age. (A) cMΦ subpopulations of adult CX3CR1GFP/+ mice were analyzed for BrdU incorporation and expression of Ki67 by flow cytometry four hours after BrdU injection (i.p.). Bars show median (n = 12–13). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; Wilcoxon test. (B and C) Total (B) or CX3CR1+MHCII− (C) cMΦ of CX3CR1GFP/+ mice of indicated age were analyzed for BrdU incorporation and Ki67 expression by flow cytometry 4 h after BrdU injection (i.p.). Bars show median (n = 4–15). ***, P ≤ 0.005, Mann-Whitney test. Data in all panels were pooled from 2–3 independent experiments.
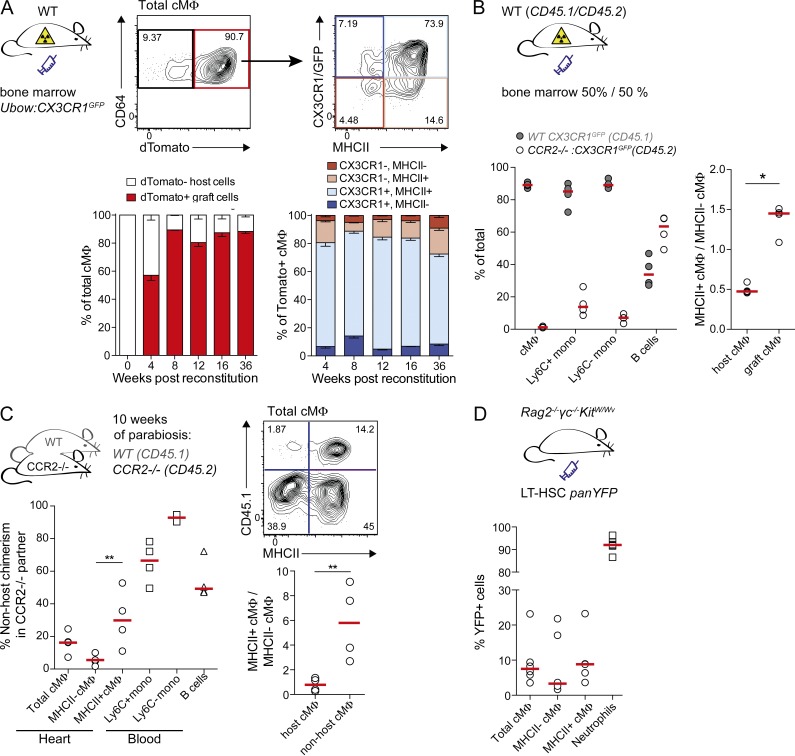
We next addressed the question of how the age-dependent loss of embryo-derived macrophages is compensated. To test the ability of monocytes to contribute to cMΦ populations, we generated BM chimeras using WT hosts and BM from transgenic mice expressing dTomato from a ubiquitously expressed human Ubiquitin-C promoter (Ubow mice; Ghigo et al., 2013) and GFP from the CX3CR1 locus that makes it possible to monitor grafted cells in all four cMΦ populations. Graft-derived dTomato+ cMΦ contributed to all four cMΦ populations from 4 wk after transplantation and almost completely replaced host cMΦ populations within 8 wk (Fig. 4 A). However, a small radio-resistant population (∼10%) persisted for 36 wk without monocyte contribution. To further analyze whether cMΦ replacement was monocyte-dependent, we generated mixed chimeras with BM from WT (CD45.1) and CCR2−/− (CD45.2) CX3CR1GFP/+ mice in CD45.1/CD45.2 double-positive WT hosts (Fig. 4 B). CCR2−/− mice have reduced numbers of Ly6C+ monocytes in the blood (Serbina and Pamer, 2006) and can therefore be used to analyze the contribution of circulating monocytes and CCR2-dependent progenitors to tissue macrophage populations (Yona et al., 2013; Tamoutounour et al., 2013). Blood analysis indeed confirmed that the vast majority of monocytes but not B cells were of WT origin. Graft-derived cMΦ were almost completely derived from WT cells (Fig. 4 B). Collectively these two experiments established that HSC-derived CCR2-dependent cells, most likely Ly6C+ monocytes, have the capacity to differentiate into all cMΦ subsets. Residual host-derived cMΦ included both MHCII+ and MHCII− cMΦ but showed a lower proportion of MHCII+ cells than grafted monocyte-derived cMΦ (Fig. 4 B).
Figure 4.
Monocytes contribute to four cMΦ subpopulations in adult mice. (A) WT mice were lethally irradiated and reconstituted with BM from Ubow:CX3CR1GFP/+ mice. cMΦ were analyzed for contribution of dTomato+ cells at indicated time points after reconstitution and graft-derived cells were analyzed for CX3CR1 and MHCII expression. Data are presented as mean percentage ± SEM (n = 3–7) and derived from two independent experiments. (B) Mixed BM chimeras were generated by reconstituting lethally irradiated WT mice (CD45.1/CD45.2) with BM from CCR2−/−:CX3CR1GFP/+ (CD45.2) and WT CX3CR1GFP/+ (CD45.1) mice. cMΦ, circulating CD11b+CD115+ monocytes (Ly6C+ and Ly6C−) and B220+ B cells were analyzed for WT (CD45.1) and CCR2−/− (CD45.2) contribution. Host- and graft-derived cMΦ were analyzed for the ratio of MHCII+/MHCII− cMΦ. Bars show median (n = 4). *, P ≤ 0.05, Mann-Whitney test. (C) Parabiosis was established between adult CCR2−/− (CD45.2) and WT (CD45.1) mice. After 10 wk, CCR2−/− mice were analyzed for contribution of CD45.1+ non-host cells to cMΦ (total, MHCII+, and MHCII−), circulating monocytes (Ly6C+ and Ly6C−), and B cells. For CD45.1− host and CD45.1+ non-host cMΦ, the ratio of MHCII+/MHCII− cMΦ was calculated and compared. Bars show median (n = 4). **, P ≤ 0.01, Mann-Whitney test. (D) BM chimeras were generated by transfer of LT-HSC isolated from panYFP mice into nonirradiated Rag2−/−γc−/−KitW/Wv mice. cMΦ (total, MHCII+, and MHCII−) and neutrophils (Gr1hi and SSChi) were analyzed for contribution of grafted YFP+ cells 8 wk after transplantation. Bars show median (n = 6). **, P ≤ 0.01, Mann-Whitney test. Data in all panels are representative of two independent experiments.
To examine monocyte contribution to cMΦ populations under homeostatic conditions, we generated parabiotic couples of WT (CD45.1) and CCR2−/− (CD45.2) mice and analyzed non-host contributions after 10 wk of parabiosis in CCR2−/− partners (Fig. 4 C). Circulating B cells were equally distributed between the partner mice. In the CCR2-deficient host Ly6C+ monocytes had reached ∼70% non-host chimerism. About 16% of total cMΦ in CCR2-deficient mice were of non-host origin. Given a 70% chimerism in Ly6C+ monocytes, the likely cells of origin, and the possibility that depending on lifetime and exchange rate cMΦ may not be fully equilibrated after 10 wk, these observations indicated a significant monocyte contribution to tissue-resident cMΦ. Again, non-host contribution to MHCII+ cMΦ (∼30%) was significantly higher compared with MHCII− cMΦ (∼5%) as highlighted by MHCII+/MHCII− cMΦ ratios for non-host and host-derived cells (Fig. 4 C).
As a complimentary experimental approach that does not depend on myeloablative conditioning, we used Rag2−/−γc−/−KitW/Wv mice, which are universal HSC recipients that accept grafts without prior irradiation (Waskow et al., 2009), and generated BM chimeras by transplantation of HSC from panYFP mice (Luche et al., 2013). More than 90% of neutrophils were YFP+ after 8 wk, demonstrating efficient reconstitution (Fig. 4 D). Analysis of cMΦ revealed ∼7% YFP+ cells in the total population. Again, donor cells showed a higher contribution to MHCII+ (∼8% YFP+) than to MHCII− cMΦ (∼4% YFP+) (Fig. 4 D). In the same mice, no donor contribution to microglia cells was observed (K. Klapproth and H.R. Rodewald, personal communication). Together, these experiments clearly demonstrated a slow but significant replacement of tissue-resident cMΦ by infiltrating monocyte-derived macrophages, which preferentially contributed to MHCII+ cMΦ.
In the present study, we have analyzed homeostasis, origin, and turnover of resident macrophages in the heart during postnatal development. We have shown that the cMΦ compartment is undergoing dynamic changes with age. Embryo-derived macrophages present in newborn mice are all CX3CR1+MHCII− and at least partially YS-derived. After birth, the cMΦ compartment diversifies into four subpopulations defined by MHCII and CX3CR1 expression. Our results show that this diversification is due both to differentiation of persisting embryo-derived CX3CR1+MHCII− cMΦ and infiltrating monocyte-derived macrophages, which both can contribute to all four cMΦ subpopulations. However, monocytes preferentially give rise to MHCII+ cMΦ, whereas embryo-derived or radio-resistant cMΦ had a higher proportion of MHCII− cells, explaining the relative increase of MHCII+ cMΦ and the reduction of the CX3CR1+MHCII− cMΦ subpopulation with age. The dynamic change of the cMΦ subpopulation distribution therefore appears to reflect the gradual replacement of embryo-derived cMΦ by monocyte-derived macrophages. The increasing contribution of monocyte-derived macrophages to the cMΦ pool with age may be explained by the decreasing self-renewal of embryo-derived cMΦ. Although CX3CR1+MHCII− cMΦ that contain the majority of embryo-derived macrophages have a higher proliferative rate than the other subpopulations, overall cMΦ self-renewal is decreasing with age and is insufficient to sustain the cMΦ pool without input from infiltrating monocytes. It will be interesting to determine whether the loss of self-renewal capacity in cMΦ is permanent or might be reactivated. The observed mechanism of cMΦ homeostasis differs from other macrophage populations such as microglia, Langerhans cells, or alveolar macrophages, which in the absence of tissue damage or inflammation appear to self-maintain in tissues without replacement from circulating cells. It is also distinct from the high turnover rate of dermal tissue macrophages or intestinal lamina propria macrophages that are constantly replenished from circulating monocytes (Zigmond et al., 2012; Tamoutounour et al., 2013). In contrast to what was proposed elsewhere (Epelman et al., 2014), our results also show that inflammation or injury are not necessarily required for replacement of embryo-derived cardiac tissue macrophages by monocytes. It will be interesting to determine whether dynamic age-dependent changes in resident tissue macrophage populations also occur in other tissues and whether they may be important for the differences in repair mechanisms and regenerative capacity observed between young and old individuals.
MATERIALS AND METHODS
Mice.
CD45.1 and CD45.2 C57BL/6 mice were obtained from Charles River. All transgenic mice used in this study have a C57BL/6 background (>7 backcrosses). CX3CR1GFP/+ mice (Jung et al., 2000), CCR2−/−, Rosa-26-yfp, CX3CR1cre (JAX Stock No. 25524 B6.C-Cx3cr1<tm1.1(cre)Jung>/J), and CX3CR1creER mice (JAX Stock No. 20940 B6J.129-Cx3cr1<tm1.1(cre/ERT2)Jung>/J; Yona et al., 2013), Ubow mice (Ghigo et al., 2013), Rag2−/−γc−/−KitW/Wv mice (Waskow et al., 2009), and panYFP mice (Luche et al., 2013) were described elsewhere. Experiments were performed on 6–10-wk-old mice, if not stated otherwise. Experiments involving CX3CR1cre and CX3CR1creER mice included littermate controls. For other experiments C57BL/6 mice served as controls. All mouse experiments were performed under specific pathogen-free conditions. Animals were handled according to protocols approved by the Weizmann Institute Animal Care Committee (Israel), the Federal Ministry for Nature, Environment and Consumers’ Protection of the state of Baden-Württemberg (Germany) or the animal ethics committee of Marseille (France) and were performed in accordance to the respective international, national, and institutional regulations.
Parabiosis.
9-wk-old C57BL/6 CD45.1 mice were sutured together with 9-wk-old B6 CCR2−/− CD45.2 and subsequently kept under Bactrim for 10 wk before analysis.
BM chimeras.
7–10-wk-old host animals were lethally irradiated and were reconstituted with donor BM by i.v. injection of minimum 106 BM cells. Donor BM was isolated from femurs and tibias of donor mice, filtered through a 70-µm mesh and resuspended in PBS for i.v. injection. Rag2−/−γc−/−KitW/Wv mice were reconstituted with long-term (LT)–HSC from panYFP BM without prior conditioning as previously described (Waskow et al., 2009). LT-HSCs were defined as lineage-negative (B220−, CD3e−, CD4−, CD8a−, CD11b−, CD19−, Gr1−, Ter119−, NK1.1−), Kit+, Sca1+, CD48−, and CD150+.
Tamoxifen treatment.
Tamoxifen (TAM; Sigma-Aldrich) was dissolved in 100% ethanol to get a 1 g/ml solution and was 10-fold diluted in corn oil (Sigma-Aldrich) to get the 100 mg/ml final solution for oral or i.p. administration. To induce Cre-mediated gene recombination in 5- to 7-wk-old CX3CR1creER mice, 5 mg TAM was administrated orally for 5 consecutive days (25 mg total). For Cre induction in the embryo, 200 µl of 20 mg/ml TAM and 10 mg/ml Progesterone dissolved in corn oil was injected i.p. into pregnant females at 9 or 13 days post coitum (dpc).
BrdU pulsing.
BrdU was purchased from Sigma-Aldrich. Mice were injected with 0.1 mg BrdU/g body weight. Newborn mice were injected subcutaneously, older mice intraperitoneally. cMΦ isolated from BrdU injected mice were analyzed 4 h after injection for BrdU incorporation using the BrdU Flow kit (BD).
Flow cytometry.
Cells were acquired on FACSCanto, LSRII, and LSRFortessa systems (BD) and analyzed with FlowJo software (Tree Star). The following antibodies were used for staining cells: anti-CD11b (clone M1/70; eBioscience), anti-CD11c (clone N418; eBioscience), anti-CD14 (clone Sa2-8; eBioscience), anti-F4/80 (clone BM8; eBioscience), anti-MHCII (clone M5/114.15.2; eBioscience), anti-CD45.1 (clone A20; eBioscience), anti-CD45.2 (clone 104; eBioscience), anti-B220 (clone RA3-6B2; eBioscience), anti-CD115 (clone AFS98; eBioscience), anti-CD117 (clone 2B8; eBioscience), anti-Sca1 (clone D7; eBioscience), anti-CD48 (clone JM48-1; eBioscience), anti-Gr1 (clone RB6-8C5; BioLegend), anti-CD64 (clone X54-5/7.1; BioLegend), anti-Ly6G (clone 1A8; BioLegend), anti-CD150 (clone TC15-12F12.2; BioLegend), anti-Ly6C (clone AL21; BD), anti-Ki67 (clone B56; BD), anti-CD4 (clone H129.19; BD), anti-CD8 (clone 53–6.7; BD), anti-CD19 (clone ID3; BD), Ter119 (clone Ter119; BD), NK1.1 (clone PK136; BD), anti-F4/80, anti-MerTK (clone BAF591; R&D Systems), anti-BrdU (clone MoBU-1; Life Technologies). Before staining, cells were preincubated with Fc receptor blocking antibody (clone 2.4 G2; BD). LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Life Technologies) was used to stain dead cells. Absolute cell numbers were determined by using Flow-Count Fluorosperes (Beckman Coulter) according to the manufacturer’s instructions.
Preparation of tissue samples.
Peripheral blood leukocytes were isolated by density separation using Lympholite-M solution (Cedarlane) according to manufacturer’s instructions. For preparing single cell suspension from heart and brain, the mice were perfused with PBS before collecting the tissue. For cMΦ, the heart was macerated and incubated with 1 mg/ml collagenase-2 (Worthington Biochemical Corporation) and 0.15 mg/ml DNase1 (Sigma-Aldrich) at 37°C for 30 min during constant agitation. Resulting cell suspension was filtered through a 70-µm mesh and erythrocytes were removed by ACK lysis. For microglia isolation a gradient of 70, 37, 30% Percoll was used. The brain tissue was homogenized and incubated with HBSS solution containing 2% BSA (Sigma-Aldrich), 1 mg/ml collagenase d (Roche), and 0.15 mg/ml DNase1, filtered through a 70-µm mesh, and resuspended in 70% Percoll, before density centrifugation (800 g for 30 min at 20°C with low acceleration and no brake).
Online supplemental material.
Fig. S1 depicts the full gating strategy and population characterization of cMΦ. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140639/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Sandrine Sarrazin for critical reading of the manuscript and help with statistical analysis, L. Razafindramanana for animal handling, and M. Barad, A. Zouine, and S. Bigot for cytometry support.
K. Molawi was supported by an HFSP long-term fellowship. The work was supported by grants to M.H. Sieweke from Aviesan ITMO IHP exploratory Project A12194AS and CNRS PICS program No. 5730. M.H. Sieweke is a Fondation pour la Recherche Médicale (DEQ. 20071210559 and DEQ. 20110421320) and INSERM-Helmholtz group leader. The work was further supported by the Israel Science Foundation (ISF), the Deutsche Forschungsgemeinschaft (DFG) Research Unit (FOR) 1336 (S. Jung and M. Prinz), and DFG-SFB 938-project L (H.-R. Rodewald and K. Klapproth).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- cMΦ
- cardiac macrophages
- HSC
- hematopoietic stem cell
- YS
- yolk sac
References
- Ajami, B., Bennett J.L., Krieger C., Tetzlaff W., and Rossi F.M.V.. 2007. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10:1538–1543. 10.1038/nn2014 [DOI] [PubMed] [Google Scholar]
- Aurora, A.B., Porrello E.R., Tan W., Mahmoud A.I., Hill J.A., Bassel-Duby R., Sadek H.A., and Olson E.N.. 2014. Macrophages are required for neonatal heart regeneration. J. Clin. Invest. 124:1382–1392. 10.1172/JCI72181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, A., Soucie E., Sarrazin S., and Sieweke M.H.. 2009. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 326:867–871. 10.1126/science.1176056 [DOI] [PubMed] [Google Scholar]
- Chorro, L., Sarde A., Li M., Woollard K.J., Chambon P., Malissen B., Kissenpfennig A., Barbaroux J.-B., Groves R., and Geissmann F.. 2009. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J. Exp. Med. 206:3089–3100. 10.1084/jem.20091586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, L.C., Jenkins S.J., Allen J.E., and Taylor P.R.. 2013. Tissue-resident macrophages. Nat. Immunol. 14:986–995. 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman, S., Lavine K.J., Beaudin A.E., Sojka D.K., Carrero J.A., Calderon B., Brija T., Gautier E.L., Ivanov S., Satpathy A.T., et al. 2014. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 40:91–104. 10.1016/j.immuni.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. Immunological Genome Consortium. 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann, F., Manz M.G., Jung S., Sieweke M.H., Merad M., and Ley K.. 2010. Development of monocytes, macrophages, and dendritic cells. Science. 327:656–661. 10.1126/science.1178331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo, C., Mondor I., Jorquera A., Nowak J., Wienert S., Zahner S.P., Clausen B.E., Luche H., Malissen B., Klauschen F., and Bajénoff M.. 2013. Multicolor fate mapping of Langerhans cell homeostasis. J. Exp. Med. 210:1657–1664. 10.1084/jem.20130403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux, F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann, T., Wieghofer P., Müller P.F., Wolf Y., Varol D., Yona S., Brendecke S.M., Kierdorf K., Staszewski O., Datta M., et al. 2013. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 16:1618–1626. 10.1038/nn.3531 [DOI] [PubMed] [Google Scholar]
- Hashimoto, D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., et al. 2013. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 38:792–804. 10.1016/j.immuni.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel, G., Wang Y., Greter M., See P., Teo P., Malleret B., Leboeuf M., Low D., Oller G., Almeida F., et al. 2012. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 209:1167–1181. 10.1084/jem.20120340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, S.J., Ruckerl D., Cook P.C., Jones L.H., Finkelman F.D., van Rooijen N., MacDonald A.S., and Allen J.E.. 2011. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 332:1284–1288. 10.1126/science.1204351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf, K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E.G., Wieghofer P., Heinrich A., Riemke P., Hölscher C., et al. 2013. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16:273–280. 10.1038/nn.3318 [DOI] [PubMed] [Google Scholar]
- Luche, H., Nageswara Rao T., Kumar S., Tasdogan A., Beckel F., Blum C., Martins V.C., Rodewald H.-R., and Fehling H.J.. 2013. In vivo fate mapping identifies pre-TCRα expression as an intra- and extrathymic, but not prethymic, marker of T lymphopoiesis. J. Exp. Med. 210:699–714. 10.1084/jem.20122609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf, M., Swirski F.K., Aikawa E., Stangenberg L., Wurdinger T., Figueiredo J.L., Libby P., Weissleder R., and Pittet M.J.. 2007. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J. Exp. Med. 204:3037–3047. 10.1084/jem.20070885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, A.R., Paolicelli R., Salimova E., Gospocic J., Slonimsky E., Bilbao-Cortes D., Godwin J.W., and Rosenthal N.A.. 2012. An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLoS ONE. 7:e36814. 10.1371/journal.pone.0036814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello, E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., and Sadek H.A.. 2011. Transient regenerative potential of the neonatal mouse heart. Science. 331:1078–1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E.W., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Serbina, N.V., and Pamer E.G.. 2006. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 7:311–317. 10.1038/ni1309 [DOI] [PubMed] [Google Scholar]
- Sieweke, M.H., and Allen J.E.. 2013. Beyond stem cells: self-renewal of differentiated macrophages. Science. 342:1242974. 10.1126/science.1242974 [DOI] [PubMed] [Google Scholar]
- Swirski, F.K., Nahrendorf M., Etzrodt M., Wildgruber M., Cortez-Retamozo V., Panizzi P., Figueiredo J.L., Kohler R.H., Chudnovskiy A., Waterman P., et al. 2009. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 325:612–616. 10.1126/science.1175202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoutounour, S., Guilliams M., Montanana Sanchis F., Liu H., Terhorst D., Malosse C., Pollet E., Ardouin L., Luche H., Sanchez C., et al. 2013. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 39:925–938. 10.1016/j.immuni.2013.10.004 [DOI] [PubMed] [Google Scholar]
- van Furth, R., and Cohn Z.A.. 1968. The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 128:415–435. 10.1084/jem.128.3.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskow, C., Madan V., Bartels S., Costa C., Blasig R., and Rodewald H.R.. 2009. Hematopoietic stem cell transplantation without irradiation. Nat. Methods. 6:267–269. 10.1038/nmeth.1309 [DOI] [PubMed] [Google Scholar]
- Yona, S., Kim K.-W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., Mack M., Shpigel N., Boneca I.G., Murphy K.M., et al. 2012. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 37:1076–1090. 10.1016/j.immuni.2012.08.026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.