Using Dicer-deficient CD4 T cells, Marcais et al. show that microRNAs regulate the expression of mTOR components that are needed to discriminate between activating and anergy-inducing stimuli.
Abstract
T cell receptor (TCR) signals can elicit full activation with acquisition of effector functions or a state of anergy. Here, we ask whether microRNAs affect the interpretation of TCR signaling. We find that Dicer-deficient CD4 T cells fail to correctly discriminate between activating and anergy-inducing stimuli and produce IL-2 in the absence of co-stimulation. Excess IL-2 production by Dicer-deficient CD4 T cells was sufficient to override anergy induction in WT T cells and to restore inducible Foxp3 expression in Il2-deficient CD4 T cells. Phosphorylation of Akt on S473 and of S6 ribosomal protein was increased and sustained in Dicer-deficient CD4 T cells, indicating elevated mTOR activity. The mTOR components Mtor and Rictor were posttranscriptionally deregulated, and the microRNAs Let-7 and miR-16 targeted the Mtor and Rictor mRNAs. Remarkably, returning Mtor and Rictor to normal levels by deleting one allele of Mtor and one allele of Rictor was sufficient to reduce Akt S473 phosphorylation and to reduce co-stimulation–independent IL-2 production in Dicer-deficient CD4 T cells. These results show that microRNAs regulate the expression of mTOR components in T cells, and that this regulation is critical for the modulation of mTOR activity. Hence, microRNAs contribute to the discrimination between T cell activation and anergy.
CD4 T cells can respond to TCR signaling with full activation and the acquisition of effector functions or with anergy, a state of unresponsiveness characterized by the inability to proliferate and display effector functions, including cytokine secretion in response to secondary stimulation (Schwartz, 2003). Two-signal models of T cell activation state that to elicit full T cell activation, TCR engagement must be accompanied by co-stimulation (Schwartz, 2003). Full T cell activation and induction of Il2 transcription is promoted by co-ligation of TCR and CD28 (Thompson et al., 1989; Linsley et al., 1991; Harding et al., 1992) through activation of phospholipase Cγ (PLCγ)-1, Ras, and protein kinase Cθ (PKCθ), activation of the MAPK, JNK, PI3K/Akt, and IκB kinase (IKK) pathways, mobilization of intracellular calcium, and activation of the transcription factors NFAT, AP-1, CREB, and NF-κB, resulting in Il2 transcription (Wells, 2009). TCR engagement in the absence of CD28 co-stimulation results in limited AP-1 and NF-κB activity, defective transactivation of the Il2 promoter, and induction of anergy (Schwartz, 2003). The early secretion of IL-2 is a key event that discriminates productive activation from anergy (Thompson et al., 1989; Linsley et al., 1991; Harding et al., 1992). IL-2 is necessary (DeSilva et al., 1991) and sufficient (Zheng et al., 2007) to avoid anergy in response to TCR engagement through signaling pathways that include PI3K and mTOR (Powell and Delgoffe, 2010; Liou and Smith, 2011), a PI3K-related Ser/Thr kinase that integrates signals from several pathways including TCR signaling and cellular metabolism (Wells, 2009; Powell and Delgoffe, 2010; Araki et al., 2011).
Anergy-inducing stimuli may act in part by inducing the degradation of signaling molecules (Heissmeyer et al., 2004), and evidence that the activation versus anergy decision is affected by the abundance of signaling components comes from the involvement in this process of E3 ubiquitin ligases, enzymes that mediate the proteolytic turnover of signaling molecules: Cbl-b, Itch, and GRAIL are up-regulated in T cells under anergizing stimuli and required for anergy induction (Paolino and Penninger, 2010). Similarly, caspase 3 promotes anergy by degrading GADS (Grb2-related adaptor of downstream of Shc) and Vav (Puga et al., 2008). Hence, several negative regulators contribute to activation versus anergy discrimination by accelerating the turnover of signaling molecules downstream of the TCR. In addition to their turnover, the abundance of signaling components is determined by the transcriptional and posttranscriptional regulation of their production. microRNAs regulate gene expression at the posttranscriptional level through mRNA stability and translation (Selbach et al., 2008). microRNAs control multiple aspects of T cell differentiation and activation, from initial signaling events (Li et al., 2007) to the acquisition of effector functions and cytokine production (Muljo et al., 2005; Steiner et al., 2011), the resolution of T cell responses (Zhang and Bevan, 2010; Yang et al., 2012) and the choice of T cell fates including T helper cell lineage (Muljo et al., 2005; Steiner et al., 2011; Baumjohann et al., 2013; Kang et al., 2013; Khan et al., 2013), the formation of memory cells (Khan et al., 2013), and regulatory T cell differentiation (Cobb et al., 2006; Liston et al., 2008; Zhou et al., 2008; Lu et al., 2010).
Because microRNAs can tune gene expression rather than switching expression on or off, they may preferentially affect signaling pathways that are sensitive to the dosage of their components (Inui et al., 2010). In line with this idea, microRNA miR-181a promotes TCR sensitivity in developing thymocytes by targeting phosphatases that counteract TCR signaling (Li et al., 2007). The microRNA effector Ago2 is degraded in response to sustained T cell activation (Bronevetsky et al., 2013) and many 3′ UTRs are shortened in activated T cells (Sandberg et al., 2008), suggesting that successful T cell activation may temporarily remove a layer of microRNA control. Conversely, T cell activation is impaired when microRNA expression is increased by the deletion of the RNase Eri1 (Thomas et al., 2012).
To gain insight into the role of posttranscriptional regulation of signaling components we have examined CD4 T cell activation in the absence of the RNseIII enzyme Dicer, which is a key component of the microRNA biogenesis pathway (Cobb et al., 2005, 2006; Muljo et al., 2005). We found that Dicer-deficient CD4 T cells had an intrinsically low threshold for TCR signals and were competent to produce IL-2 in the absence of co-stimulatory signals. Co-stimulation–independent IL-2 production perpetuated T cell activation signals in an autocrine and paracrine fashion, abrogated the discrimination between activating and anergizing signals, and enabled T cell proliferation in response to TCR signals alone.
mRNA and protein expression profiling and analysis of TCR-induced signaling showed elevated phosphorylation of Akt S473 and S6 ribosomal protein, indicating hyperactivity of the mechanistic target of rapamycin (mTOR) complexes mTORC1 (which phosphorylates S6) and mTORC2 (which phosphorylates Akt on S473; Sarbassov et al., 2006) and overexpression of the mTOR complex components Mtor and Rictor. Mtor and Rictor transcripts were up-regulated at the posttranscriptional level, and targeted by microRNAs, including miR-16 and Let-7. Remarkably, we found that the key signaling abnormalities of microRNA-deficient CD4 T cells, namely increased Akt S473 phosphorylation and co-stimulation–independent Il2 transcription were dependent on elevated Mtor and Rictor expression, and could be reversed by returning Mtor and Rictor to WT levels.
RESULTS
Dicer-deficient CD4 T cells show heightened sensitivity to TCR signaling, IL-2 production, and proliferation in the absence of co-stimulation
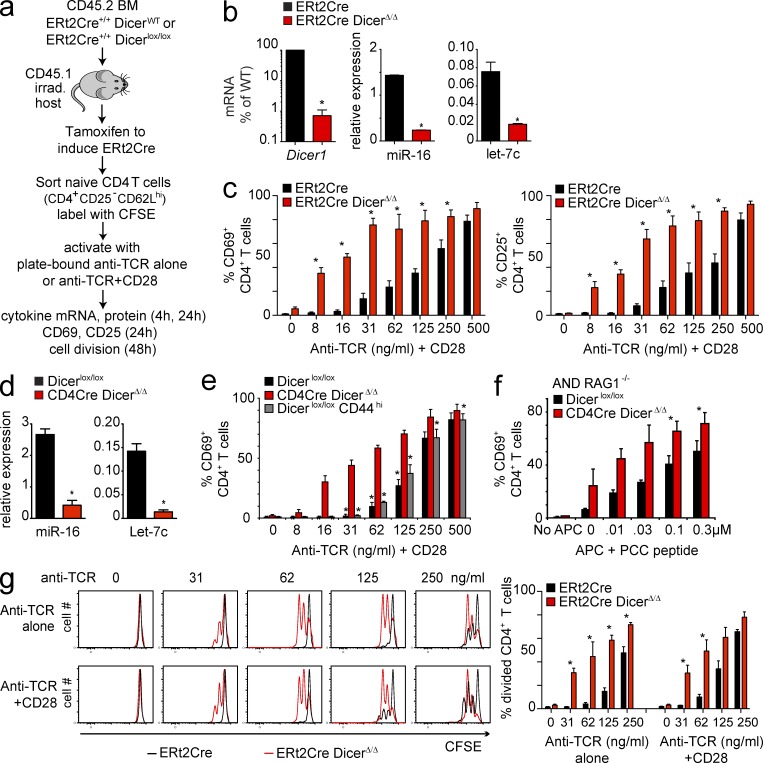
To deplete microRNAs from peripheral T cells, we reconstituted irradiated (CD45.1) hosts with allotypically marked CD45.2 ERt2Cre Dicerwt/wt or ERt2Cre Dicerlox/lox BM, and administered tamoxifen to fully reconstituted chimeras (Fig. 1 a). Naive CD25− CD62Lhi CD45.2+ CD4 T cells isolated from these mice showed near-complete loss of Dicer1 mRNA and a substantial reduction in the expression of Dicer-dependent microRNAs, including miR-16 and Let-7c (Fig. 1 b). Naive ERt2Cre Dicerwt/wt and ERt2Cre DicerΔ/Δ CD25− CD62Lhi CD4 T cells were activated with graded concentrations of plate-bound TCR antibody (H57) alone or in the presence of anti-CD28 as a source of co-stimulation. The percentage of cells expressing CD69 and CD25 was determined by flow cytometry 24 h later. In the presence of co-stimulation, both control (black bars) and Dicer-deficient CD4 T cells (red bars) initiated the expression of the activation markers CD69 (Fig. 1 c, left) and CD25 (Fig. 1 c, right). At limiting concentrations of TCR antibody, Dicer-deficient CD4 T cells were markedly more sensitive to TCR signals than control CD4 T cells, as indicated by a higher percentage of cells that expressed CD69 (Fig. 1 c, left) and CD25 (Fig. 1 c, right).
Figure 1.
microRNA deficiency confers increased sensitivity to TCR signals. (a) Irradiated CD45.1 hosts were reconstituted with CD45.2 ERt2Cre Dicerwt/wt or ERt2Cre Dicerlox/lox BM. After 8–12 wk, to allow for complete reconstitution, chimeras were given 1 mg tamoxifen i.p. on 5 consecutive days and naive CD45.2+ CD4 T cells were isolated 3 wk later. (b) Expression of microRNAs in peripheral CD4 T cells with ERt2Cre-mediated deletion of Dicer. Tamoxifen-induced deletion of Dicer1 mRNA in CD4 T cells from ERt2Cre Dicerlox/lox chimeras was around 99% as determined by quantitative RT-PCR (mean ± SE; n = 4; normalized to Hprt1; *, P < 0.05), and the expression of Dicer-dependent microRNAs, including miR-16 and Let-7c (mean ± SE; n = 4; normalized to sno-135; *, P < 0.05). (c) Naive ERt2Cre Dicerwt/wt (black bars) and ERt2Cre DicerΔ/Δ (red bars) CD25− CD62Lhi CD4 T cells were isolated and activated with the indicated concentrations of plate-bound anti-TCR+CD28. The percentage of cells expressing CD69 (left) or CD25 (right) was determined by flow cytometry 24 h later (mean ± SE; n = 4; *, P < 0.05). (d) Expression of Dicer-dependent microRNAs, including miR-16 and Let-7c, after CD4Cre-mediated deletion of Dicer (mean ± SE; n = 4; normalized to sno-135; *, P < 0.05). (e) CD4Cre DicerΔ/Δ and Dicerlox/lox CD25− CD62Lhi or CD44hi CD4 T cells were isolated by cell sorting and activated with the indicated concentrations of plate-bound anti-TCR+CD28. The percentage of cells expressing CD69 was determined by flow cytometry 24 h later (mean ± SE; n = 3). Dicer-deficient CD4 T cells (red bars) were compared with naive (black bars) or CD44hi (gray bars) control CD4 T cells (*, P < 0.05). (f) Sensitivity of Dicer-deficient CD4 T cells to cognate peptide-MHC ligands. RAG1−/−, AND TCR transgenic CD4Cre Dicerwt/wt or Dicerlox/lox CD4 T cells were cultured with BM-derived dendritic cells loaded with the indicated concentrations of cognate peptide and the percentage of cells expressing CD69 was determined (mean ± SE; n = 3). (g) Left: ERt2Cre Dicerwt/wt and DicerΔ/Δ CD4+ CD25− CD62Lhi or cells were labeled with CFSE and activated with the indicated concentrations of plate-bound anti-TCR alone (top) or anti-TCR+CD28 (bottom). Cell division was assessed 48 h later by CFSE dilution. One representative experiment of 3 is shown. Right: mean ± SE of 3 independent biological replicates.
We used a developmentally regulated CD4Cre transgene as an additional approach to delete the Dicer locus. Although active from the double-positive stage of thymocyte development, CD4Cre only depletes microRNA from mature single-positive thymocytes and peripheral T cells (Muljo et al., 2005; Cobb et al., 2006; Fig. 1 d). We sorted naive CD25− CD62Lhi CD4 T cells Dicerlox/lox and CD4Cre DicerΔ/Δ cells. We also isolated Dicerlox/lox CD44hi CD4 T cells for comparison because CD4Cre DicerΔ/Δ CD4 T cells express slightly elevated levels of the microRNA target and activation marker CD44. At limiting concentrations of anti-TCR, a significantly higher percentage of Dicer-deficient CD4 T cells expressed CD69 (Fig. 1 e) and CD25 (not depicted) than either naive or CD44hi control CD4 T cells. Experiments comparing CD4Cre Dicerwt/wt versus CD4Cre Dicerlox/lox instead of Dicerlox/lox versus CD4Cre Dicerlox/lox CD4 T cells gave indistinguishable results (unpublished data).
To test the sensitivity of Dicer-deficient CD4 T cells to peptide–MHC complexes on antigen-presenting cells (APCs) we generated mice harboring conditional Dicer alleles, CD4Cre and the AND TCR transgene (Kaye et al., 1989) on a Rag-deficient background. BM-derived APC pulsed with graded concentrations of cognate peptide elicited CD69 expression by significantly higher percentages of Dicer-deficient than control CD4 T cells (Fig. 1 f). This result confirmed heightened sensitivity of Dicer-deficient CD4 T cells to physiological TCR ligands.
Compared with control CD4 T cells, a greater percentage of ERt2Cre DicerΔ/Δ CD4 T cells entered the cell cycle and completed cell division in response to limiting concentrations of anti-TCR in combination with CD28 as judged by the dilution of CFSE. In marked contrast to control CD4 T cells, highly purified Dicer-deficient CD4 T cells proliferated in response to plate-bound anti-TCR in the absence of co-stimulation (Fig. 1 g). ERt2Cre DicerΔ/Δ CD4 T cells proliferated equally well in the absence and in the presence of anti-CD28, whereas control CD4 T cells depended on co-stimulation for efficient proliferation (Fig. 1 g). Co-stimulation–independent proliferation of Dicer-deficient CD4 T cells was confirmed in CD4Cre DicerΔ/Δ CD4 T cells (unpublished data).
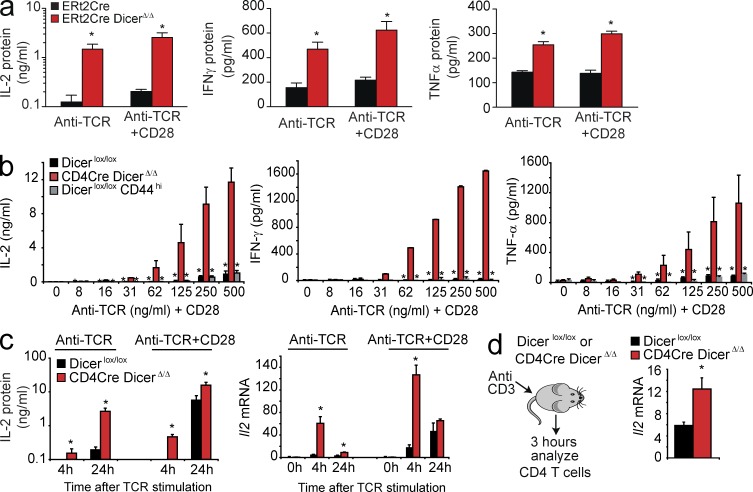
ERt2Cre DicerΔ/Δ CD4 T cells produced strikingly higher amounts of IL-2 than control ERt2Cre Dicerwt/wt CD4 T cells when they were stimulated with anti-TCR alone or limiting concentrations of anti-TCR+CD28 (Fig. 2 a, left). In addition to IL-2, Dicer-deficient CD4 T cells also produced significantly higher amounts of IFN-γ (Fig. 2 a, center) and TNF (Fig. 2 a, right). To examine cytokine production by Dicer-deficient CD4 T cells in a different experimental system we used CD4Cre-mediated deletion of Dicer (Muljo et al., 2005; Cobb et al., 2006). Over a wide range of activation conditions, CD4Cre DicerΔ/Δ CD25− CD62Lhi CD4 T cells produced significantly more IL-2 (Fig. 2 b, left), IFN-γ (Fig. 2 b, center), and TNF (Fig. 2 b, right) than naive Dicerlox/lox CD4 T cells or CD44hi Dicerlox/lox CD4 T cells. Furthermore, CD4Cre DicerΔ/Δ CD4 T cells transcribed substantial amounts of Il2 mRNA when activated by anti-TCR in the absence of co-stimulation (Fig. 2 c, left) and secreted IL-2 protein at earlier times than control Dicerlox/lox cells (Fig. 2 c, left). Dicer-deficient CD4 T cells activated transcription of the Il2 gene faster (peak at 4 h) than control cells (peak at 24 h) and in the absence of co-stimulation (Fig. 2 c, right). Experiments comparing CD4Cre Dicerwt/wt versus CD4Cre Dicerlox/lox instead of Dicerlox/lox versus CD4Cre Dicerlox/lox CD4 T cells gave indistinguishable results (unpublished data). In vivo activation of Dicer-deficient CD4 T cells also induced significantly higher levels of Il2 mRNA (Fig. 2 d). Hence, Dicer-deficient CD4 T cells showed heightened sensitivity to TCR signaling and did not require co-stimulatory signals for IL-2 production or proliferation.
Figure 2.
Dicer-deficient CD4 T cells produce IL-2 in the absence of co-stimulation. (a) Naive ERt2Cre Dicerwt/wt and ERt2Cre Dicerlox/lox CD4+ CD25− T cells were stimulated with 500 ng/ml plate-bound anti-TCR alone or anti-TCR+CD28. Production IL-2 (left), IFN-γ (middle), and TNF (right) was assessed 48 h later by ELISA (mean ± SE; n = 3). (b) Naive Dicerlox/lox or CD4Cre Dicerlox/lox CD4 T cells were activated with the indicated concentrations of plate-bound anti-TCR+CD28. Production IL-2 (left), IFN-γ (middle), and TNF (right) was assessed 48 h later by ELISA. CD44hi Dicerlox/lox CD4 T cells were included as a control (mean ± SE, n = 3). (c) Naive Dicerlox/lox or CD4Cre Dicerlox/lox CD4 T cells were activated with plate-bound anti-TCR alone or anti-TCR+CD28. Production IL-2 was assessed 48 h later by ELISA (left; mean ± SE; n = 3). The expression of Il2 mRNA was assessed by quantitative RT-PCR at 0, 4, and 24 h (right, mean ± SE; n = 5, normalized to Hprt1). (d) Dicerlox/lox and CD4Cre Dicerlox/lox mice were injected with anti-CD3 (2C11, 10 µg). 3 h later, CD4+ CD25− T cells were purified and Il2 mRNA was quantified by qRT-PCR (mean ± SE; n = 6 mice in 3 independent experiments).
Dicer-deficient CD4 T cells are resistant to the induction of anergy by TCR signaling in the absence of co-stimulation
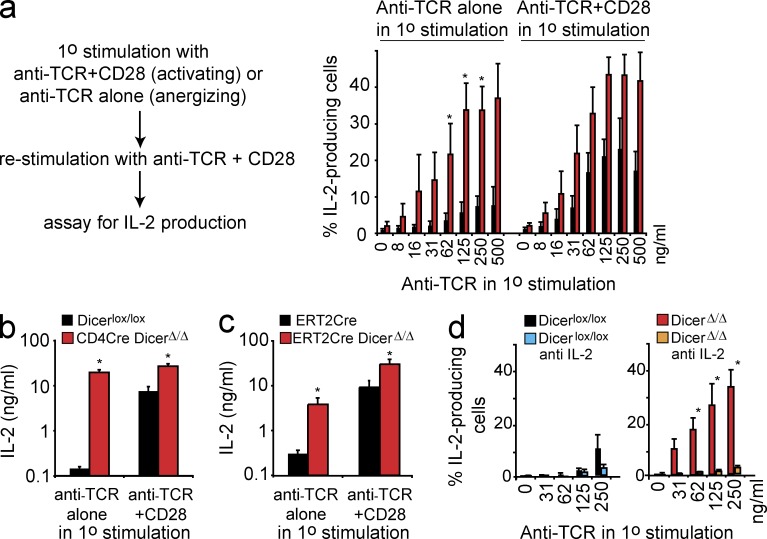
We next addressed the impact of TCR signals in the presence or absence of co-stimulation on the ability of CD4 T cells to produce IL-2 upon restimulation. After primary stimulation with anti-TCR+CD28, CD4 T cells readily produced IL-2 upon restimulation (Fig. 3 a) and a substantial amount of IL-2 was secreted (Fig. 3, b and c). Consistent with the operational definition of anergy (Schwartz, 2003), IL-2 production occurred in a reduced percentage of control CD4 T cells that had previously been exposed to TCR signals in the absence of co-stimulation (Fig. 3 a), and IL-2 secretion was much lower (Fig. 3, b and c). In contrast, CD4Cre DicerΔ/Δ (Fig. 3, a and b) and ERt2Cre DicerΔ/Δ CD4 T cells (Fig. 3 c) produced IL-2 upon restimulation regardless of whether the primary stimulus consisted of anti-TCR+CD28 or anti-TCR alone. Dicer-deficient CD4 T cells exposed to activation by anti-TCR alone remained competent to produce not only IL-2, but also IFN-γ and TNF upon restimulation (unpublished data).
Figure 3.
Dicer-deficient CD4 T cells use IL-2 to resist anergy induced by TCR signals in the absence of co-stimulation. (a) CD4Cre DicerΔ/Δ and Dicerlox/lox CD4+ CD25− T cells were subjected to primary stimulation with plate-bound anti-TCR alone or anti-TCR+CD28. The cells were restimulated with anti-TCR+CD28 and IL-2 production was determined 5h later by intracellular staining and flow cytometry (mean ± SE, n = 3 independent experiments). (b) CD4Cre DicerΔ/Δ and Dicerlox/lox CD4+ CD25− T cells were subjected to primary stimulation with plate-bound anti-TCR alone or anti-TCR+CD28. The cells were restimulated with anti-TCR+CD28 and IL-2 production was assessed 48 h later by ELISA (mean ± SE, n = 3 independent experiments). (c) ERt2Cre Dicerwt/wt and ERt2Cre DicerΔ/Δ CD4+ CD25− T cells were subjected to primary stimulation with plate-bound anti-TCR alone or anti-TCR+CD28. The cells were restimulated with anti-TCR+CD28 and IL-2 production was assessed 48 h later by ELISA (mean ± SE, n = 4 independent experiments). (d) CD4Cre DicerΔ/Δ and Dicerlox/lox CD4+ CD25− T cells were stimulated with graded concentrations of plate-bound anti-TCR alone in the presence or the absence of neutralizing anti–IL-2. Cells were restimulated with anti-TCR+CD28 and IL-2 production was determined 5 h later (mean ± SE; n = 2 independent experiments).
Anergy resistance of Dicer-deficient CD4 T cells is mediated by IL-2
IL-2 receptor signals are required for CD4 T cells to avoid anergy (Wells, 2009). We therefore asked whether resistance of Dicer-deficient CD4 T cells to anergy relied on IL-2 production. When Dicer-deficient CD4 T cells were stimulated with anti-TCR in the presence of a neutralization antibody to IL-2, subsequent restimulation failed to elicit substantial IL-2 production, indicating that anergy had been induced (Fig. 3 d). We conclude that Dicer-deficient CD4 T cells escape from anergy by producing IL-2 independently of co-stimulatory signals.
Signaling events in Dicer-deficient CD4 T cells indicate a role for deregulated expression of mTOR components
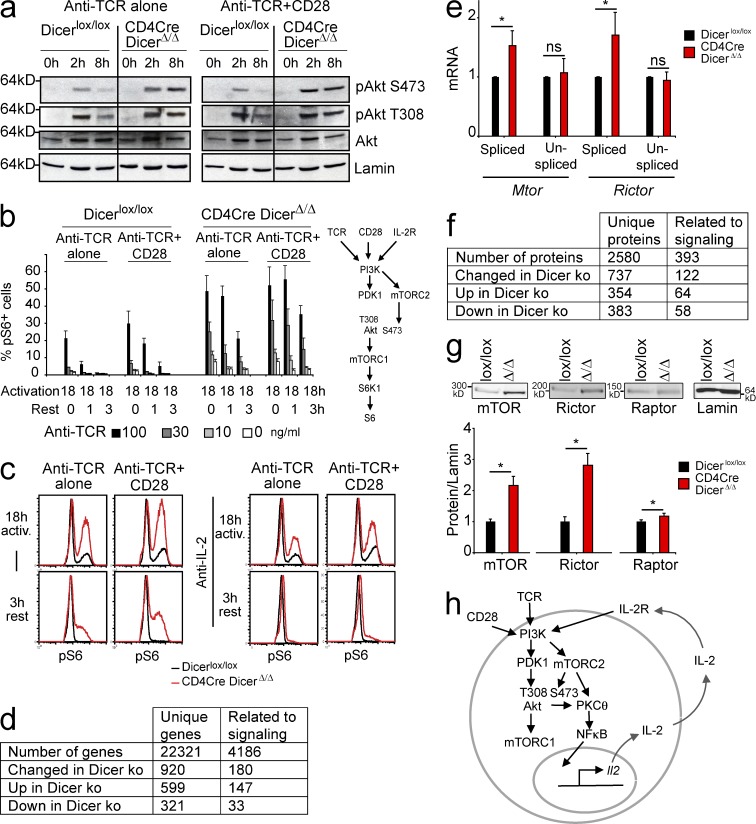
To examine the mechanisms that enable Dicer-deficient CD4 T to produce IL-2 in the absence of co-stimulation we investigated signaling events downstream of the TCR. Time course analysis of tyrosine phosphorylation 2, 5, and 15 min after TCR ligation revealed no abnormalities (unpublished data). Ca2+ fluxes appeared slightly blunted in Dicer-deficient CD4 T cells, but the subsequent dephosphorylation of NFATc1 and NFATc2 was not substantially reduced or delayed (unpublished data). The TCR-induced phosphorylation of ERK1 and ERK2 appeared unchanged (unpublished data). Notably, however, the phosphorylation of Akt S473 was increased in Dicer-deficient CD4 T cells compared with control CD4 T cells after 2 h of activation with anti-TCR alone (Fig. 4 a, left) or with anti-TCR+CD28 (Fig. 4 a, right). pAkt S473 persisted for extended periods in Dicer-deficient CD4 T cells (Fig. 4 a). In contrast, pAkt T308 appeared to be unaffected (Fig. 4 a). TCR-induced PI3K activation results in Akt phosphorylation of threonine 308 (pAkt T308) by PDK1, whereas serine 473 (pS473) is phosphorylated by the mTORC2 complex (Sarbassov et al., 2006). The elevated and prolonged phosphorylation of Akt on S473 therefore indicates increased and persistent activation of mTORC2 in Dicer-deficient CD4 T cells.
Figure 4.
Posttranscriptional deregulation of the mTOR components Mtor and Rictor and increased mTOR activity in Dicer-deficient CD4 T cells. (a) CD4Cre DicerΔ/Δ and Dicerlox/lox CD4+ CD25− T cells were stimulated in the absence (left) or in the presence (right) of anti-CD28 and immunoblotted for pAkt S473, pAkt T308, total Akt and LaminB as a loading control (one representative experiment of 3). (b) CD4Cre DicerΔ/Δ and Dicerlox/lox CD4+ CD25− T cells were stimulated with anti-TCR in the presence or in the absence of anti-CD28. After 18 h of activation, the percentage of pS6+ cells was analyzed either immediately or after resting the cells for 1 or 3 h in the absence of anti-TCR (mean ± SE; n = 3 independent experiments). The inset shows pathways to Akt and S6 phosphorylation. (c) CD4Cre DicerΔ/Δ and Dicerlox/lox CD4+ CD25− T cells were stimulated for 18 h with anti-TCR (100 ng/ml) and the phosphorylation of S6 ribosomal protein (pS6) was determined by intracellular staining and flow cytometry (representative of 3 independent experiments). (d) Statistically significant mRNA expression changes in Dicer-deficient CD4 T cells affect signaling molecules (Storey’s q-value < 0.05). (e) RNA was subjected to quantitative RT-PCR using primers specific for spliced (mature) or unspliced (primary) Mtor and Rictor transcripts (mean ± SE; n = 5 independent experiments, normalized to Hprt1). (f) Quantitative proteomic analysis shows signaling-related protein expression changes in Dicer-deficient CD4 T cells. (g) CD4Cre DicerΔ/Δ and Dicerlox/lox CD4+ CD25− T cells were analyzed for mTOR, Rictor, and Raptor protein expression (normalized to Lamin B, mean ± SE; n = 5 independent experiments). (h) Dicer-deficient CD4 T cells respond to TCR signals by IL-2 production and proliferation in the absence of co-stimulation and escape the induction of anergy in an IL-2–dependent manner. IL-2 sustains T cell activation even after TCR signals are withdrawn.
Because Akt is an activator of mTORC1, we analyzed the phosphorylation of S6 ribosomal protein (pS6) by S6 kinase 1 (S6K1), which is a direct downstream target of mTORC1, as a proxy for mTORC1 activation (inset, Fig. 4). S6 was phosphorylated in a higher percentage of Dicer-deficient than control CD4 T cells in response to anti-TCR alone or anti-TCR+CD28 (Fig. 4 b). When CD4 T cells were activated for 18 h and subsequently removed from anti-TCR stimulation, pS6 persisted for longer in Dicer-deficient than in control CD4 T cells (Fig. 4 b). The addition of neutralizing IL-2 antibodies terminated pS6 upon removal of TCR signals, indicating that sustained mTORC1 activity in Dicer-deficient CD4 T cells was driven by IL-2 (Fig. 4 c). Hence, mTORC1 and mTORC2 activation was increased in Dicer-deficient CD4 T cells, and was maintained in response to co-stimulation–independent IL-2 production. Consistent with data that Akt can activate NF-κB via the degradation of IκB (Kane et al., 1999; Ozes et al., 1999), TCR stimulation resulted in reduced IκBα expression and increased nuclear cRel translocation in Dicer-deficient CD4 T cells (unpublished data and this study).
mRNA expression profiling showed that several signaling-related transcripts were deregulated in Dicer-deficient CD4 T cells (Fig. 4 d and Table S1). Transcripts encoding the mTOR subunits Mtor and Rictor were up-regulated (although not statistically significant after correction for multiple testing), and up-regulation was confirmed by quantitative RT-PCR analysis, which showed increased expression of mature (spliced) Mtor and Rictor transcripts in Dicer-deficient CD4 T cells (Fig. 4 e). Quantitative proteomics (Fig. 4 f) showed significantly increased Mtor and Rictor protein expression in Dicer-deficient CD4 T cells (2.2- and 2.6-fold, respectively; Table S2), which was confirmed by Western blotting (Fig. 4 g).
Interestingly, quantitative RT-PCR analysis showed that only spliced but not unspliced Mtor and Rictor transcripts were up-regulated in Dicer-deficient CD4 T cells (Fig. 4 e). This indicates that the elevated expression of Mtor and Rictor was not explained by increased transcription, but rather by the loss of post-transcriptional regulation in Dicer-deficient CD4 T cells.
Summarizing the data so far, we find that Dicer-deficient CD4 T cells respond to TCR engagement in the absence of co-stimulatory signals by producing IL-2, which protects them from the induction of anergy and sustains T cell activation even after TCR signals are withdrawn (Fig. 4 h). Expression profiling at the mRNA and protein levels showed overexpression of the mTOR complex components Mtor and Rictor, which occurred as a result of posttranscriptional, rather than transcriptional, deregulation. This was consistent with signaling abnormalities detected in Dicer-deficient T cells; the activity of mTORC1 and mTORC2 was elevated as reflected by the increased phosphorylation of S6 ribosomal protein and Akt S473, respectively.
Elevated expression of mTOR components contributes to co-stimulation–independent Il2 expression by microRNA-deficient CD4 T cells
Consistent with aberrant S6 and Akt S473 phosphorylation described above, the expression of the mTOR components Mtor and Rictor was deregulated in Dicer-deficient CD4 T cells. mTOR and Rictor deregulation occurred at the posttranscriptional level, suggesting a potential involvement of microRNAs. Both Mtor and Rictor are validated microRNA targets in cancer cells (Fornari et al., 2010; Nagaraja et al., 2010; Uesugi et al., 2011; Tsuruta et al., 2011) and Ago2 CLIP experiments had shown that the 3′ UTR sequences of Mtor and, in particular, Rictor mRNAs are extensively targeted by microRNA-containing RISC complexes in activated CD4 T cells (Loeb et al., 2012; Fig. 5, a and b, left). Based on this information and on data from heterologous reporter assays in 293T cells (Fig. 5, a and b, right) we devised a microRNA sensor to probe the impact of endogenous microRNAs on the Mtor and Rictor 3′UTR in CD4 T cells. This sensor contains a GFP reporter linked to the entire Mtor 3′UTR or a portion of Rictor 3′UTR (nt 3,000–4,200) and Cherry as an internal control for the normalization of GFP fluorescence. GFP-Mtor was de-repressed by the mutation of predicted binding sites for Let-7 and miR-16 (Fig. 5 c). GFP-Rictor 3′UTR expression was de-repressed by mutation of a conserved miR-16–binding site and by cotransfection locked nucleic acid (LNA) designed to inhibit miR-16, but not a control LNA (Fig. 5 d). Mtor and Rictor protein expression were reduced in Dicer-deficient CD4 T cells transfected with miR-16 and Let-7c microRNA mimics (Fig. 5 e) and increased in control CD4 T cells transfected with LNAs designed to antagonize miR-16 and the Let-7 family of microRNAs (Fig. 5 f). Given the length and extensive targeting of the Rictor 3′ UTR by Ago2 (Loeb et al., 2012), these experiments are unlikely to be exhaustive, but they support a role for endogenous microRNAs including miR-16 and Let-7 in modulating Mtor and Rictor expression in CD4 T cells.
Figure 5.
microRNA regulation of the mTOR components Mtor and Rictor. (a, left) Schematic of the Mtor 3′UTR with Ago2 CLIP results (Loeb et al., 2012), predicted microRNA-binding sites, and mutations introduced into predicted miR-16 and Let-7 sites (red, numbers indicate nucleotide positions). (right) 293T cells were cotransfected with firefly luciferase reporter constructs containing the Mtor 3′UTR, Renilla luciferase plasmids, and plasmids driving the expression of microRNAs predicted to target the Mtor 3′UTR (miRecords; Xiao et al., 2009). Firefly over Renilla luciferase ratios were determined (mean ± SE; n = 3 independent experiments with triplicate wells) and miR-195, miR-16, and Let-7c had statistically significant effects (P < 10−4; P < 0.001; and P < 0.01. (b) Schematic of the Rictor 3′UTR with predicted microRNA-binding sites, mutations introduced into a conserved Rictor miR-16 site (red), and Ago2 CLIP. 293T cells were transfected with firefly luciferase reporter constructs containing 1-kb sections of the Rictor 3′UTR along with control Renilla luciferase plasmids and plasmids driving the expression of the indicated microRNAs. Firefly over Renilla luciferase ratios were determined (mean ± SE; n = 3 independent experiments with triplicate wells) and showed statistically significant effects for miR-16 and Let-7c, the p-values are, respectively, for the 1,000–2,000 part: P < 0.05 and P < 0.01; for the 2,000–3,000 part: P < 0.01; and for the 3,000–4,200 part: P < 10−5. (c) CD4 T cells were transfected with GFP constructs containing the WT Mtor 3′UTR or Let-7 and/or miR-16–binding site mutants along with Cherry control constructs. The ratio of GFP and Cherry fluorescence was determined by flow cytometry (mean ± SE; n = 3 independent experiments). (d) CD4 T cells were transfected with GFP constructs containing WT or miR-16 binding site mutant of the Rictor 3′UTR (nucleotides 3,000–4,200) in the presence or absence of control or mIR-16 family directed LNA as indicated along with Cherry control constructs. The ratio of GFP and Cherry fluorescence was determined by flow cytometry (mean ± SE; n = 3 independent experiments). (e) Dicer-deficient (left) or control (right) CD4 T cells were transfected with the indicated microRNA mimics and, 30 h later, the expression of the mTOR components Mtor, Rictor, and Raptor was assessed by immunoblotting in comparison to Dicerlox/lox control CD4 T cells (mean ± SE, n = 3 independent experiments, one representative blot is shown). (f) Control (left) or Dicer-deficient (right) CD4 T cells were transfected with the indicated LNA oligonucleotides to antagonize endogenous microRNAs and, 30 h later, the expression of mTOR components was assessed by immunoblotting in comparison to CD4Cre DicerΔ/Δ CD4 T cells (mean ± SE; n = 4 independent experiments, one representative blot is shown).
Collectively, the results of our signaling studies, protein and mRNA expression data, and evidence that mTOR components are targeted by microRNAs in CD4 T cells, implicated Mtor and Rictor as potential mediators of aberrant responses to TCR signaling in microRNA-deficient CD4 T cells.
As a genetic approach to address the contribution of Mtor and Rictor overexpression, we examined Dicer-deficient CD4 T cells that lacked one copy of Mtor and Rictor. The expression of Mtor and Rictor was elevated in Dicer-deficient CD4 T cells, but expression levels were returned to near normal in CD4Cre Dicerlox/lox Mtorlox/wt Rictorlox/wt CD4 T cells (Fig. 6, a and b). Interestingly, heterozygosity in both Mtor and Rictor corrected the activation-induced phosphorylation of Akt S473 in Dicer-deficient CD4 T cells to near WT levels (Fig. 6 c). Remarkably, the co-stimulation–independent expression of Il2, Ifng, and Tnf mRNA in Dicer-deficient CD4 T cells and the overproduction of IL-2, IFN-γ, and TNF protein were at least partially corrected by returning Mtor and Rictor to near-normal levels (Fig. 6 d). Heterozygosity in Mtor and Rictor also reduced the heightened responsiveness of Dicer-deficient CD4 T cells to limiting concentrations of anti-TCR as assessed by the induction of the activation markers CD69 and CD25 (P = 0.0063 for CD69; P = 0.0053 for CD25; unpublished data). Hence, despite the deregulated expression of numerous signaling components in Dicer-deficient CD4 T cells, correcting the overexpression of Mtor and Rictor was sufficient to significantly redress the key signaling abnormalities in these cells.
Figure 6.
The deregulated expression of Mtor and Rictor contributes to increased Akt S473 phosphorylation and co-stimulation–independent IL-2 production by Dicer-deficient CD4 T cells. (a) Outline of rationale for generating Mtor and Rictor heterozygous CD4 T cells on a Dicer-deficient background. (b) Expression of mTOR, Rictor, and Raptor protein in Dicerlox/lox, CD4Cre DicerΔ/Δ, and CD4Cre DicerΔ/Δ RictorΔ/WT MtorΔ/WT CD4+ CD25− T cells (mean ± SE; n = 4 independent experiments). (c) Dicerlox/lox, CD4Cre DicerΔ/Δ, and CD4Cre DicerΔ/Δ RictorΔ/WT MtorΔ/WT CD4+ CD25− T cells were activated with plate-bound anti-TCR (500 ng/ml) in the absence (top) or in the presence (bottom) of anti-CD28 for 0, 2, or 8 h and immunoblotted for pAkt S473 and LaminB as a loading control (representative of three independent experiments). (d) Dicerlox/lox, CD4Cre DicerΔ/Δ, and CD4Cre DicerΔ/Δ RictorΔ/WT MtorΔ/WT CD4+ CD25− T cells were activated with anti-TCR (500 ng/ml). The expression of cytokine mRNA was assessed by quantitative RT-PCR (left; mean ± SE; n = 3, normalized to Hprt1) and cytokine secretion was determined by ELISA (right; mean ± SE; n = 3 independent experiments). (top) IL-2; (middle) IFN-γ; (bottom) TNF.
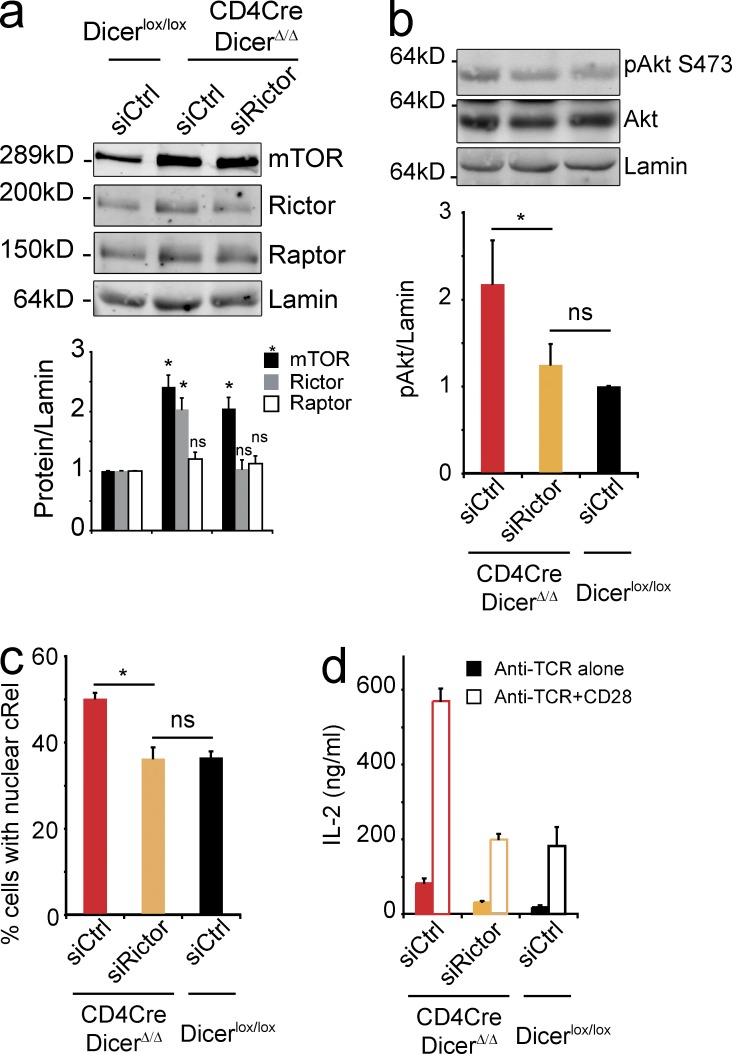
Although heterozygosity in either Mtor or Rictor alone was not sufficient to correct co-stimulation–independent IL-2 production (data not shown) we noticed that Raptor protein expression was elevated in CD4Cre Dicerlox/lox that were heterozygous for Rictor (Rictorlox/wt) but WT for Mtor (Mtorwt/wt; unpublished data). This compensatory up-regulation of Raptor was not seen in CD4Cre Dicerlox/lox Mtorwt/wt Rictorwt/wt CD4 T cells (Fig. 4 h), in CD4Cre Dicerlox/lox Mtorlox/wt Rictorlox/wt CD4 T cells (Fig. 6 b), or in mature Dicer-deficient CD4 T cells during acute siRNA-mediated knockdown of Rictor (Fig. 7 a). We titrated siRNA oligonucleotides so that Rictor protein expression in Dicer-deficient CD4 T cells was reduced to levels resembling control CD4 T cells (Fig. 7 a). This was sufficient to significantly reduce elevated Akt S473 phosphorylation (Fig. 7 b), NF-κB translocation (Fig. 7 c), and co-stimulation–independent IL-2 expression (Fig. 7 d) in Dicer-deficient CD4 T cells.
Figure 7.
Partial correction of Akt S473 phosphorylation, nuclear translocation of NF-κB, and co-stimulation–independent IL-2 production by transient knockdown of Rictor in Dicer-deficient CD4 T cells. (a) siRNA-mediated knockdown of Rictor in Dicer-deficient CD4 T cells affects Rictor but not Raptor expression (mean ± SE; n = 4 independent experiments). (b) Naive CD4Cre DicerΔ/Δ and control Dicerlox/lox CD4 T cells were activated with anti-TCR for 2 h and analyzed by immunoblotting with antibodies to Akt, pAkt Ser473; and Lamin B for normalization (mean ± SE; n = 3 independent experiments; *, P < 0.05). (c) Naive CD4Cre DicerΔ/Δ and control Dicerlox/lox CD4 T cells were activated with anti-TCR for 3 h and the nuclear translocation cRel was determined (ImageStream similarity scores > 0.5; mean ± SE; n = 3 independent experiments). (d) Naive CD4Cre DicerΔ/Δ and control Dicerlox/lox CD4 T cells treated as in a) were activated with anti-TCR or anti-TCR+CD28 and secreted IL-2 was quantified by ELISA after 18 h (mean ± SE; n = 3 independent experiments).
Biological significance
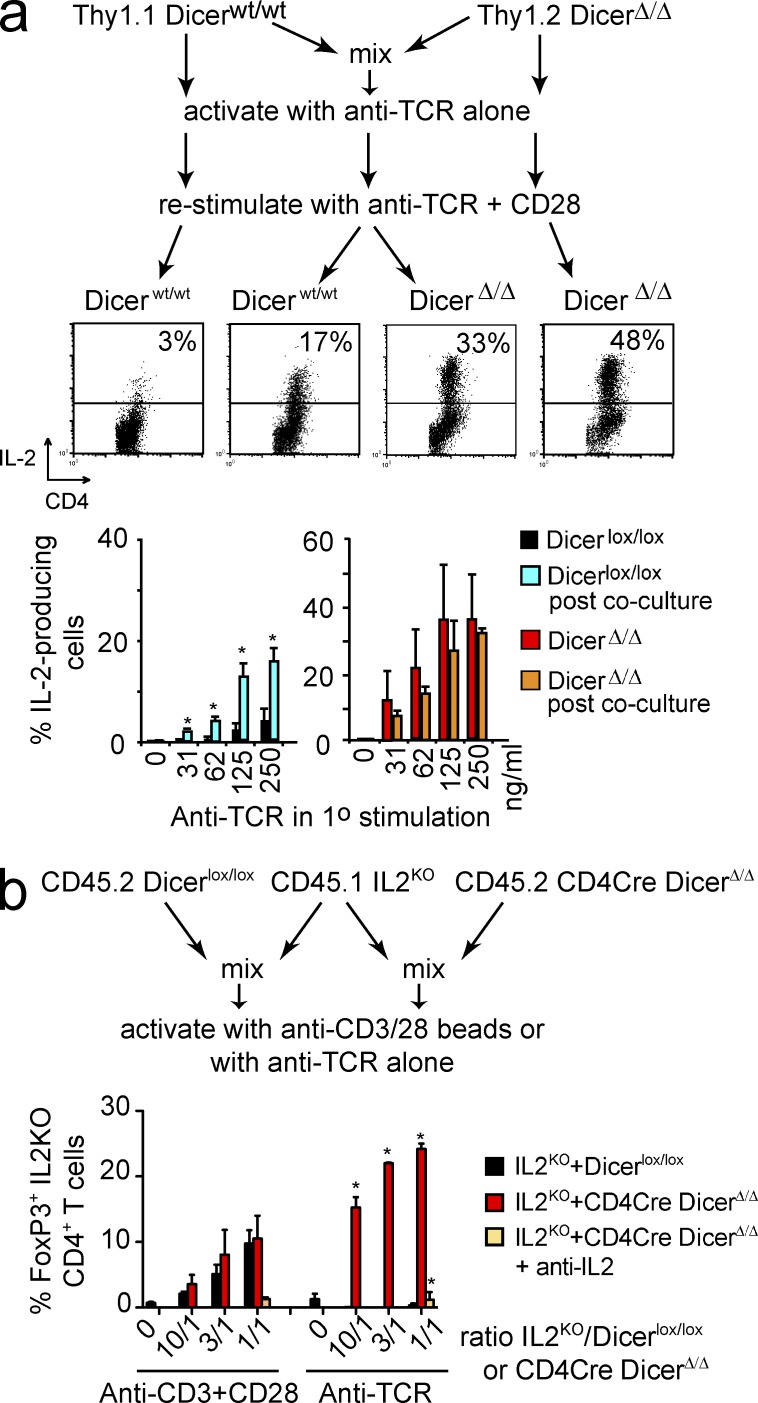
On their own, Dicer-deficient CD4 T cells have limited capacity as effector cells, as indicated by observations that in the absence of T reg cells, Dicer-deficient CD4 T cells cause immunopathology with delayed onset compared with WT CD4 T cells (Cobb et al., 2006). We therefore analyzed the extent to which IL-2 produced by Dicer-deficient CD4 T cells can influence the decision making by other T cells in a paracrine fashion. To determine whether the anergy resistance of Dicer-deficient CD4 T cells was cell-autonomous we performed co-culture experiments. When Thy1.1+ control CD4 T cells and Thy1.2+ CD4Cre DicerΔ/Δ CD4 T cells were stimulated separately with anti-TCR, Dicer-deficient, but not control CD4 T cells produced IL-2 upon restimulation (Fig. 8 a). Interestingly, when Thy1.1+ control and Thy1.2+ Dicer-deficient CD4 T cells were stimulated with anti-TCR in co-culture, both Dicer-deficient and control cells produced IL-2 upon restimulation (Fig. 8 a). This indicated that Dicer-deficient CD4 T cells not only escaped anergy, but also could prevent anergy induction in WT CD4 T cells.
Figure 8.
Functional relevance of IL-2 produced by Dicer-deficient CD4 T cells in the absence of co-stimulation. (a) Resistance of Dicer-deficient CD4 T cells to anti-TCR–induced anergy is contagious. Thy1.1+ control and Thy1.2+ CD4Cre DicerΔ/Δ CD4+ CD25− T cells were cultured either alone or as a mixture (mix) on plate-bound anti-TCR (500 ng/ml, left; graded concentrations, right). IL-2 production was determined 5 h after restimulation with anti-TCR+CD28 using Thy1.1 to distinguish control CD4 T cells from Dicer-deficient CD4 T cells (mean ± SE; n = 2 independent experiments). (b) CD45.1 Il2-deficient CD4 T cells were activated with anti-TCR alone or with anti-CD3+CD28 in the presence of TGF-β (1 ng/ml). Where indicated, cultures were supplemented with CD45.2 control or Dicer-deficient CD4 T cells. Il2-deficient CD4 T cells alone were unable to induce Foxp3. In the presence of co-stimulatory signals, both control and Dicer-deficient CD4 T cells could rescue Foxp3 induction in Il2-deficient CD4 T cells. In the absence of co-stimulation, only Dicer-deficient CD4 T cells were able to facilitate Foxp3 induction in Il2-deficient CD4 T cells (mean ± SEM; n = 3 independent experiments).
To assess the potential relevance of co-stimulation–independent IL-2 secretion by Dicer-deficient CD4 T cells by an independent approach, we addressed its impact on Il2-deficient CD4 T cells, which have a severe defect in the expression of inducible Foxp3 in response to activation in the presence of TGFβ (Fig. 8 b). When activated on their own, Il2-deficient CD4 T cells were unable to induce Foxp3, regardless of co-stimulatory signals (Fig. 8 b). Co-culture with control CD4 T cells restored Foxp3 induction in Il2-deficient CD4 T cells in response to CD3+CD28, but not anti-TCR alone. In contrast, the presence of Dicer-deficient CD4 T cells enabled Il2-deficient CD4 T cells to induce Foxp3 expression in the absence of co-stimulation (Fig. 8 b). Hence, IL-2 derived from Dicer-deficient CD4 T cells can override the induction of anergy in control CD4 T cells and restore inducible Foxp3 expression in Il2-deficient CD4 T cells.
DISCUSSION
Our findings indicate that the ability of CD4 T cells to discriminate between activating and anergizing stimuli requires microRNA-mediated post-transcriptional regulation of the mTOR components Mtor and Rictor. To the best of our knowledge, mTOR components had not previously been shown to be regulated by microRNAs in the immune system. However, Mtor (Nagaraja et al., 2010; Fornari et al., 2010) and Rictor (Uesugi et al., 2011; Tsuruta et al., 2011) are validated microRNA targets in cancer, where deregulated mTOR activity contributes to cellular survival and proliferation. The 3′UTR of Rictor in particular is extensively targeted by Ago-2 containing RISC complexes in CD4 T cells (Loeb et al., 2012) and our experiments show that miR-16 and Let-7 family members are among the microRNAs that regulate Mtor and Rictor expression.
Co-stimulation–independent IL-2 production is central to the phenotype of Dicer-deficient CD4 T cells, and linked to increased phosphorylation of Akt by overexpression of Rictor and Mtor. Transcription of Il2 is regulated by integration of signals that emanate from the TCR and co-stimulatory receptors such as CD28 and converge on the Il2 locus through NF-κB, NF-AT, and AP-1 (Kane et al., 2001; Mondino and Mueller, 2007). Akt is upstream of mTORC1 and downstream of mTORC2 (Sarbassov et al., 2006) and in its activated state may replace CD28 signals in the transcriptional regulation of Il2 through the Carma1–Bcl10–Malt1 (CBM) complex (Kane et al., 1999). In addition to Akt, mTORC2 regulates PKCθ (Lee et al., 2010), and together PKCθ and Akt facilitate the nuclear translocation of NF-κB, which drives Il2 expression in association with NF-AT and AP-1 (Kane et al., 2001; Mondino and Mueller, 2007). Once initiated, IL-2 production can perpetuate T cell activation, and we have shown that IL-2 prolongs the pS6 and pAkt S473 phosphorylation in Dicer-deficient CD4 T cells, even when TCR signals are withdrawn. Persistent mTORC1 activation correlates with successful T cell activation and supports the increased metabolic demands of T cell activation, proliferation, and the acquisition of effector functions (Thompson and Jones, 2007). Importantly, co-stimulation–independent IL-2 production not only allows Dicer-deficient CD4 T to escape anergy and to proliferate in an autocrine fashion, but also acts in a paracrine fashion to enable WT CD4 T cells to avoid anergy in the absence of co-stimulatory signals, and to allow Foxp3 induction in Il2-deficient CD4 T cells.
Regulation by microRNAs is often combinatorial, with multiple microRNAs regulating multiple targets. In some cases, however, individual microRNA targets can be identified that account for micro-RNA-dependent phenotypes (Li et al., 2007; Xiao et al., 2007). Mtor and Rictor are by no means the only signaling components deregulated in Dicer-deficient CD4 T cells (Fig. 6, a and b). Nevertheless, our experiments with Rictor and Mtor compound heterozygous, Dicer-deficient CD4 T cells provide a striking demonstration that the microRNA targets Rictor and Mtor are key to co-stimulation–independent IL-2 production. Correcting the elevated expression of Mtor and Rictor was sufficient to alleviate increased Akt S473 phosphorylation and co-stimulation–independent IL-2 production in Dicer-deficient CD4 T cells. This result indicates that the deregulated expression of mTOR components is a major contributor to co-stimulation–independent IL-2 production in Dicer-deficient CD4 T cells.
In signaling networks, individual components, or nodes, can be critical for network output, in particular nodes with integrator function such as mTOR complexes. microRNAs and their target mRNAs frequently participate in feed-forward regulation (Tsang et al., 2007). In the scenario described here, a feed-forward loop exists between the intracellular PI3K–Akt–mTOR signaling network and the production of the extracellular signaling molecule IL-2. The result of this feed-forward loop is self-sustaining network activity, which is interrupted when extracellular IL-2 is neutralized.
A fundamental insight of the work presented here is that the deregulated expression of Mtor and Rictor contributes significantly to the increased mTOR activity observed in response to TCR stimulation of Dicer-deficient CD4 T cells. Hence, the biochemical and functional activity of mTOR, especially mTORC2, is restrained by microRNA-mediated regulation of Mtor and Rictor expression. Increased mTORC2 activity led to increased IL-2 production in Dicer-deficient cells. To what extent mTOR becomes biochemically rate limiting when the expression of mTOR components is reduced to lower than normal levels may be condition dependent. Published results suggest either a decrease or no apparent change in IL-2 production when Rictor expression is reduced genetically (Lee et al., 2010; Delgoffe et al., 2011) and the up-regulation of Raptor we observe after CD4Cre-mediated deletion of Rictor suggests the potential for compensatory mechanisms during T cell development.
Although previous work on the regulation of T cell anergy had emphasized the importance of E3 ligases that connect the activation of signaling components to their proteolytic degradation in the immune system (Paolino and Penninger, 2010), our data makes the point that the controlled production of signaling components, not just their degradation, contributes to activation versus anergy discrimination.
Autoimmunity and immune pathology in microRNA-deficient mice may be attributable not just to T cell activation in the absence of co-stimulation, but also to reduced regulatory T cell numbers and activity (Cobb et al., 2006; Chong et al., 2008; Liston et al., 2008; Zhou et al., 2008; Lu et al., 2010), overproduction of IFN-γ linked to the T-box transcription factors and miR-29 targets T-bet and Eomes (Steiner et al., 2011), and TNF, which is controlled by miR-146a via TRAF6, IRAK, RelB, and NF-κB pathway activation (Taganov et al., 2006). The severity of the resulting phenotypes is limited because microRNA-deficient T cells show poor long-term effector function and survival (Cobb et al., 2005; Muljo et al., 2005; Chong et al., 2008; Liston et al., 2008; Zhou et al., 2008; Steiner et al., 2011).
Previous studies had linked mTOR activation to anergy by demonstrating that the mTOR inhibitor rapamycin causes CD4 T cell anergy irrespective of co-stimulation (Powell et al., 1999; Mondino and Mueller, 2007; Powell and Delgoffe, 2010). In considering the interplay between Dicer and the mTOR pathway in CD4 T cells, it is noteworthy that rapamycin treatment can restore an anergic state to Dicer-deficient CD4 T cells (unpublished data) despite the elevation of Mtor and Rictor protein levels. Of note, prolonged (>8 h) rapamycin treatment of WT CD4 T cells eliminated their capacity for TCR/CD28-induced Akt S473 phosphorylation, which is indicative of reduced mTORC2 activity (Sarbassov et al., 2006; Lee et al., 2010; Delgoffe et al., 2011). Rapamycin also diminished the phosphorylation of Akt on S473 phosphorylation in human Jurkat T cells in vitro and in mouse thymocytes in vivo (Sarbassov et al., 2006), suggesting that rapamycin could exert its effect as an immunosuppressant in part by establishing an anergic state in vivo. However, the overall impact of rapamycin-induced anergy in conventional CD4 T cells may be counterbalanced by a decrease in mTOR-dependent T reg cell activity (Procaccini et al., 2010; Zeng et al., 2013).
MATERIALS AND METHODS
Mice, flow cytometry, cell sorting, and culture.
Animal work was performed under a UK Home Office Project Licence according to the Animals (Scientific Procedures) Act, UK. Mice carrying floxed Dicer (Cobb et al., 2005) or mTOR (Risson et al., 2009) alleles in combination with CD4Cre or ERt2Cre transgenes on a mixed C57BL/129 background and Thy1.1 congenic C57BL/6 mice were maintained under specific pathogen–free conditions.
For the generation of BM chimeras, C57BL/6 CD45.1 x CD45.2 mice were lethally irradiated and reconstituted i.v. with a mixture of T cell–depleted IL2KO CD45.1 and either Dicer lox/lox or CD4Cre Dicerlox/lox CD45.2 BM cells. In a separate set of experiments, C57BL/6 CD45.1 irradiated recipient were reconstituted with ERt2Cre+ Dicerwt/wt or ERt2Cre+ Dicerlox/lox CD45.2 BM cells. Chimeras were analyzed 8–12 wk after reconstitution. Deletion of Dicer was induced by injection of ERt2Cre+ Dicerwt/wt or ERt2Cre+ Dicerlox/lox mice, ERt2Cre+ Dicerwt/wt or ERt2Cre+ Dicerlox/lox chimeras 1 mg tamoxifen i.p. twice daily for 5 d. Analysis was done on day 21.
Cells were stained, analyzed, and sorted on Calibur, DIVA, or Aria flow cytometers (BD) as previously described (Cobb et al., 2005). In some experiments, cells were sorted using magnetic beads (Miltenyi Biotec). Antibodies used were as follows: CD25-PE, CD25-APC, CD69-FITC, CD62L-PE Thy1.1-biotin, anti–IL-2 PE, anti–IFN-γ APC, anti-TNF FITC (BD), CD4-TC (Invitrogen); pS6 (Cell Signaling Technology) anti-Rabbit Ig FITC (Invitrogen).
Anergy was induced by stimulation on anti-TCR–coated plates in the absence of anti-CD28. Cells were replated after 24 h, allowed to rest for 24 h, and restimulated for 5 h. Cytokine production was measured by intracellular staining or by ELISA.
T cell activation in vivo was induced by intraperitoneal injection of anti-CD3 (10 µg, 2C11; eBioscience). For CFSE labeling, 5–10 × 106 cells/ml were incubated for 8 min at 37°C with 10 µg/ml of CFSE in PBS.
For intracellular staining, 1–2 × 105 T cells were stimulated for 5 h in flat bottom 96-well plates coated with 500 ng/ml anti-TCR (H57) and 2 µg/ml soluble anti-CD28 (BD) in the presence of Golgi-stop, permeabilized with Cytofix/Cytoperm (BD) and stained with the indicated antibodies. Where indicated, recombinant IL-2 (R&D Systems) was used at 5 ng/ml and anti–IL–2 (S4B6; BD) at 10 µg/ml.
ELISA.
IL-2, IFN-γ, and TNF were measured by ELISA according to the supplier’s instructions (R&D Systems).
Immunoblotting.
Standard immunoblotting techniques were used with the following antibodies: phosphotyrosine (4G10; EMD Millipore), Lamin B (C20; Santa Cruz Biotechnology, Inc.), phosphoAkt Thr308 (244F9, 4056), phosphoAkt Ser473 (9271), Akt1 (2H10, 2967), mTOR (7C10), Rictor (53A2), and Raptor (24C12; Cell Signaling Technology). Blots were quantified using fluorescent secondary antibodies and an Odyssey system (Licor).
Proteomics.
The mouse T cell line EL4 was labeled for five passages in Dulbecco’s modified Eagle’s medium (Invitrogen) devoid of arginine and lysine and supplemented with 10% dialyzed fetal bovine serum (Invitrogen, 26400–044); 3.5 mg/ml glucose (to a final concentration of 4.5 mg/ml); 0.1 mm nonessential amino acids without arginine, lysine, and proline; 100 U/ml penicillin/streptomycin (Invitrogen); 2 mm Glutamax (Invitrogen); 100 β-mercaptoethanol (Sigma-Aldrich) and arginine and lysine with heavy (13C) carbons and (15N) nitrogens (Arg10; Cambridge Isotope Laboratories; Lys8, Cambridge Isotope Laboratories) at 28 µg/ml for arginine and 49 µg/ml for lysine. CD4Cre DicerΔ/Δ and Dicerlox/lox control CD4 T cells were freshly isolated ex vivo and therefore contained arginine and lysine with normal light (12C) carbons and (14N) nitrogens. Incorporation of labeled amino acids was verified as previously described (Graumann et al., 2008). Cell lysates were prepared as previously described (Graumann et al., 2008) and EL4 cell lysates were mixed 1:1 with either CD4Cre DicerΔ/Δ or Dicerlox/lox control CD4 T cell extracts, processed, gel fractionated, in-gel digested, and subjected to isoelectric focusing of peptides and high resolution analysis on a linear ion trap-orbitrap instrument (LTQ-Orbitrap) as previously described (Graumann et al., 2008). Mass spectra were acquired using Xcalibur software and analyzed using MaxQuant (Cox and Mann, 2008; version 1.0.4.11) with a false positive rate of 5% at the peptide level and 1% at the protein level. Peptide ratios between heavy EL4 cells and either light CD4Cre DicerΔ/Δ or Dicerlox/lox control CD4 T cells to derive a ratio of ratios between CD4Cre DicerΔ/Δ and Dicerlox/lox control CD4 T cells. Annotations were extracted from the European Bioinformatics Institute Gene Ontology Annotation (GOA) Mouse 36.0 release containing 34,888 proteins.
Quantitative RT-PCR.
Total RNA was isolated using RNAbee (AMS Biotech) and reverse transcribed using Superscript III (Invitrogen). Real-time PCR analysis was performed on an Opticon DNA engine (MJ Research Inc.). The dissociation curve was analyzed for each sample. Relative level of the target sequence against the reference sequence was calculated using the ΔΔ cycle threshold method. Primer sequences were as follows: Hprt1 forward, 5′-TCAGTCAACGGGGGACATAAA-3′, Hprt1 reverse 5′-GGGGCTGTACTGCTTAACCAG-3′; Il2 forward, 5′-GTGCCTAGAAGATGAACTTGGA-3′, Il2 R 5′-AAATGTGTTGTCAGAGCCCT-3′, Dicer1 forward, 5′-TATCGCCTTCACTGCCTTCT-3′ Dicer1 reverse, 5′-TTTTCCACCCGAAGTCTAAGTT-3′; spliced Mtor forward 5′-ACCGGCACACATTTGAAGAAG-3′ Mtor reverse, 5′-CTCGTTGAGGATCAGCAAGG-3′; spliced Rictor forward, 5′-ACCGACACCATCACCATGAAG-3′ Rictor reverse, 5′-GACACCATAGACCTAACTGAGGA-3′; unspliced Mtor forward, 5′-TACCGGGTGAGAGATGGGTC-3′ Mtor reverse, 5′-CACAGTGAGCAGGAGAGAG-3′; unspliced Rictor, forward 5′-TGGTGGTAAAAGAGTGGC-3′ Rictor reverse 5′-GTGTAAGTCAGAGGACGG-3′. microRNAs were quantified using miScript RT and qPCR kits according to the manufacturer’s instructions (QIAGEN).
Reporter constructs.
The full-length 3′UTRs of mTOR and Rictor were amplified from genomic DNA (Mtor forward 5′-CTGAGGCCTGGAAAACCACGTCGTCTCC-3′, reverse 5′-TATGCTTTTAAAATTCTGATGTCATTTATTGG-3′; Rictor forward 5′-CCTCATGCTTATGACGTTTATAGCTGG-3′, reverse 5′-AAGAATTTTAAGTACATTTTATTAACAATG-3′) and cloned into pmaxGFP (Amaxa) or pRL (Promega). Mutants were made by site directed mutagenesis using the primers: Mtor let-7 site 5′-CCACTATCCTGTTtggagaACCCGTCCCTGG-3′, miR-16 site 5′-CCAACCTCCTAGCTcgacgaGAAAAGACACTGTC-3′, Rictor miR-16 site in 3,000–4,200 fragment 5′-GACCTTTTTTTTTTTTTTTTTAGTAATACcgacgatCATTTTTGGAGG-3′. A pmaxCherry plasmid was used as a transfection control.
Luciferase assays.
Control or 3′UTR bearing Firefly constructs alongside Renilla control plasmids were transfected into 293T cells. Cells were cotransfected with empty or microRNA containing MIG plasmid. Luciferase luminescence was determined 36 h later. Ratios between Firefly and Renilla were normalized to the ratios obtained when the empty MIG plasmid was used.
miRNA mimics and LNAs.
miRNA mimics (miRIDIAN) supplied by Thermo Scientific and used according to manufacturer instructions. LNAs targeting miRNA family seed sequences (Exiqon) were designed and used as described (Obad et al., 2011).
Online supplemental material.
Table S1 shows differentially expressed signaling-related mRNAs in Dicer-deficient CD4 T cells identified by gene expression arrays. Table S2 shows differentially expressed signaling-related proteins in Dicer-deficient CD4 T cells identified by quantitative proteomics. Table S3 shows densitometry values of scanned western blots. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132059/DC1.
Supplementary Material
Acknowledgments
We thank Dr. S Kozma for Mtorlox/lox mice, Dr. G Loeb for sharing information on Ago2 binding to the 3′ UTR of Mtor and Rictor before publication, Drs P. Hexley and J. Elliott for cell sorting, Dr J. Marvel for critical reading of the manuscript, and the members of the Lymphocyte Development Group for discussion and advice.
This work was supported by the Medical Research Council UK and a post-doctoral fellowship from Fondation Pour la Recherche Médicale (AM). The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- IKK
- IκB kinase
- LNA
- locked nucleic acid
- PLCγ
- phospholipase Cγ
References
- Araki, K., Ellebedy A.H., and Ahmed R.. 2011. TOR in the immune system. Curr. Opin. Cell Biol. 23:707–715. 10.1016/j.ceb.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann, D., Kageyama R., Clingan J.M., Morar M.M., Patel S., de Kouchkovsky D., Bannard O., Bluestone J.A., Matloubian M., Ansel K.M., and Jeker L.T.. 2013. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat. Immunol. 14:840–848. 10.1038/ni.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky, Y., Villarino A.V., Eisley C.J., Barbeau R., Barczak A.J., Heinz G.A., Kremmer E., Heissmeyer V., McManus M.T., Erle D.J., et al. 2013. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 210:417–432. 10.1084/jem.20111717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, M.M., Rasmussen J.P., Rudensky A.Y., and Littman D.R.. 2008. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205:2005–2017. 10.1084/jem.20081219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, B.S., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., and Merkenschlager M.. 2005. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201:1367–1373. 10.1084/jem.20050572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, B.S., Hertweck A., Smith J., O’Connor E., Graf D., Cook T., Smale S.T., Sakaguchi S., Livesey F.J., Fisher A.G., and Merkenschlager M.. 2006. A role for Dicer in immune regulation. J. Exp. Med. 203:2519–2527. 10.1084/jem.20061692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Delgoffe, G.M., Pollizzi K.N., Waickman A.T., Heikamp E., Meyers D.J., Horton M.R., Xiao B., Worley P.F., and Powell J.D.. 2011. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 12:295–303. 10.1038/ni.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva, D.R., Urdahl K.B., and Jenkins M.K.. 1991. Clonal anergy is induced in vitro by T cell receptor occupancy in the absence of proliferation. J. Immunol. 147:3261–3267 [PubMed] [Google Scholar]
- Fornari, F., Milazzo M., Chieco P., Negrini M., Calin G.A., Grazi G.L., Pollutri D., Croce C.M., Bolondi L., and Gramantieri L.. 2010. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 70:5184–5193. 10.1158/0008-5472.CAN-10-0145 [DOI] [PubMed] [Google Scholar]
- Graumann, J., Hubner N.C., Kim J.B., Ko K., Moser M., Kumar C., Cox J., Schöler H., and Mann M.. 2008. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics. 7:672–683. 10.1074/mcp.M700460-MCP200 [DOI] [PubMed] [Google Scholar]
- Harding, F.A., McArthur J.G., Gross J.A., Raulet D.H., and Allison J.P.. 1992. CD28-mediated signalling co-stimulates murine T cells and prevents induction of anergy in T-cell clones. Nature. 356:607–609. 10.1038/356607a0 [DOI] [PubMed] [Google Scholar]
- Heissmeyer, V., Macián F., Im S.H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y.C., Dustin M.L., and Rao A.. 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 5:255–265. 10.1038/ni1047 [DOI] [PubMed] [Google Scholar]
- Inui, M., Martello G., and Piccolo S.. 2010. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 11:252–263. 10.1038/nrm2868 [DOI] [PubMed] [Google Scholar]
- Jones, R.G., and Thompson C.B.. 2007. Revving the engine: signal transduction fuels T cell activation. Immunity. 27:173–178. 10.1016/j.immuni.2007.07.008 [DOI] [PubMed] [Google Scholar]
- Kane, L.P., Shapiro V.S.S., Stokoe D., and Weiss A.. 1999. Induction of NF-kappaB by the Akt/PKB kinase. Curr. Biol. 9:601–604. 10.1016/S0960-9822(99)80265-6 [DOI] [PubMed] [Google Scholar]
- Kane, L.P., Andres P.G., Howland K.C., Abbas A.K., and Weiss A.. 2001. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-γ but not TH2 cytokines. Nat. Immunol. 2:37–44. 10.1038/83144 [DOI] [PubMed] [Google Scholar]
- Kang, S.G., Liu W.H., Lu P., Jin H.Y., Lim H.W., Shepherd J., Fremgen D., Verdin E., Oldstone M.B., Qi H., et al. 2013. MicroRNAs of the miR-17∼92 family are critical regulators of T(FH) differentiation. Nat. Immunol. 14:849–857. 10.1038/ni.2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye, J., Hsu M.L., Sauron M.E., Jameson S.C., Gascoigne N.R., and Hedrick S.M.. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 341:746–749. 10.1038/341746a0 [DOI] [PubMed] [Google Scholar]
- Khan, A.A., Penny L.A., Yuzefpolskiy Y., Sarkar S., and Kalia V.. 2013. MicroRNA-17∼92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood. 121:4473–4483. 10.1182/blood-2012-06-435412 [DOI] [PubMed] [Google Scholar]
- Lee, K., Gudapati P., Dragovic S., Spencer C., Joyce S., Killeen N., Magnuson M.A., and Boothby M.. 2010. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 32:743–753. 10.1016/j.immuni.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q.J., Chau J., Ebert P.J., Sylvester G., Min H., Liu G., Braich R., Manoharan M., Soutschek J., Skare P., et al. 2007. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 129:147–161. 10.1016/j.cell.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Linsley, P.S., Brady W., Grosmaire L., Aruffo A., Damle N.K., and Ledbetter J.A.. 1991. Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation. J. Exp. Med. 173:721–730. 10.1084/jem.173.3.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, H.C., and Smith K.A.. 2011. The roles of c-rel and interleukin-2 in tolerance: a molecular explanation of self-nonself discrimination. Immunol. Cell Biol. 89:27–32. 10.1038/icb.2010.120 [DOI] [PubMed] [Google Scholar]
- Liston, A., Lu L.F., O’Carroll D., Tarakhovsky A., and Rudensky A.Y.. 2008. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205:1993–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, G.B., Khan A.A., Canner D., Hiatt J.B., Shendure J., Darnell R.B., Leslie C.S., and Rudensky A.Y.. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell. 48:760–770. 10.1016/j.molcel.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L.F., Boldin M.P., Chaudhry A., Lin L.L., Taganov K.D., Hanada T., Yoshimura A., Baltimore D., and Rudensky A.Y.. 2010. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 142:914–929. 10.1016/j.cell.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondino, A., and Mueller D.L.. 2007. mTOR at the crossroads of T cell proliferation and tolerance. Semin. Immunol. 19:162–172. 10.1016/j.smim.2007.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo, S.A., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., and Rajewsky K.. 2005. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 202:261–269. 10.1084/jem.20050678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja, A.K., Creighton C.J., Yu Z., Zhu H., Gunaratne P.H., Reid J.G., Olokpa E., Itamochi H., Ueno N.T., Hawkins S.M., et al. 2010. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol. Endocrinol. 24:447–463. 10.1210/me.2009-0295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad, S., dos Santos C.O., Petri A., Heidenblad M., Broom O., Ruse C., Fu C., Lindow M., Stenvang J., Straarup E.M., et al. 2011. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 43:371–378. 10.1038/ng.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozes, O.N., Mayo L.D., Gustin J.A., Pfeffer S.R., Pfeffer L.M., and Donner D.B.. 1999. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 401:82–85. 10.1038/43466 [DOI] [PubMed] [Google Scholar]
- Paolino, M., and Penninger J.M.. 2010. Cbl-b in T-cell activation. Semin. Immunopathol. 32:137–148. 10.1007/s00281-010-0197-9 [DOI] [PubMed] [Google Scholar]
- Powell, J.D., and Delgoffe G.M.. 2010. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 33:301–311. 10.1016/j.immuni.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J.D., Lerner C.G., and Schwartz R.H.. 1999. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. J. Immunol. 162:2775–2784 [PubMed] [Google Scholar]
- Procaccini, C., De Rosa V., Galgani M., Abanni L., Calì G., Porcellini A., Carbone F., Fontana S., Horvath T.L., La Cava A., and Matarese G.. 2010. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 33:929–941. 10.1016/j.immuni.2010.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga, I., Rao A., and Macian F.. 2008. Targeted cleavage of signaling proteins by caspase 3 inhibits T cell receptor signaling in anergic T cells. Immunity. 29:193–204. 10.1016/j.immuni.2008.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risson, V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A., et al. 2009. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 187:859–874. 10.1083/jcb.200903131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg, R., Neilson J.R., Sarma A., Sharp P.A., and Burge C.B.. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 320:1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov, D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., and Sabatini D.M.. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 22:159–168. 10.1016/j.molcel.2006.03.029 [DOI] [PubMed] [Google Scholar]
- Schwartz, R.H.2003. T cell anergy. Annu. Rev. Immunol. 21:305–334. 10.1146/annurev.immunol.21.120601.141110 [DOI] [PubMed] [Google Scholar]
- Selbach, M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., and Rajewsky N.. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature. 455:58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Steiner, D.F., Thomas M.F., Hu J.K., Yang Z., Babiarz J.E., Allen C.D., Matloubian M., Blelloch R., and Ansel K.M.. 2011. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 35:169–181. 10.1016/j.immuni.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov, K.D., Boldin M.P., Chang K.J., and Baltimore D.. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 103:12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M.F., Abdul-Wajid S., Panduro M., Babiarz J.E., Rajaram M., Woodruff P., Lanier L.L., Heissmeyer V., and Ansel K.M.. 2012. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and antiviral function. Blood. 120:130–142. 10.1182/blood-2011-11-394072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, C.B., Lindsten T., Ledbetter J.A., Kunkel S.L., Young H.A., Emerson S.G., Leiden J.M., and June C.H.. 1989. CD28 activation pathway regulates the production of multiple T-cell-derived lymphokines/cytokines. Proc. Natl. Acad. Sci. USA. 86:1333–1337. 10.1073/pnas.86.4.1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, J., Zhu J., and van Oudenaarden A.. 2007. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 26:753–767. 10.1016/j.molcel.2007.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruta, T., Kozaki K., Uesugi A., Furuta M., Hirasawa A., Imoto I., Susumu N., Aoki D., and Inazawa J.. 2011. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 71:6450–6462. 10.1158/0008-5472.CAN-11-0364 [DOI] [PubMed] [Google Scholar]
- Uesugi, A., Kozaki K., Tsuruta T., Furuta M., Morita K., Imoto I., Omura K., and Inazawa J.. 2011. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 71:5765–5778. 10.1158/0008-5472.CAN-11-0368 [DOI] [PubMed] [Google Scholar]
- Wells, A.D.2009. New insights into the molecular basis of T cell anergy: anergy factors, avoidance sensors, and epigenetic imprinting. J. Immunol. 182:7331–7341. 10.4049/jimmunol.0803917 [DOI] [PubMed] [Google Scholar]
- Xiao, C., Calado D.P., Galler G., Thai T.H., Patterson H.C., Wang J., Rajewsky N., Bender T.P., and Rajewsky K.. 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 131:146–159. 10.1016/j.cell.2007.07.021 [DOI] [PubMed] [Google Scholar]
- Xiao, F., Zuo Z., Cai G., Kang S., Gao X., and Li T.. 2009. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37:D105-D110. 10.1093/nar/gkn851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Boldin M.P., Yu Y., Liu C.S., Ea C.K., Ramakrishnan P., Taganov K.D., Zhao J.L., and Baltimore D.. 2012. miR-146a controls the resolution of T cell responses in mice. J. Exp. Med. 209:1655–1670. 10.1084/jem.20112218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H., Yang K., Cloer C., Neale G., Vogel P., and Chi H.. 2013. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 499:485–490. 10.1038/nature12297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., and Bevan M.J.. 2010. Dicer controls CD8+ T-cell activation, migration, and survival. Proc. Natl. Acad. Sci. USA. 107:21629–21634. 10.1073/pnas.1016299107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Collins S.L., Lutz M.A., Allen A.N., Kole T.P., Zarek P.E., and Powell J.D.. 2007. A role for mammalian target of rapamycin in regulating T cell activation versus anergy. J. Immunol. 178:2163–2170. 10.4049/jimmunol.178.4.2163 [DOI] [PubMed] [Google Scholar]
- Zhou, X., Jeker L.T., Fife B.T., Zhu S., Anderson M.S., McManus M.T., and Bluestone J.A.. 2008. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 205:1983–1991. 10.1084/jem.20080707 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.