CARD9 is dispensable for NF-κB activation induced by Dectin-1 ligands in mice. However, Dectin-1–induced H-Ras activation is mediated by a complex with CARD9, which leads to ERK activation for host innate immune responses to Candida albicans infection.
Abstract
Dectin-1 functions as a pattern recognition receptor for sensing fungal infection. It has been well-established that Dectin-1 induces innate immune responses through caspase recruitment domain-containing protein 9 (CARD9)–mediated NF-κB activation. In this study, we find that CARD9 is dispensable for NF-κB activation induced by Dectin-1 ligands, such as curdlan or Candida albicans yeast. In contrast, we find that CARD9 regulates H-Ras activation by linking Ras-GRF1 to H-Ras, which mediates Dectin-1–induced extracellular signal-regulated protein kinase (ERK) activation and proinflammatory responses when stimulated by their ligands. Mechanistically, Dectin-1 engagement initiates spleen tyrosine kinase (Syk)–dependent Ras-GRF1 phosphorylation, and the phosphorylated Ras-GRF1 recruits and activates H-Ras through forming a complex with CARD9, which leads to activation of ERK downstream. Finally, we show that inhibiting ERK activation significantly accelerates the death of C. albicans–infected mice, and this inhibitory effect is dependent on CARD9. Together, our studies reveal a molecular mechanism by which Dectin-1 induces H-Ras activation that leads to ERK activation for host innate immune responses against fungal infection.
The immune system uses germline-encoded pattern recognition receptors (PRRs) to sense various pathogens including bacteria, viruses, and fungi, or endogenous danger signals, initiating intracellular signal cascades that result in the activation of transcription factors, up-regulation of defense-associated target genes, and release of cytokines (Medzhitov, 2009). During fungal infection, the initial innate immune response is determined by the recognition of PAMPs (pathogen-associated molecular patterns) by PRRs of innate immune cells (Romani, 2004). Several types of PRRs with signaling capacity include TLRs, NLRs (nucleotide-oligomerization domain-like receptors), RLRs (retinoic acid-inducible gene I–like helicases), and the recently emerging family of spleen tyrosine kinase (Syk)–coupled C-type lectin receptors (CLRs; Kerrigan and Brown, 2011). The ability of these PRRs to elicit inflammation and innate immunity is their capability to activate multiple signaling cascades, leading to activation of various transcription factors including NF-κB (Vallabhapurapu and Karin, 2009; Cargnello and Roux, 2011).
Dectin-1 is a Syk-coupled CLR and recognizes β-glucan carbohydrates on various fungi including Candida albicans, Aspergillus fumigatus, and Pneumocystis carinii (Brown and Gordon, 2001; Steele et al., 2005; Saijo et al., 2007), whereas Dectin-2 and Dectin-3, two Syk-coupled CLRs, form heterodimeric PRR to recognize α-mannans on the surface of C. albicans hyphae (Sato et al., 2006; Saijo et al., 2010; Zhu et al., 2013). As the main nonopsonic receptor involved in fungal uptake (Heinsbroek et al., 2008), Dectin-1 binds to β-glucans from different fungal species (Brown and Gordon, 2001) and forms a phagocytic synapse complex on the cytoplasm membrane (Goodridge et al., 2011). Dectin-1 collaborates with TLR2 (Ferwerda et al., 2008), SIGNR1 (Takahara et al., 2011), and galectin-3 (Esteban et al., 2011) in recognizing β-glucans on the surface of C. albicans yeast cells leading to induction of proinflammatory cytokines.
The caspase recruitment domain-containing protein 9 (CARD9) is the critical adaptor protein that operates downstream of several immunoreceptor tyrosine-based activation motif (ITAM)–associated CLRs including Dectin-1, Dectin-2, and Mincle (Gross et al., 2006; Robinson et al., 2009; Bi et al., 2010; Saijo et al., 2010; Schoenen et al., 2010). After receptor engagement and Syk kinase phosphorylation, CARD9 forms a complex with B cell leukemia-lymphoma 10 (Bcl10) and mucosa-associated lymphoid tissue 1 (Malt1), which transduces non–TLR signaling to the canonical NF-κB pathway (Kingeter and Lin, 2012), leading to inducing the expression of proinflammatory cytokines, including TNF, IL-1β, and IL-6. These proinflammatory cytokines control antifungal immune responses (Netea et al., 1999; Vonk et al., 2006). More importantly, the role of Dectin-1 and CARD9 in antifungal host defense has been confirmed by in vivo studies (Gross et al., 2006; Saijo et al., 2007; Taylor et al., 2007), together with the discovery that host mutations in Dectin-1 or CARD9 lead to primary immunodeficiencies associated with fungal infections (Ferwerda et al., 2009; Glocker et al., 2009). Thus, CARD9 plays a central role in antifungal defense by receiving signals from Dectin-1 and Dectin-2 and stimulating proinflammatory responses.
However, most of the in vitro studies on Dectin-1 signaling have used β-glucan–containing particles, such as zymosan from Saccharomyces cerevisiae, curdlan from nonpathogenic bacteria, or heat-inactivated C. albicans yeasts as stimuli (Gross et al., 2006) because β-glucans only become accessible at the site of budding scars in living C. albicans yeast cells (Gantner et al., 2005). In addition, serum can induce a dimorphic transition of C. albicans from yeast to hyphae forms (Sevilla and Odds, 1986), which complicates the data interpretation of in vitro studies of β-glucan signaling through Dectin-1. Recent studies showed that β-glucans on the surface of C. albicans can be unmasked by several experimental manipulations, such as heat-killing (Gantner et al., 2005), treatment with the antifungal drug caspofungin (Wheeler and Fink, 2006; Wheeler et al., 2008), or deletion of certain Candida genes including extracellular signal-regulated kinase gene CEK (Galán-Díez et al., 2010), the histidine kinase gene CHK1 (Klippel et al., 2010), the predicted glucosyltransferase gene KRE5 (Wheeler and Fink, 2006), or the glycosidase gene PHR2 (Wheeler and Fink, 2006). Furthermore, unmasked β-glucans of C. albicans can be specifically recognized by Dectin-1 and lead to more potent activation of NF-κB and extracellular signal-regulated protein kinase (ERK) in human DCs (Wheeler and Fink, 2006; Galán-Díez et al., 2010).
In contrast to the model that Dectin-1–induced NF-κB is mediated through the CARD9-dependent pathway, our previous study shows that stimulation of Dectin-1 with zymosan, curdlan, or HI C. albicans yeasts induces NF-κB activation through a CARD9-independent pathway in macrophages, whereas Dectin-2 stimulation by C. albicans hyphae triggers NF-κB activation through the CARD9-dependent pathway (Bi et al., 2010). This study suggests that Dectin-1–induced signaling may be more complicated than originally proposed (Gross et al., 2006), and the molecular mechanism by which CARD9 is involved in Dectin-1 signaling pathway may need to be further determined. Furthermore, a recent study also shows that Dectin-1–induced signaling can module NF-κB through Raf-1–dependent phosphorylation and acetylation of p65 subunit of NF-κB (Gringhuis et al., 2009), suggesting that multiple signaling pathways may modulate Dectin-1–induced NF-κB activation.
In this study, using Curdlan, a β-glucan ligand and a mutant C. albicans strain mnn5, which has the highly exposed surface β-glucans, we were able to examine the requirement of CARD9 for β-glucan–induced signaling in innate immune cells and found that although CARD9 is dispensable for Dectin-1–induced NF-κB activation, it is required for Dectin-1–induced ERK activation by linking Ras-GRF1 to H-Ras. Together, our studies reveal a molecular mechanism by which CARD9 mediates Dectin-1-induced ERK activation for host anti-fungal immune responses.
RESULTS
β-glucans exposed on the surface of C. albicans yeast activate NF-κB and ERK through Dectin-1
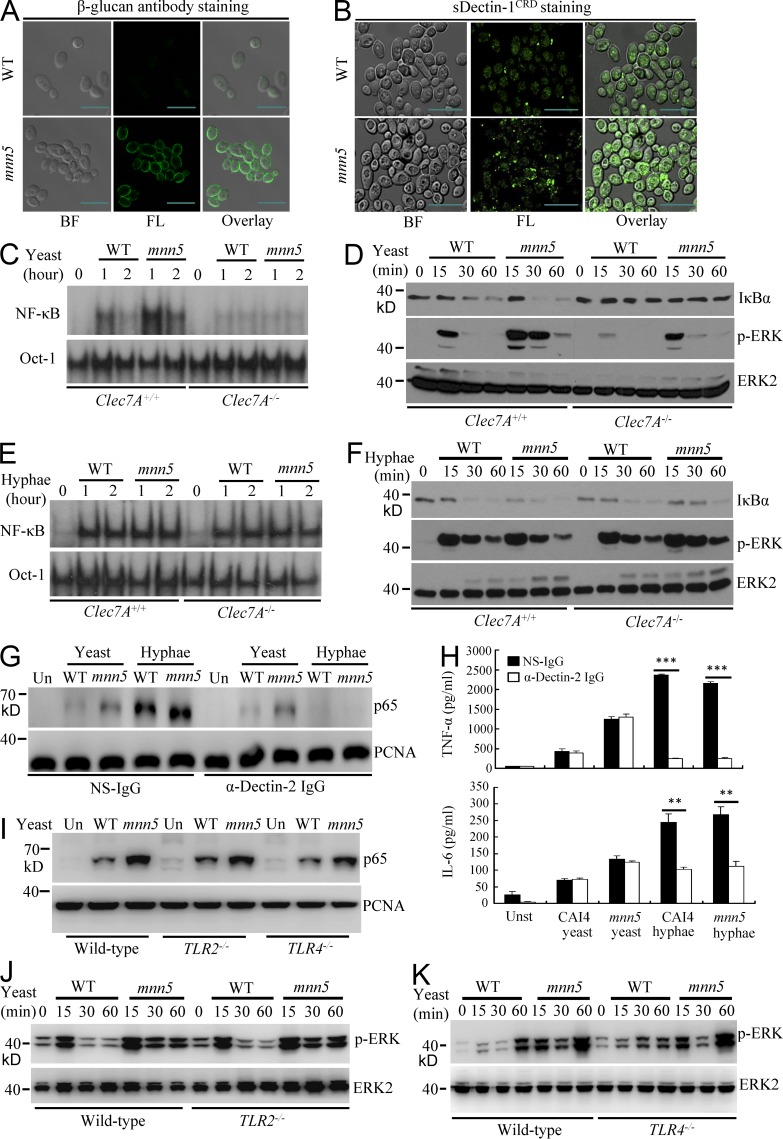
C. albicans is coated with mannoproteins that prevent β-glucan moieties from being recognized by Dectin-1 on innate immune cells (Gantner et al., 2005; Wheeler and Fink, 2006). To study the effect of surface β-glucans of live fungi on Dectin-1–mediated innate immune responses, we generated a mutant strain of C. albicans, mnn5, in which the α-1,2-mannosyltransferanse gene MNN5 (Bai et al., 2006) was disrupted by homologous recombination, thereby causing the increased exposure of β-glucans on the cell wall of C. albicans yeasts (Fig. 1 A). Consistently, the β-glucan moiety on the surface of mnn5 yeast was more accessible to soluble GFP-tagged extracellular carbohydrate-binding domain (CRD) of Dectin-1 (sDectin-1CRD) than that on WT C. albicans yeast cells (Fig. 1 B).
Figure 1.
Surface β-glucans on C. albicans yeasts activate NF-κB and ERK through Dectin-1. (A and B) Surface β-glucan accessibility on WT or mnn5 yeast C. albicans strains, which were stained with anti–β-1,3-glucan monoclonal antibodies (β-glucan), followed by staining with FITC-labeled secondary antibodies (A) or soluble GFP-tagged Dectin-1 carbohydrate recognition domain (sDectin-1CRD; B). Bright field (BF), fluorescence (FL), and overlay (right) are shown individually. Bars, 10 µm. (C and D) WT and Dectin-1 (Clec7A)–deficient BMDMs were stimulated with UV-inactivated WT yeasts or mnn5 yeasts (MOI = 5) for indicated times. Nuclear extracts were subjected to EMSA using 32P-labeled NF-κB or Oct-1 probe (C). Cell lysates were subjected to immunoblotting analysis using the indicated antibodies (D). (E and F) WT and Dectin-1 (Clec7A)–deficient BMDMs were stimulated with hyphae of WT yeasts or mnn5 (MOI = 1) for the indicated times. Nuclear extracts were subjected to EMSA using 32P-labeled NF-κB or Oct-1 probe (E). Cell lysates were subjected to immunoblotting analysis using the indicated antibodies (F). (G) WT BMDMs were stimulated with C. albicans WT or mnn5 yeast (MOI = 5) or hyphae (MOI = 1) in the absence or presence of blocking antibodies against Dectin-2 (α-Dectin-2, 20 µg/ml) for 60 min. Nuclear extracts were prepared from these cells and subjected to immunoblotting analysis using indicated antibodies. (H) WT BMDMs were stimulated with the UV-inactivated yeast (MOI = 5) or hyphae (MOI = 1) form of CAI4 or mnn5 mutant in the absence or presence of blocking antibodies against Dectin-2 (20 µg/ml) for 6 h. The amount of TNF and IL-6 in the cultured media was determined using ELISA. **, P < 0.01; ***, P < 0.001. Data are means ± SD of triplicate wells and are representative of three independent experiments. (I–K) WT, TLR2-, and TRL4-deficient BMDMs were stimulated with UV-inactivated WT (CAI4) or mnn5 strains of C. albicans yeast cells (MOI = 5) for 1 h for preparing nuclear extracts (I) or for the indicated times for preparing cell lysates (J and K). Samples were subjected to immunoblotting analysis using indicated antibodies. Data shown are representative of three independent and reproducible experiments.
To further confirm that the exposed β-glucan on the surface of mnn5 yeast activates the signaling pathway through Dectin-1, we stimulated BMDMs from WT (Clec7A+/+) or Dectin-1–deficient (Clec7A−/−) mice with WT or mnn5 yeast cells, and found that mnn5 yeasts induced more potent NF-κB DNA binding than WT yeast (Fig. 1 C), together with more IκBα degradation (Fig. 1 D). Consistently, Dectin-1 deficiency impaired the NF-κB activation (Fig. 1 C) and IκBα degradation (Fig. 1 D). Moreover, mnn5 yeasts triggered stronger and sustained activation of ERK than WT yeast in BMDMs (Fig. 1 D), but Dectin-1 deficiency significantly impaired yeast-induced ERK activation in BMDMs (Fig. 1 D). In contrast, when these cells were stimulated with hyphal forms of C. albicans, NF-κB activation (Fig. 1 E) and IκBα degradation and ERK activation (Fig. 1 F) were no different between WT and Dectin-1–deficient cells stimulated with WT or mnn5 mutant. These data indicate that mnn5 mutation does not increase the exposure of β-glucan on the surface of hyphae form of C. albicans; thereby Dectin-1 deficiency does not affect mnn5 hyphal form stimulation-induced signaling.
We also found that blockade of Dectin-2 using its specific monoclonal antibodies (Zhu et al., 2013) in BMDMs had almost no effect on NF-κB activation induced by WT and mnn5 yeast stimulation (Fig. 1 G) and the production of proinflammatory cytokine TNF and IL-6 (Fig. 1 H). However, when challenged with WT and mnn5 hyphae, blocking Dectin-2 completely inhibited NF-κB activation (Fig. 1 G) and significantly reduced secretion of TNF and IL-6 (Fig. 1 H). Furthermore, deficiency of TRL2 or TLR4 had no influences on WT and mnn5 yeast stimulation-induced activation of NF-κB (Fig. 1 I) and ERK (Fig. 1, J and K). Together, these results indicate that the exposed β-glucan on yeast surface can induce potent proinflammatory responses through Dectin-1, but not Dectin-2, TLR2, or TLR4.
CARD9 is required for the C. albicans yeast stimulation-induced ERK but not NF-κB activation
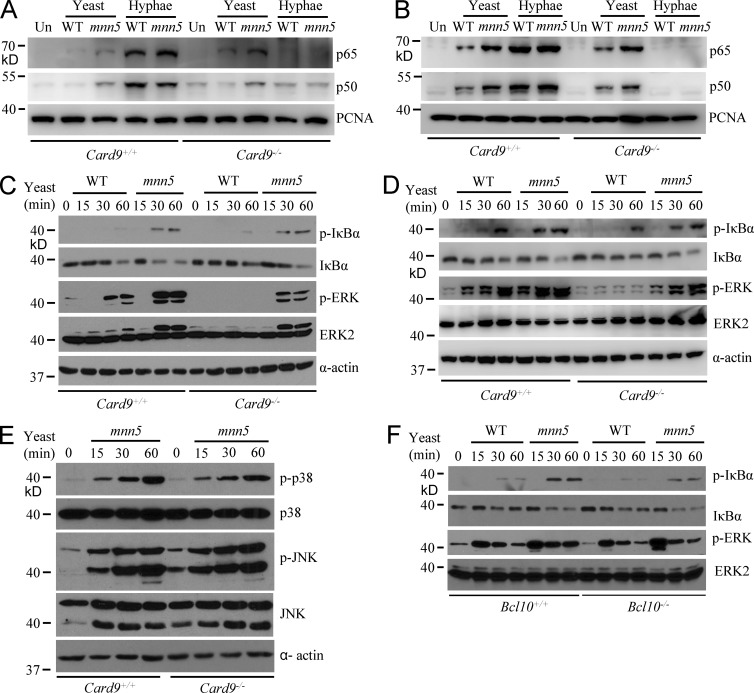
Previous studies suggest that β-glucans from C. albicans activate Dectin-1–mediated NF-κB activation through a CARD9-dependent pathway (Gross et al., 2006). To further investigate the role of CARD9 in Dectin-1–induced signaling pathways in response to fungal infection, BM-derived macrophages (BMDMs) or BM-derived DCs (BMDCs) from WT (Card9+/+) or CARD9-deficient (Card9−/−) mice were stimulated with WT or mnn5 strain of C. albicans yeast. We found that CARD9 deficiency in BMDMs and BMDCs had no influences on the yeast stimulation-induced nuclear translocation of NF-κB (p65 and p50 subunits; Fig. 2, A and B), as well as IκBα phosphorylation and degradation (Fig. 2, C and D, top). In contrast, when these cells were stimulated by C. albicans hyphae that activate Dectin-2 but not Dectin-1 signaling (Saijo et al., 2010), the nuclear translocation of NF-κB subunits was defective in CARD9-deficient BMDMs (Fig. 2 A) and BMDCs (Fig. 2 B). Unlike the yeast stimulation-induced NF-κB activation, we surprisingly found that C. albicans yeast stimulation-induced ERK phosphorylation was defective in CARD9-deficient BMDMs (Fig. 2 C, middle) and BMDCs (Fig. 2 D, middle), suggesting that CARD9 deficiency affects Dectin-1–induced ERK activation. In addition, CARD9 deficiency had no significant impact on Dectin-1–induced activation of p38 and JNK MAP kinases (Fig. 2 E). Together, these results suggest that CARD9 may be dispensable for Dectin-1–induced activation of NF-κB, p38, and JNK, but it is required for Dectin-1–induced ERK activation.
Figure 2.
CARD9 engages the yeast-induced ERK, but not NF-κB activation. (A and B) WT and CARD9-deficient BMDMs (A) or BMDCs (B) were stimulated with UV-inactivated WT or mnn5 strains of C. albicans yeasts (MOI = 5) or hyphae (MOI = 1) for 1 h. Nuclear extracts were prepared and subjected to immunoblotting analysis using indicated antibodies. (C and D) WT and CARD9-deficient BMDMs (C) or BMDCs (D) were stimulated with UV-inactivated WT or mnn5 mutant strain of C. albicans yeast for the indicated times. Cell lysates were prepared and subjected to immunoblotting analysis using indicated antibodies. (E) WT and CARD9-deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells (MOI = 5) for the indicated times. Cell lysates were prepared and subjected to immunoblotting analysis using indicated antibodies. (F) WT and Bcl10-deficient BMDMs were stimulated with UV-inactivated WT or mnn5 yeast cells (MOI = 5) for the indicated times. Cell lysates were prepared and subjected to immunoblotting analysis using indicated antibodies. Data shown are representative of three independent and reproducible experiments.
Because CARD9 forms a complex with Bcl10 (Bertin et al., 2000), we examined whether Bcl10 deficiency affected Dectin-1–induced ERK activation using BMDMs from WT or Bcl10-deficient (Bcl10−/−) mice, and found that Bcl10 deficiency affected neither C. albicans yeast stimulation-induced IκBα phosphorylation and degradation nor ERK activation (Fig. 2 F). This result suggests that although Dectin-1–induced ERK activation is dependent on CARD9, it is independent of the CARD9–Bcl10–Malt1 (CBM) complex (Kingeter and Lin, 2012).
CARD9 is required for β-glucan–induced ERK, but not NF-κB, activation
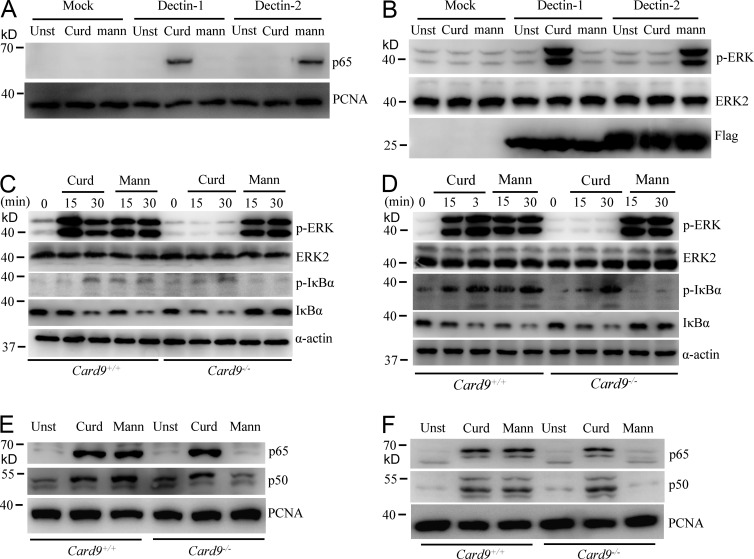
It has been shown that Dectin-1 activates NF-κB through CARD9-dependent pathway (Gross et al., 2006), and curdlan, a mixture of β-glucan from nonpathogenic bacteria, is one of the ligands for Dectin-1 (Palma et al., 2006). To further determine the role of CARD9 in Dectin-1 signaling, we used plate-bound curdlan or plate-bound α-mannans, which potently activated Dectin-1 or Dectin-2, respectively, leading to induction of NF-κB nuclear translocation and ERK phosphorylation (Fig. 3, A and B). Interestingly, we found that curdlan-induced ERK activation was defective in CARD9-deficient BMDMs and BMDCs (Fig. 3, C and D), whereas α-mannan–induced ERK activation was intact (Fig. 3, C and D). Consistent with our previous findings (Bi et al., 2010), curdlan-induced nuclear translocation of NF-κB was intact (Fig. 3, E and F). In contrast, α-mannan–induced nuclear translocation of NF-κB was defected in CARD9-deficient cells (Fig. 3, E and F), which is consistent with our previous studies that CARD9 is required for Dectin-2–induced NF-κB activation (Bi et al., 2010). Together, these results suggest that CARD9 is required for Dectin-1–induced ERK activation but is dispensable for NF-κB activation. In contrast, CARD9 is required for Dectin-2–induced NF-κB activation.
Figure 3.
CARD9 mediates Curdlan-induced ERK activation but is not dispensable for NF-κB. (A and B) RAW264.7 cells stably expressing human Dectin-1, Dectin-2, or mock were stimulated with plate-coated curdlan (50 µg/ml) or α-mannans (40 µg/ml) for 1 h for preparing nuclear extract (A) or 30 min for preparing cell lysate (B), and then subjected to immunoblotting analysis using the indicated antibodies. (C–F) WT and CARD9-deficient BMDMs (C and E) or BMDCs (D and F) were stimulated with plate-coated curdlan (50 µg/ml) or α-mannans (40 µg/ml) for the indicated times for preparing cell lysate (C and D) and 1 h for preparing nuclear extract (E and F). The cell lysates or nuclear extracts were subjected to immunoblotting analysis using the indicated antibodies. Data shown are representative of three independent and reproducible experiments.
CARD9 links Ras-GRF1 to H-Ras in response to C. albicans yeast stimulation
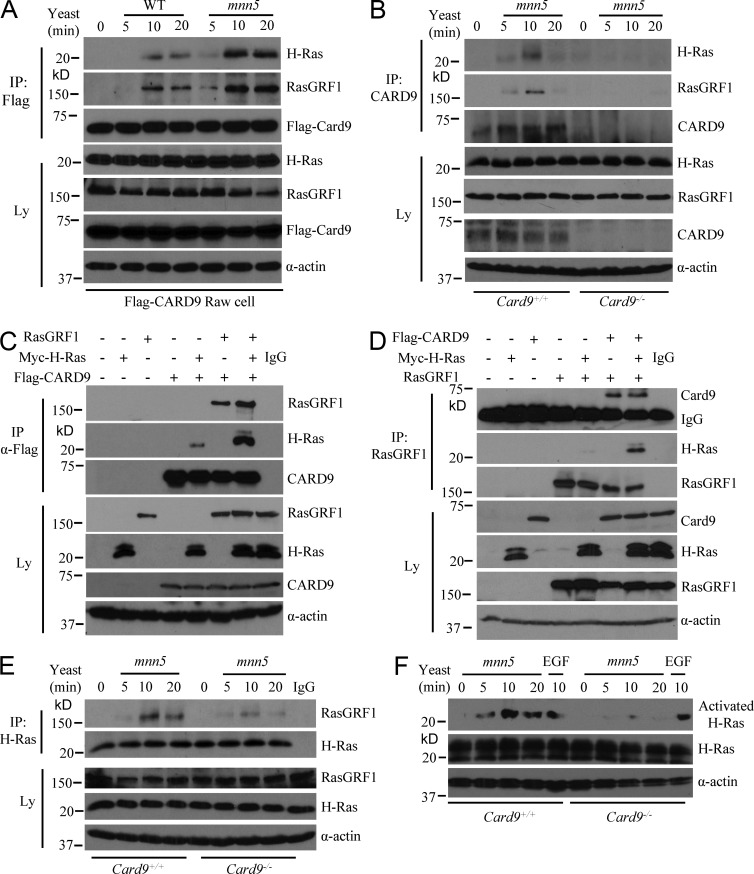
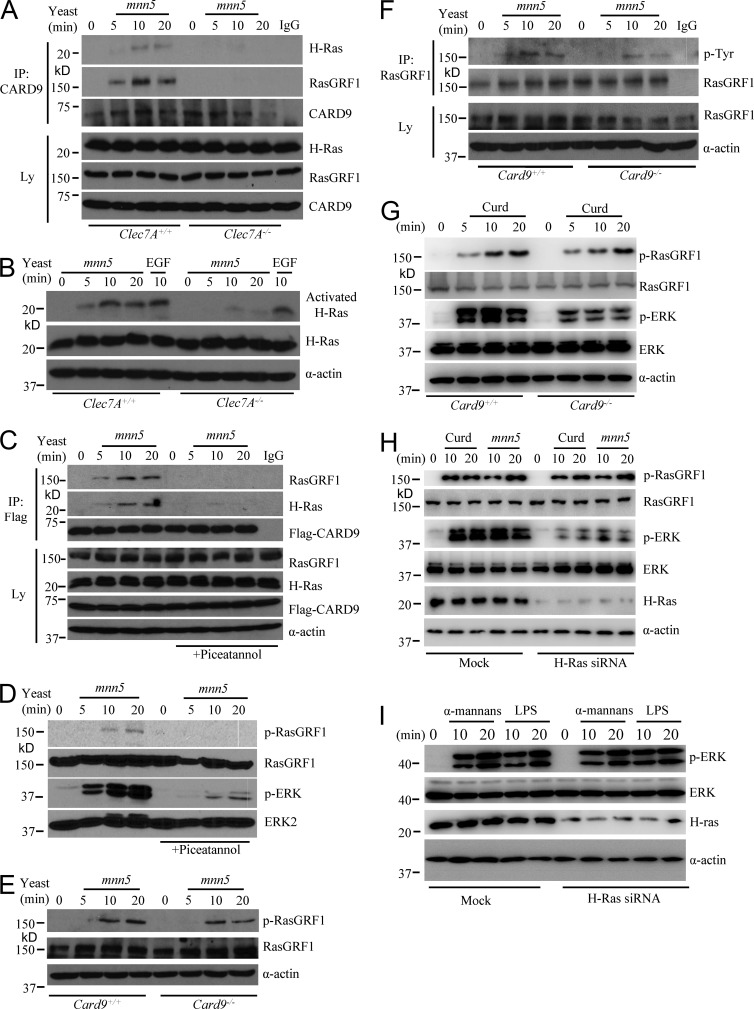
To determine the molecular mechanism by which CARD9 is linked to the signaling pathway leading to activation of ERK, we performed a Yeast Cyto-trap two-hybrid screening. Multiple positive clones, which encode H-Ras (a member of Ras family of small guanosine triphosphatases [GTPases]) and Ras-GRF1 (Ras-guanine-nucleotide-releasing factor 1), were identified in this screening (unpublished data), suggesting that CARD9 may associate with H-Ras and Ras-GRF1, which are upstream signaling components, leading to activation of ERK (Imamura et al., 2009; Wang et al., 2013). To examine whether CARD9 is associated with H-Ras and Ras-GRF1, we immunoprecipitated CARD9 from RAW264.7 cells expressing Flag-tagged CARD9 and found that immunoprecipitation of Flag-CARD9 effectively coprecipitated H-Ras and Ras-GRF1 after the stimulation by WT C. albicans yeast (Fig. 4 A). Consistently, mnn5 yeast stimulation induced a stronger association of CARD9 with both H-Ras and Ras-GRF1 (Fig. 4 A), suggesting that CARD9 inducibly forms a complex with H-Ras and Ras-GRF1 in Dectin-1–induced signaling cascade.
Figure 4.
CARD9 links Ras-GRF1 to H-Ras in response to C. albicans yeast stimulation. (A) RAW264.7 cells stably expressing Flag-CARD9 were stimulated with UV-inactivated WT or mnn5 yeast cells for indicated times. Cell lysates (Ly) were immunoprecipitated (IP) with anti-Flag antibody. (B) WT and CARD9-deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells for indicated times. Cell lysates were immunoprecipitated with α-CARD9 antibody. The cell lysate or immunoprecipitates were subjected to immunoblots using indicated antibodies. (C and D) HEK293T cells were transfected with expression vectors encoding Ras-GRF1, Myc-H-Ras, and Flag-CARD9 at different combinations. Cell lysates were subjected to immunoprecipitation with anti-Flag (C) or anti-Ras-GRF1 antibody (D). (E) WT and CARD9-deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells for indicated times. Cell lysates were immunoprecipitated with anti–H-ras antibody. Immunoprecipitated (IP) and lysate (Ly) fractions were analyzed by immunoblotting using the indicated antibodies. (F) WT and CARD9-deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells or EGF (as a control) for indicated times. Ras activation was determined by pull-down assay using a Ras activation assay Biochem kit according to the manufacturer’s instructions. Immunoassay of the total and activated Ras was performed for determining the Ras activation. Data shown are representative of three independent and reproducible experiments.
To further confirm the inducible association of CARD9 with H-Ras and Ras-GRF1, endogenous CARD9 was immunoprecipitated from BMDMs after stimulation by C. albicans yeasts. We found that CARD9 inducibly coimmunoprecipitated with both H-Ras and Ras-GRF1 after stimulation (Fig. 4 B), and this association was specific because CARD9 deficiency abolished this association (Fig. 4 B). Together, these results suggest that CARD9 associates with H-Ras and Ras-GRF1 in response to the stimulation of C. albicans yeasts.
Interestingly, CARD9 constitutively associated with H-Ras or Ras-GRF1 when they were coexpressed in HEK293T cells (Fig. 4 C). However, the association of CARD9 with H-Ras was significantly enhanced when three of them were coexpressed in 293T cells (Fig. 4 C). Similarly, although Ras-GRF1 weakly associated with H-Ras, this association was significantly enhanced upon coexpression of them with CARD9 (Fig. 4 D). Consistently, CARD9 deficiency impaired the yeast-induced association of Ras-GRF1 with H-Ras (Fig. 4 E). These results suggest that CARD9 bridges the interaction between Ras-GRF1 and H-Ras, which leads to activation of downstream signaling. Because Ras-GRF1 activates H-Ras (Zhu et al., 2007), we examined H-Ras activation and found that H-Ras could be effectively activated by C. albicans stimulation, whereas CARD9 deficiency impaired this activation (Fig. 4 F). Together, these results indicate that CARD9 mediates H-Ras activation through bridging Ras-GRF1 to H-Ras.
Stimulation of Dectin-1 by C. albicans yeast induces ERK activation though CARD9-bridged Ras-GRF1–H-Ras complex
To determine whether the formation of CARD9-bridged Ras-GRF1 and H-Ras complex is dependent on Dectin-1-induced signaling, we examined this complex formation after C. albicans yeast stimulation in WT (Clec7A+/+) and Dectin-1–deficient (Clec7A−/−) BMDMs, and found that this complex was completely defective in Dectin-1–deficient cells (Fig. 5 A). Consistently, C. albicans yeast stimulation-induced H-Ras activation was completely defective in Dectin-1–deficient BMDMs (Fig. 5 B). In addition, treatment of BMDMs with Syk inhibitor Piceatannol also blocked this complex formation (Fig. 5 C), and the yeast-induced phosphorylation of Ras-GRF1 as well as ERK activation (Fig. 5 D). In contrast, CARD9 deficiency did not significantly impair the yeast-induced Ras-GRF1 phosphorylation (Fig. 5, E and F), indicating that CARD9 functions downstream of Ras-GRF1. Similarly, CARD9 deficiency significantly impaired curdlan-induced phosphorylation of ERK, but not Ras-GRF1 (Fig. 5 G), indicating that CARD9 functions downstream of Ras-GRF1. Consistently, knockdown of endogenous H-Ras in BMDMs specifically inhibited curdlan- or yeast-induced phosphorylation of ERK (Fig. 5 H) but had no effect on α-mannans– or LPS-induced ERK activation (Fig. 5 I). Together, these results demonstrate that Dectin-1 signaling activated by β-glucans induces Syk-dependent phosphorylation of Ras-GRF1, and then the phosphorylated Ras-GRF1 activates H-Ras through scaffold protein CARD9, leading to the activation of ERK pathway.
Figure 5.
Dectin-1/Syk stimulation by C. albicans yeast induces the formation of a Ras-GRF1–CARD9–H-Ras complex. (A) WT and Dectin-1–deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells (MOI = 5) for the indicated times. Cell lysates were immunoprecipitated with anti-CARD9 antibody and immunoprecipitated (IP) and lysate (Ly) fractions were analyzed by immunoblotting using indicated antibodies. (B) WT and Dectin-1–deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells (MOI = 5) or EGF for indicated times. Ras activation was determined by pull-down assay using a Ras activation assay Biochem kit. Immunoassay of the total and activated Ras was performed for determining the Ras activation. (C) RAW264.7 cells stably expressing Flag-Card9 were pretreated with or without Syk inhibitor (piceatannol) for 30 min before stimulation with UV-inactivated mnn5 yeast cells (MOI = 5) for indicated times. Cell lysates were immunoprecipitated with anti-Flag antibody and analyzed by immunoblotting using indicated antibodies. (D) WT BMDMs were pretreated with or without piceatannol for 30 min before stimulation with UV-inactivated mnn5 yeast cells (MOI = 5) for the indicated times. (E) WT and CARD9-deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells (MOI = 5) for the indicated times. (F) WT and CARD9-deficient BMDMs were stimulated with UV-inactivated mnn5 yeast cells for indicated times. Cell lysates were immunoprecipitated with anti-Ras-GRF1 antibody and immunoprecipitated (IP) and lysate (Ly) fractions were analyzed by immunoblotting using indicated antibodies. (G) WT and CARD9-deficient BMDMs were stimulated with plate-coated curdlan (50 µg/ml) for the indicated times. (H and I) Knockdown of endogenous H-Ras by RNA interference in BMDMs, which were transfected with siRNA against murine H-Ras and nontargeting control siRNA using Trans IT-TKO transfection reagent (Mirus). Cells were cultured for 48 h after transfection and then stimulated with plate-coated curdlan (50 µg/ml), UV-inactivated mnn5 yeast cells (MOI = 5), plate-coated α-mannans (50 µg/ml), or LPS (100ng/ml) for the indicated times. Cell lysates were subjected to immunoblotting analysis using indicated antibodies. Data shown are representative of three independent and reproducible experiments.
The CARD9/H-Ras/ERK signaling cascade is critical for immune responses triggered by C. albicans yeast
To determine the functional effect of CARD9 on Dectin-1–induced cytokine production, we first treated C. albicans yeasts with UV light that fixes C. albicans at the yeast stage and prevents them from transforming into hyphae, and then stimulated WT or CARD9-deficient BMDMs with UV-inactivated C. albicans yeast cells. We found that secretion of cytokines including TNF, IL-6, and IL-1β had a significant reduction, whereas IL-12 production was only slightly affected in CARD9-deficient BMDMs (Fig. 6 A). In contrast, when BMDMs were stimulated with C. albicans hyphae, which induces proinflammatory signaling through the Dectin-2– and Dectin-3–dependent pathways (Saijo et al., 2010; Zhu et al., 2013), CARD9 deficiency was significantly impaired in the expression of these cytokines (Fig. 6 B). Furthermore, we examined the effect of inhibiting ERK signaling on cytokine production by WT or CARD9-deficient BMDMs, which were treated with or without U0126, an inhibitor for ERK activation. We found that U0126 significantly suppressed TNF, IL-6, and IL-1β production by WT BMDMs in response to Dectin-1 agonists C. albicans yeast (Fig. 6 C) and curdlan (Fig. 6 D). However, U0126 treatment did not significantly affect the expression of these cytokines in CARD9-deficient BMDMs after the stimulation by C. albicans yeast (Fig. 6 C) or curdlan (Fig. 6 D), suggesting that the inhibited signaling by U0126 may be largely overlapped with CARD9-induced signaling. Consistent with the effect on CARD9-deficient on TNF and IL-6 production, knockdown of endogenous H-Ras in BMDMs also significantly reduced curdlan- or yeast-induced production of TNF and IL-6 (Fig. 6 E). As controls, U0126 treatment and knockdown of endogenous H-Ras in BMDMs had no effect on α-mannans– or LPS-induced secretions of TNF, IL-6, IL-1β, and IL-12 (Fig. 6, F and G). Together, these results suggest that the activation of CARD9-mediated H-Ras/ERK cascade induced by Dectin-1 agonist is critical for the production of proinflammatory cytokines.
Figure 6.
CARD9-mediated H-Ras activation is critical for immune responses triggered by C. albicans yeast and curdlan. (A and B) ELISA results for cytokines TNF, IL-1β, IL-6, and IL-12p40 in supernatants of WT and CARD9-deficient BMDMs, which were stimulated for 6 h with UV-inactivated WT or mnn5 yeast (MOI = 5; A) or hyphae (MOI = 0.1; B). (C and D) ELISA results for cytokines TNF, IL-1β, IL-6, and IL-12p40 in supernatants of WT and CARD9-deficient BMDMs, which were pretreated with or without 5 µM U0126 for 30 min and then stimulated with UV-inactivated mnn5 yeast cells (MOI = 5; C) or plate-coated curdlan (50 µg/ml; D) for 6 h. (E and F) ELISA results for cytokines TNF, IL-1β, IL-6, and IL-12p40 in supernatants of WT BMDMs, which were transfected with siRNA against murine H-Ras and nontargeting control siRNA using Trans IT-TKO transfection reagent (Mirus). Cells were cultured for 48 h after transfection and then stimulated with plate-coated curdlan (50 µg/ml), UV-inactivated mnn5 yeast cells (MOI = 5), plate-coated α-mannans (50 µg/ml), or LPS (100 ng/ml) for 6 h. (G) ELISA results for cytokines TNF, IL-1β, IL-6, and IL-12p40 in supernatants of WT and CARD9-deficient BMDMs, which were pretreated with or without 5 µM U0126 for 30 min and then stimulated with plate-coated α-mannans (50 µg/ml) or LPS (100 ng/ml) for 6 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data are means ± SD of triplicate samples and are representative of three independent experiments.
ERK activation is critical for CARD9-mediated innate immunity against C. albicans
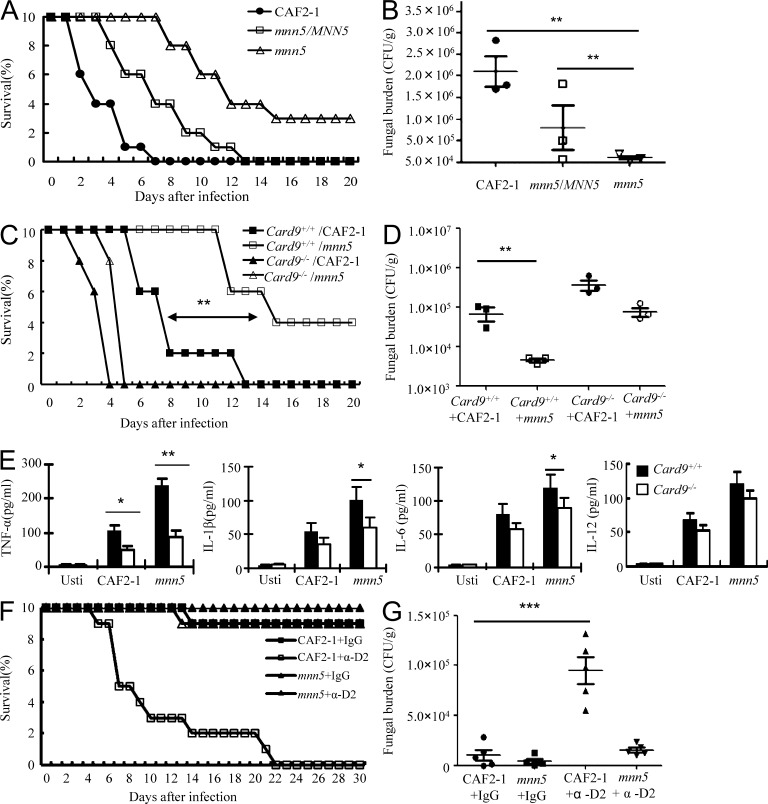
To examine the effect of β-glucan exposed on the surface of C. albicans yeast on host immune responses, we compared the pathogenicity of WT, mnn5 mutant, and MNN5-reconstituted mnn5 strains (mnn5/MNN5) of C. albicans in a murine model of systemic candidiasis. Consistent with a previous report (Bai et al., 2006), mnn5 mutant-infected mice survived significantly longer than CAF2-1 (the parental strain of C. albicans)– or mnn5/MNN5-infected mice with a lethal dose (1 × 106) of C. albicans (Fig. 7 A). The fungal burdens of kidneys in mice infected with mnn5 mutant were significantly lower than those infected with either CAF2-1 or mnn5/MNN5 strains (P < 0.01, Fig. 7 B). However, we found that all CARD9-deficient mice died within 5 d after infection with a sublethal dose (3 × 105) of WT (CAF2-1) or mnn5 mutant strains of C. albicans (P > 0.05, Fig. 7 C) and, consistently, fungal burden in the kidney of CARD9-deficient mice had no significant difference when challenging with CAF2-1 or mnn5 strains (P > 0.05, Fig. 7 D).
Figure 7.
Surface β-glucans exposed on C. albicans significantly enhance CARD9-mediated proinflammation responses against systemic candidiasis. (A and B) Infected mice survival and kidney CFU assay. Groups of C57B/L6 female mice were injected via lateral tail vein with 1 × 106 CFU of C. albicans WT (CAF2-1), mnn5 mutant, and its restored strain (mnn5/MNN5) in 200 µl sterile saline. Survival of these mice (n = 10 per group) was monitored and plotted (A). Kidney CFU assay (n = 5 per group) was performed at day 2 after infection (B). Shown are means and SD for n = 3. (C and D) WT (Card9+/+; squares) and CARD9-deficient mice (Card9−/−; triangles) were infected intravenously with 3 × 105 CFU of C. albicans WT (CAF2-1) and mnn5 mutant strains in 200 µl sterile saline. Survival of these mice (n = 10 per group) was monitored and plotted (C). Kidney CFU assay (n = 5 per group) was performed at day 2 after infection (D). Shown are means and SD for n = 3. (E) ELISA results for TNF, IL-6, IL-1β, and IL-12p40 in mouse sera at day 2 after infection. WT (Card9+/+) and CARD9-deficient (Card9−/−) mice (n = 5 per group) were challenged with UV-inactivated C. albicans WT (CAF2-1) and mnn5 mutant yeast cells (1 × 106) in 200 µl sterile saline. Data shown are representative of three independent experiments. SDs are indicated. (F and G) Infected mice survival and kidney CFU assay. Groups of C57B/L6 female mice infected with 6 × 104 CFU of C. albicans WT (CAF2-1) and mnn5 mutant strains, which were treated with 200 µg anti–Dectin-2 monoclonal antibodies (α-D2) or nonspecific control IgG per mouse for four times at 6 h before or 2, 4, or 6 d after injection of C. albicans. Survival of these mice (n = 10 per group) was monitored and plotted (F). Kidney CFU assay (n = 5 per group) was performed at day 2 after infection (G). Shown are means and SD for n = 5. Two independent experiments were conducted (n = 10 in each group). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further confirm the effect of CARD9-mediated immune responses on the clearance ability to systemic C. albicans infections, we challenged WT or CARD9-deficient mice with UV-inactivated WT (CAF2-1) or mnn5 mutant C. albicans. Similar to the in vitro challenging of macrophages by UV-inactivated C. albicans (Fig. 6 A), CARD9-deficient mice had significantly lower levels of TNF, IL-6, and IL-1β production in their serum at day 2 after infected with mnn5 mutant, but only TNF production had a significant reduction when infecting CARD9-deficient mice with CAF2-1 strain (Fig. 7 E). To determine the role of Dectin-2–mediated host defense against systemic infections with mnn5 mutant, mice receiving anti–Dectin-2 or nonspecific control IgG were intravenously injected with a sublethal dose (6 × 104) of WT (CAF2-1) or mnn5 mutant strains. Blocking Dectin-2 significantly decreased the survival rate of CAF2-1–infected mice (P < 0.01; Fig. 7 F) but had very little impact on mnn5 mutant-infected mice (Fig. 7 F). The fungal burden in kidney of these mice consistently showed similar phenotypes (Fig. 7 G). Together, these data suggest that mnn5 mutant C. albicans is virulent to CARD9-deficient mice, and that the faster clearance of mnn5 mutant of C. albicans by WT mice may be due to a stronger activation of β-glucan–induced Dectin-1–, but not Dectin-2–, and CARD9-dependent inflammatory responses.
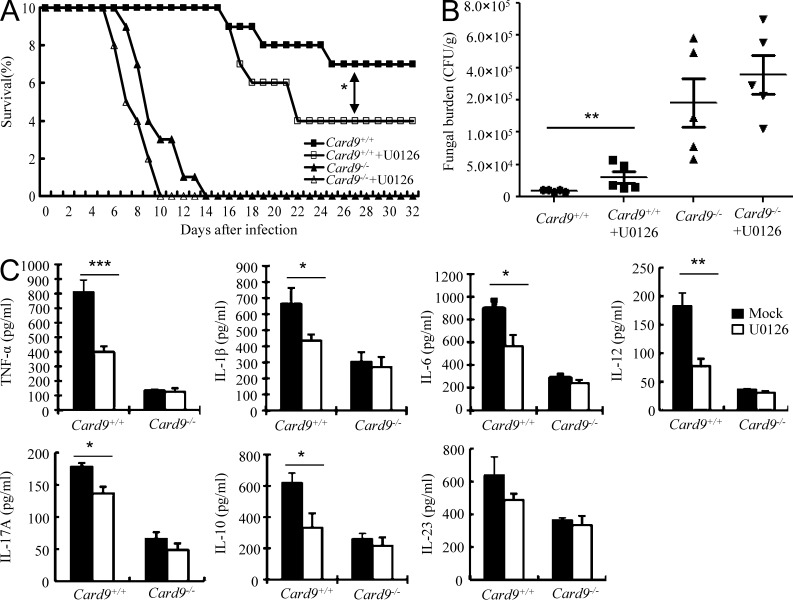
To determine the contribution of CARD9-mediated ERK activation on antifungal immunity in vivo, we pretreated WT or CARD9-deficient mice with or without U0126, and then intravenously infected these mice with a low dose (5 × 104) of mnn5 mutant. We found that administration of U0126 reduced the survival rate of WT mice (P < 0.05; Fig. 8 A). However, administration of U0126 did not have a significant effect on the survival of CARD9-defecient mice (P > 0.05; Fig. 8 A). Consistently, U0126 treatment only significantly affected the fungal load (P < 0.01; Fig. 8 B) and cytokine production (Fig. 8 C) in the kidney of WT but not CARD9-defecient mice after mnn5 infection, suggesting that the contribution of ERK activation on antifungal immunity is largely dependent on CARD9-mediated signaling.
Figure 8.
ERK activation is critical for CARD9-mediated innate immunity against C. albicans. (A and B) WT and CARD9-deficient mice were intravenously injected with 5 × 104 CFU of mnn5. Infected mice were treated with or without U0126 (50 µg per mouse) at days 0, 2, 4, and 6 after infection. Survival of infected mice (n = 10 per group) was monitored (A) and kidney CFU assay (n = 5 per group) was performed at day 2 after infection (B). *, P < 0.05; **, P < 0.01. (C) ELISA results of TNF, IL-1β, IL-6, IL-12p40, IL-17A, IL-10, and IL-23 in the extracts of homogenized kidneys from WT and CARD9-deficient mice 4 d after infection with 5 × 104 CFU of mnn5. Infected mice were treated with or without 50 µg U0126 per mouse at days 0 and 2 after infection. *, P < 0.05; **, P < 0.01; ***, P < 0.001, n = 5 per group. Data shown are representative of three independent experiments. SDs are indicated.
DISCUSSION
During C. albicans infection, both yeast and hyphae forms can be found in infected tissues, and innate immune cells discriminate them through different PRRs, especially CLRs, which elicit protective immune responses against this infection. Previous studies have shown that CARD9, a scaffold protein, is required to mediate different CLR-induced NF-κB activation after fungal infection. In this study, we found that CARD9 is dispensable for Dectin-1–induced NF-κB activation but it is required for Dectin-1–induced ERK activation with stimulation of C. albicans yeasts, whereas Dectin-2–induced NF-κB activation is completely dependent on CARD9 with stimulation of C. albicans hyphae. Mechanistically, we found that CARD9 links Ras-GRF1 to H-Ras and regulates the activation of H-Ras, leading to activation of downstream ERK activation. Together, we have revealed a previously unknown mechanism by which Dectin-1 induces H-Ras activation that, in turn, induces downstream ERK activation.
In this study, we have also found that CARD9-mediated signaling had more significant contributions to Dectin-2–mediated proinflammatory responses than those mediated by Dectin-1, including induction of IL-1β and IL-6, two pivotal cytokines for priming downstream Th17 differentiation. Consistent with our observations, CARD9 has been shown to be indispensable for development of Th17 responses to C. albicans in mice, mostly through Dectin-2, and, to a lesser extent, Dectin-1 signaling (Robinson et al., 2009). Therefore, the CARD9-mediated different transcriptional activation programs are at the convergence of innate and adaptive antifungal immune responses downstream of Dectin-1 and Dectin-2.
Signaling from Dectin-1 after ligand binding is mediated through the cytoplasmic ITAM-like motif that becomes phosphorylated by Src family kinases, providing a docking site for Syk (Brown, 2006). Therefore, Syk is a pivotal kinase mediating Dectin-1–induced downstream cellular responses, such as cytokine production and induction of the respiratory burst (Brown, 2006). Although the components of the Dectin-1–Syk signaling pathway have yet to be fully elucidated, CARD9, which assembles with BCL10 and MALT1 (Bertin et al., 2000), has been identified as an essential downstream adaptor linking Syk-coupled CLRs to the canonical NF-κB pathway (Gross et al., 2006; Hara et al., 2007). However, in our study, we have found that NF-κB activation induced by Dectin-1 ligands, both curdlan and mutant C. albicans cells with highly exposed β-glucans, are intact in CARD9-deficient cells. These results indicate that CARD9 is dispensable for Dectin-1–induced NF-κB activation. One possible explanation for our observations is that these Dectin-1 ligands may be able to activate NF-κB through both CARD9-dependent and CARD9-independent pathways. In this case, these Dectin-1 ligands may activate another undefined coreceptor leading to activation of NF-κB via CARD9-independent pathway. In contrast, because this undefined coreceptor cannot induce ERK activation, ERK activation by Dectin-1 ligands is only dependent on CARD9-mediated signaling pathway.
It has been shown that Dectin-1 engagement by the cell wall extract zymosan or C. albicans yeasts in DCs triggers Syk and ERK pathway activation (Rogers et al., 2005; Dillon et al., 2006; Slack et al., 2007; Galán-Díez et al., 2010). However, it has not been shown how Dectin-1 signaling pathway leads to ERK activation. Our studies provide a molecular mechanism by which Dectin-1 activates ERK. In this case, stimulation of Dectin-1 with C. albicans yeasts induces Syk-dependent phosphorylation of Ras-GRF1, and the phosphorylated Ras-GRF1, in turn, forms a complex with CARD9 that further recruits H-Ras, leading to activation of H-Ras and subsequent activation of downstream ERK pathway.
It has been well documented that the small G protein H-Ras is a key regulator of many signal transduction pathways (Rebollo and Martínez-A, 1999). In its active form, H-Ras can activate a variety of downstream targets, including the ERK cascade (Takai et al., 2001). H-Ras is activated by GEFs (guanine nucleotide exchange factors) that promote the release of guanosine diphosphate (GDP) bound to H-Ras and exchange for the activating GTP (Shou et al., 1992). Ras-GRF1 catalyzes the GDP/GTP exchange needed for H-Ras activation (Quilliam et al., 2002). Our findings indicate that CARD9 mediates H-Ras activation through linking Ras-GRF1 to H-Ras in response to C. albicans yeast stimulation.
It is interesting to note that CARD9 regulates the activation of another small G protein Rac1 through dissociating LyGDI (lymphoid-specific GDP dissociation inhibitor) from Rac1 after Listeria monocytogenes stimulation (Wu et al., 2009). The activation of Rac1 induces reactive oxygen species (ROS) production, which contributes to the killing of intracellular bacteria L. monocytogenes. Therefore, CARD9-deficient macrophages are defective in ROS production in response to L. monocytogenes infection and are impaired in intracellular bacterial killing (Wu et al., 2009). Consistently, CARD9-deficient mice exhibit hyper-susceptibility to infection of L. monocytogenes (Hara et al., 2007; Hsu et al., 2007). Future studies should also determine whether CARD9 regulates Rac1 GEFs besides Rac1 GDI.
Up to now, there have only been limited studies about the roles of ERK cascade on host defense against fungal infections. It has been shown that the ERK cascade plays a minor role in modulating the killing of C. albicans by neutrophil (Hii et al., 1999) but plays a more important role for promoting the killing of C. albicans by macrophages (Ibata-Ombetta et al., 2001). However, inhibition of the ERK signaling pathway abolished neutrophil migration induced by C. albicans hyphae and selectively impaired their ability to kill C. albicans (Wozniok et al., 2008). Another study shows that activation of the ERK cascade by C. albicans hyphae, but not yeasts, is necessary to induce a cytokine response by oral epithelial cells (Moyes et al., 2010). In this study, our data shows that β-glucan exposed on the surface of C. albicans yeast can induce strong activation of both NF-κB and ERK pathways through Dectin-1 signaling. However, Dectin-1–induced ERK activation is completely dependent on the adaptor protein CARD9. Furthermore, our in vivo data shows that treatment with an ERK inhibitor significantly accelerates the death of C. albicans–infected mice in a murine systemic candidiasis model, and this inhibitory effect is largely dependent on the presence of CARD9. Therefore, we conclude that Dectin-1–mediated antifungal immune responses to C. albicans infection are mainly through CARD9-mediated ERK activation both in vitro and in vivo.
Dectin-1 genes are highly conserved across mammals, including human and mouse, indicating that the roles of Dectin-1 in innate immunity are conserved among different species. The impact of Dectin-1 deficiency on antifungal immunity has been shown in Dectin-1–deficient mice (Taylor et al., 2007). Moreover, the importance of Dectin-1 in human antifungal immunity has shown that human patients with Dectin-1 Y238X mutation, in which the protein translation of Dectin-1 gene is prematurely terminated at Tyrosine 238, are highly susceptible to fungal infection (Ferwerda et al., 2009; Cunha et al., 2010; Chai et al., 2011). Homozygotes of this mutation are associated with mucocutaneous infections, including vaginal candidiasis and onychomycosis, even though fungal killing and phagocytosis occur normally (Plantinga et al., 2009; van der Velden et al., 2010). The higher susceptibility of patients with Dectin-1 mutations is likely due to the defect of antifungal innate immune responses induced by Dectin-1–mediated NF-κB and MAPK activation.
Previous studies indicate that CARD9 is required for NF-κB activation induced by several CLRs including Dectin-1 and Dectin-2 in response to fungal infection (Gross et al., 2006; Robinson et al., 2009; Bi et al., 2010; Saijo et al., 2010). Our findings that CARD9 also regulates H-Ras activation and mediates Dectin-1–induced ERK activation further indicate the importance of CARD9 in antifungal immunity. Consistent with the critical role of CARD9 in NF-κB activation induced by multiple CLRs and ERK activation by Dectin-1, patients with CARD9 mutations (Glocker et al., 2009; Lanternier et al., 2013; Wang et al., 2014) are susceptible to the infection by a broad range of different fungal species and appear to have more severe fungal infection than those found in patients with Dectin-1 mutations (Ferwerda et al., 2009; Cunha et al., 2010; Chai et al., 2011). Considering the emerging information of the complexity of CLR signaling, further investigating CARD9-associated signaling cascades may provide novel therapeutic insight for designing antifungal agents.
MATERIALS AND METHODS
Plasmids.
Human Dectin-1 and Dectin-2 were amplified by PCR using full-length cDNA of human peripheral blood cells as a template. All PCR amplifying fragments, including the Flag-encoding DNA sequence, were inserted into a lentivirus vector, pRV3, between the SalI and BglII sites. To obtain GFP/Dectin-1CRD fusion gene, the method of overlapping PCR was used as described previously (Guan et al., 2007). Four primers for PCR were designed as follows based on the GFP and the Dectin-1CRD encoding sequence: GFP (sense), 5′-CAGGACATATGGTGAGCAAGGGCGAG-3′; GFP (anti-sense), 5′-GCTGCCACCTCCACCGCTACCGCCGCCTCCCTTGTACAGCTC-3′; Dectin-1CRD (sense), 5′-GGTGGAGGTGGCAGCTTTTGGCGACACAAT-3′; and Dectin-1CRD (anti-sense), 5′-ACTGAGGGATCCCAGTTCCTTCTCA-3′. The GFP (anti-sense) and Dectin-1CRD (sense) primers contained a (Gly4Ser)2 linker encoding sequence (boxed). The fusion PCR product was digested by NdeI and BamHI and was inserted in frame into pET28a expression vector (EMD Millipore).
Reagents and antibodies.
Antibodies against phospho-IKKα/β, phospho-IκBα, phospho-ERK, phospho-p38, and p38 were purchased from Cell Signaling Technology; antibodies against p65, IKKα, IκBα, ERK, H-Ras, Ras-GRF1, α-actin, and PCNA were from Santa Cruz Biotechnology. Curdlan and α-mannans were from Wako and Sigma-Aldrich, respectively.
Generation of mnn5 null mutant and its revertant strains.
To construct mnn5 null mutant, the fragment containing 5′ and 3′ ends of MNN5 gene for homologous recombination and the 4-kb hisG-URA3-hisG fragment from the plasmid p5921 were subsequently cloned into plasmid pUCm-t (Sangon). The XhoI digested fragment of pUC-MNN5-URA3 was transformed into the ura3Δ/Δ mutant (CAI4) by standard methods (Sanglard et al., 1996). To construct mnn5 mutant revertant strain, MNN5 containing their ORFs and 1 kb up/downstream were amplified by PCR with Pyrobest polymerase (Takara Bio Inc.) and cloned into plasmid pBes116 to obtain plasmid pBes-MNN5. Then, the AscI digested fragment of pBes-MNN5 was transformed into mnn5 null mutant as previously (Sanglard et al., 1996).
C. albicans growth and preparation.
C. albicans yeast cells were grown overnight in YPD rich medium at 30°C. For hyphae, yeast cells were counted and then resuspended in RPMI 1640 to grow at 37°C for 3 h.
Inactivation of C. albicans strains.
Inactivation of C. albicans strains was performed as described previously (Wheeler and Fink, 2006). For UV inactivation of C. albicans, the equivalent of 2.5 × 107 cells from a culture were washed and resuspended in 1 ml of PBS in a 6-well plate. The fungi were exposed to four doses of 100,000 µjoules/cm2 in a CL-1000 UV-cross-linker (UVP; Upland), with agitation between each dose to treat cells evenly.
Screening for β-glucan exposure.
C. albicans yeast cells or hyphae were stained with anti-β-1,3-glucan primary antibody (Biosupplies Inc.) and FITC-labeled goat anti–mouse secondary. Stained cells were adhered to clear-bottom plates with Concanavalin A (Sigma-Aldrich) and scanned under an oil immersion objective at 100× magnification on a confocal microscope (TCS SP5; Leica).
BMDM preparation.
Dectin-1−/− mice were provided by G. Brown (University of Aberdeen, Aberdeen, Scotland). Primary cultures of BMDMs from WT or Decitn-1−/−, CARD9−/−, or Bcl10−/−mice were prepared as previously described (Bi et al., 2010; Gorjestani et al., 2011). In brief, BM cells were harvested from the femurs and tibias of mice. Erythrocytes were removed from cells samples by subjecting the samples to hypotonic solution. Cells were cultured for 7 d in DMEM containing 20% FBS, 55 µM β-mercaptoethanol, 100 µg/ml streptomycin, 100 U/ml penicillin, and 30% conditioned media from L929 cells expressing macrophage CSF (M-CSF). Non-adherent cells were removed and cells were passed every 3 d. After 1 wk of cultures, flow cytometry analysis indicated that the harvested cell population contained 86–95% CD11b+ F4/80+cells as assessed.
BMDC preparation.
BMDCs were prepared from mouse femurs and tibias by culture of BM cells for 6 d in complete RPMI 1640 (10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 55 µM β-mercaptoethanol) supplemented with 25 ng/ml GM-CSF (PeproTech) and 1 ng/ml IL-4 (PeproTech). After 6 d of cultures, BMDCs were purified by magnetic selection with anti-CD11c microbeads. This routinely gave purities of >98%.
Yeast cyto-trap two-hybrid system screening.
Human CARD9 coding sequence was inserted into pSOS vector and used to screen its binding partners by using CytoTrap XR Mouse Spleen cDNA Library according to the manufacture protocol (Agilent Technologies).
Knockdown of endogenous H-Ras by RNA interference.
BMDMs were plated in a 6-well plate (1 × 106 cells/well) in 1 ml DMEM containing 10% FBS. Cells at 50–80% knockdown efficiency were transfected with different concentrations of siRNA against murine H-Ras. Trans IT-TKO transfection reagent was used to transfect siRNA and nontargeting control siRNA according to the manufacturer’s protocol (Mirus). Cells were lysed 48 h after transfection, and relative intensities of H-Ras protein were determined by Western blotting.
Western blot and immunoprecipitation.
BMDMs were serum-starved overnight, stimulated, and lysed in lysis buffer (150 mM NaCl, 50 mM Hepes, pH 7.4, 1 mM EDTA, 1% Nonidet P-40, and protease inhibitors). The cell lysates were immunoprecipitated with the indicated antibody-conjugated agarose. The resulting immunoprecipitates and lysates were subjected to SDS-PAGE and then blotted using the indicated antibodies.
Electrophoretic mobility shift assay.
After BMDMs were stimulated with yeast cells or hyphae, their nuclear extracts were prepared. 5 µg of the resulting nuclear protein was incubated with 32P-labeled NF-κB or Oct-1 probe (Promega) for 15 min at 25°C, and then subjected to PAGE and exposed to x-ray films.
Cytokine measurements.
The enzyme-linked immunosorbent assay kit for TNF, IL-6, IL-12p40, and IL-1β ELISA Ready-SET-GO kits were purchased from eBioscience. All of the samples were measured in triplicate times according to the manufacturer’s protocol.
Ras pull-down assay.
Ras activation was determined by pull-down assay using a Ras activation assay Biochem kit (Cytoskeleton, Inc.) according to the manufacturer’s instructions. In brief, left ventricular myocardium sample (∼100 mg) was homogenized in lysis buffer. Equal amounts of protein (∼200 µg) from left ventricular samples were incubated at 4°C for 1 h with Raf-RBD (Ras binding domain) beads (40 µl), which specifically recognize the GTP-bound form of Ras. Immunoassay of the total and activated Ras was performed for determining the Ras activation.
Murine systemic candidiasis model.
For the in vivo C. albicans infection, a group of C57B/L6 and Card9−/− female mice were injected via lateral tail vein with 200 µl of a suspension containing a sublethal dose (3 × 105) or a low dose (5 × 104) of CAF2-1 or mnn5 strain of C. albicans yeast cells in sterile saline. Kaplan–Meier and Life Table analyses were used to estimate survival probabilities. All animal experiments were performed in compliance with the institutional guidelines and according to the protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center and the Animal Ethics Committee of Tongji University. For cytokine measurement, sera from infected mice or cultured media from DMEMs after fungal infections were used and detected by ELISA kits (eBioscience).
Ethics statement.
All the animal experiments were performed in compliance with the institutional guidelines and according to the protocol approved by the Institutional Animal Use and Care Committee of the University of Pittsburg, the Institutional Animal Care and Use Committee of MD Anderson Cancer Center, and the Animal Ethics Committee of Tongji University.
Statistical analysis.
At least two biological replicates were performed for all experiments unless otherwise indicated. Log-rank testing was used to evaluate the equality of survival curves. Student’s t test for paired observations was used for statistical analyses of cytokine expression levels. Statistical significance was set at a p-value of <0.05, <0.01, or <0.001.
Acknowledgments
We would like to thank Dr. G. Brown for providing Dectin-1 KO mice.
This work was supported by Shanghai Basic Research Program (12JC1408401; X.-M. Jia), National Basic Research Program of China (Program 973, 2013CB967500; G.-T. Xu), and National Institutes of Health grant R01AI050848 (X. Lin), and X. Lin is a Changjiang Lecture Professor of the Ministry of Education, China.
The authors declare no competing financial interests.
Author contributions: X.-M. Jia, B. Tang, L.-L. Z.hu, Y.-H. Liu, X.-Q. Zhao, S. Gorjestani, Y.-M.S. Hsu, L. Yang, and J.-H. Guan performed experiments. G.-T. Xu, X.-M. Jia, and X. Lin directed the project and wrote the paper.
Footnotes
Abbreviations used:
- Bcl10
- B cell leukemia-lymphoma 10
- BMDC
- BM-derived dendritic cell
- BMDM
- BM-derived macrophage
- CARD9
- caspase recruitment domain-containing protein 9
- CLR
- C-type lectin receptor
- CRD
- extracellular carbohydrate-binding domain
- ERK
- extracellular signal-regulated protein kinase
- GDP
- guanosine diphosphate
- GTPase
- guanosine triphosphatase
- ITAM
- immunoreceptor tyrosine-based activation motif
- Malt1
- mucosa-associated lymphoid tissue 1
- PRR
- pattern recognition receptor
- RLR
- retinoic acid-inducible gene I–like helicase
- Syk
- spleen tyrosine kinase
References
- Bai, C., Xu X.L., Chan F.Y., Lee R.T.H., and Wang Y.. 2006. MNN5 encodes an iron-regulated α-1,2-mannosyltransferase important for protein glycosylation, cell wall integrity, morphogenesis, and virulence in Candida albicans. Eukaryot. Cell. 5:238–247. 10.1128/EC.5.2.238-247.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin, J., Guo Y., Wang L., Srinivasula S.M., Jacobson M.D., Poyet J.-L., Merriam S., Du M.-Q., Dyer M.J., Robison K.E., et al. 2000. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-κB. J. Biol. Chem. 275:41082–41086. 10.1074/jbc.C000726200 [DOI] [PubMed] [Google Scholar]
- Bi, L., Gojestani S., Wu W., Hsu Y.M.S., Zhu J., Ariizumi K., and Lin X.. 2010. CARD9 mediates dectin-2-induced IκBα kinase ubiquitination leading to activation of NF-κB in response to stimulation by the hyphal form of Candida albicans. J. Biol. Chem. 285:25969–25977. 10.1074/jbc.M110.131300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.D.2006. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat. Rev. Immunol. 6:33–43. 10.1038/nri1745 [DOI] [PubMed] [Google Scholar]
- Brown, G.D., and Gordon S.. 2001. Immune recognition. A new receptor for β-glucans. Nature. 413:36–37. 10.1038/35092620 [DOI] [PubMed] [Google Scholar]
- Cargnello, M., and Roux P.P.. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75:50–83. 10.1128/MMBR.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, L.Y., de Boer M.G., van der Velden W.J., Plantinga T.S., van Spriel A.B., Jacobs C., Halkes C.J., Vonk A.G., Blijlevens N.M., van Dissel J.T., et al. 2011. The Y238X stop codon polymorphism in the human β-glucan receptor dectin-1 and susceptibility to invasive aspergillosis. J. Infect. Dis. 203:736–743. 10.1093/infdis/jiq102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha, C., Di Ianni M., Bozza S., Giovannini G., Zagarella S., Zelante T., D’Angelo C., Pierini A., Pitzurra L., Falzetti F., et al. 2010. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood. 116:5394–5402. 10.1182/blood-2010-04-279307 [DOI] [PubMed] [Google Scholar]
- Dillon, S., Agrawal S., Banerjee K., Letterio J., Denning T.L., Oswald-Richter K., Kasprowicz D.J., Kellar K., Pare J., van Dyke T., et al. 2006. Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J. Clin. Invest. 116:916–928. 10.1172/JCI27203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, A., Popp M.W., Vyas V.K., Strijbis K., Ploegh H.L., and Fink G.R.. 2011. Fungal recognition is mediated by the association of dectin-1 and galectin-3 in macrophages. Proc. Natl. Acad. Sci. USA. 108:14270–14275. 10.1073/pnas.1111415108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferwerda, G., Meyer-Wentrup F., Kullberg B.J., Netea M.G., and Adema G.J.. 2008. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell. Microbiol. 10:2058–2066. 10.1111/j.1462-5822.2008.01188.x [DOI] [PubMed] [Google Scholar]
- Ferwerda, B., Ferwerda G., Plantinga T.S., Willment J.A., van Spriel A.B., Venselaar H., Elbers C.C., Johnson M.D., Cambi A., Huysamen C., et al. 2009. Human dectin-1 deficiency and mucocutaneous fungal infections. N. Engl. J. Med. 361:1760–1767. 10.1056/NEJMoa0901053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán-Díez, M., Arana D.M., Serrano-Gómez D., Kremer L., Casasnovas J.M., Ortega M., Cuesta-Domínguez A., Corbí A.L., Pla J., and Fernández-Ruiz E.. 2010. Candida albicans β-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect. Immun. 78:1426–1436. 10.1128/IAI.00989-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner, B.N., Simmons R.M., and Underhill D.M.. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277–1286. 10.1038/sj.emboj.7600594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker, E.-O., Hennigs A., Nabavi M., Schäffer A.A., Woellner C., Salzer U., Pfeifer D., Veelken H., Warnatz K., Tahami F., et al. 2009. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med. 361:1727–1735. 10.1056/NEJMoa0810719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge, H.S., Reyes C.N., Becker C.A., Katsumoto T.R., Ma J., Wolf A.J., Bose N., Chan A.S., Magee A.S., Danielson M.E., et al. 2011. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature. 472:471–475. 10.1038/nature10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjestani, S., Yu M., Tang B., Zhang D., Wang D., and Lin X.. 2011. Phospholipase Cγ2 (PLCγ2) is key component in Dectin-2 signaling pathway, mediating anti-fungal innate immune responses. J. Biol. Chem. 286:43651–43659. 10.1074/jbc.M111.307389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis, S.I., den Dunnen J., Litjens M., van der Vlist M., Wevers B., Bruijns S.C., and Geijtenbeek T.B.. 2009. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-κB activation through Raf-1 and Syk. Nat. Immunol. 10:203–213. 10.1038/ni.1692 [DOI] [PubMed] [Google Scholar]
- Gross, O., Gewies A., Finger K., Schäfer M., Sparwasser T., Peschel C., Förster I., and Ruland J.. 2006. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 442:651–656. 10.1038/nature04926 [DOI] [PubMed] [Google Scholar]
- Guan, Z., Yao W., Ye J., Dan W., Shen J., and Zhang S.. 2007. The construction and characterization of a bifunctional EGFP/sAPRIL fusion protein. Appl. Microbiol. Biotechnol. 73:1114–1122. 10.1007/s00253-006-0591-3 [DOI] [PubMed] [Google Scholar]
- Hara, H., Ishihara C., Takeuchi A., Imanishi T., Xue L., Morris S.W., Inui M., Takai T., Shibuya A., Saijo S., et al. 2007. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat. Immunol. 8:619–629. 10.1038/ni1466 [DOI] [PubMed] [Google Scholar]
- Heinsbroek, S.E.M., Taylor P.R., Martinez F.O., Martinez-Pomares L., Brown G.D., and Gordon S.. 2008. Stage-specific sampling by pattern recognition receptors during Candida albicans phagocytosis. PLoS Pathog. 4:e1000218. 10.1371/journal.ppat.1000218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hii, C.S., Stacey K., Moghaddami N., Murray A.W., and Ferrante A.. 1999. Role of the extracellular signal-regulated protein kinase cascade in human neutrophil killing of Staphylococcus aureus and Candida albicans and in migration. Infect. Immun. 67:1297–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, Y.-M.S., Zhang Y., You Y., Wang D., Li H., Duramad O., Qin X.-F., Dong C., and Lin X.. 2007. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat. Immunol. 8:198–205. 10.1038/ni1426 [DOI] [PubMed] [Google Scholar]
- Ibata-Ombetta, S., Jouault T., Trinel P.-A., and Poulain D.. 2001. Role of extracellular signal-regulated protein kinase cascade in macrophage killing of Candida albicans. J. Leukoc. Biol. 70:149–154 [PubMed] [Google Scholar]
- Imamura, Y., Oda A., Katahira T., Bundo K., Pike K.A., Ratcliffe M.J., and Kitamura D.. 2009. BLNK binds active H-Ras to promote B cell receptor-mediated capping and ERK activation. J. Biol. Chem. 284:9804–9813. 10.1074/jbc.M809051200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrigan, A.M., and Brown G.D.. 2011. Syk-coupled C-type lectins in immunity. Trends Immunol. 32:151–156. 10.1016/j.it.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingeter, L.M., and Lin X.. 2012. C-type lectin receptor-induced NF-κB activation in innate immune and inflammatory responses. Cell. Mol. Immunol. 9:105–112. 10.1038/cmi.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel, N., Cui S., Groebe L., and Bilitewski U.. 2010. Deletion of the Candida albicans histidine kinase gene CHK1 improves recognition by phagocytes through an increased exposure of cell wall β-1,3-glucans. Microbiology. 156:3432–3444. 10.1099/mic.0.040006-0 [DOI] [PubMed] [Google Scholar]
- Lanternier, F., Pathan S., Vincent Q.B., Liu L., Cypowyj S., Prando C., Migaud M., Taibi L., Ammar-Khodja A., Boudghene Stambouli O., et al. 2013. Deep dermatophytosis and inherited CARD9 deficiency. N. Engl. J. Med. 369:1704–1714. 10.1056/NEJMoa1208487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov, R.2009. Approaching the asymptote: 20 years later. Immunity. 30:766–775. 10.1016/j.immuni.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Moyes, D.L., Runglall M., Murciano C., Shen C., Nayar D., Thavaraj S., Kohli A., Islam A., Mora-Montes H., Challacombe S.J., and Naglik J.R.. 2010. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe. 8:225–235. 10.1016/j.chom.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea, M.G., van Tits L.J., Curfs J.H., Amiot F., Meis J.F., van der Meer J.W., and Kullberg B.J.. 1999. Increased susceptibility of TNF-α lymphotoxin-α double knockout mice to systemic candidiasis through impaired recruitment of neutrophils and phagocytosis of Candida albicans. J. Immunol. 163:1498–1505 [PubMed] [Google Scholar]
- Palma, A.S., Feizi T., Zhang Y., Stoll M.S., Lawson A.M., Díaz-Rodríguez E., Campanero-Rhodes M.A., Costa J., Gordon S., Brown G.D., and Chai W.. 2006. Ligands for the β-glucan receptor, Dectin-1, assigned using “designer” microarrays of oligosaccharide probes (neoglycolipids) generated from glucan polysaccharides. J. Biol. Chem. 281:5771–5779. 10.1074/jbc.M511461200 [DOI] [PubMed] [Google Scholar]
- Plantinga, T.S., van der Velden W.J., Ferwerda B., van Spriel A.B., Adema G., Feuth T., Donnelly J.P., Brown G.D., Kullberg B.J., Blijlevens N.M., and Netea M.G.. 2009. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 49:724–732. 10.1086/604714 [DOI] [PubMed] [Google Scholar]
- Quilliam, L.A., Rebhun J.F., and Castro A.F.. 2002. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 71:391–444. 10.1016/S0079-6603(02)71047-7 [DOI] [PubMed] [Google Scholar]
- Rebollo, A., and Martínez-A C.. 1999. Ras proteins: recent advances and new functions. Blood. 94:2971–2980 [PubMed] [Google Scholar]
- Robinson, M.J., Osorio F., Rosas M., Freitas R.P., Schweighoffer E., Gross O., Verbeek J.S., Ruland J., Tybulewicz V., Brown G.D., et al. 2009. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J. Exp. Med. 206:2037–2051. 10.1084/jem.20082818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, N.C., Slack E.C., Edwards A.D., Nolte M.A., Schulz O., Schweighoffer E., Williams D.L., Gordon S., Tybulewicz V.L., Brown G.D., and Reis e Sousa C.. 2005. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 22:507–517. 10.1016/j.immuni.2005.03.004 [DOI] [PubMed] [Google Scholar]
- Romani, L.2004. Immunity to fungal infections. Nat. Rev. Immunol. 4:11–23. 10.1038/nri1255 [DOI] [PubMed] [Google Scholar]
- Saijo, S., Fujikado N., Furuta T., Chung S.H., Kotaki H., Seki K., Sudo K., Akira S., Adachi Y., Ohno N., et al. 2007. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat. Immunol. 8:39–46. 10.1038/ni1425 [DOI] [PubMed] [Google Scholar]
- Saijo, S., Ikeda S., Yamabe K., Kakuta S., Ishigame H., Akitsu A., Fujikado N., Kusaka T., Kubo S., Chung S.H., et al. 2010. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 32:681–691. 10.1016/j.immuni.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Sanglard, D., Ischer F., Monod M., and Bille J.. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., Yang X.L., Yudate T., Chung J.S., Wu J., Luby-Phelps K., Kimberly R.P., Underhill D., Cruz P.D. Jr, and Ariizumi K.. 2006. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor γ chain to induce innate immune responses. J. Biol. Chem. 281:38854–38866. 10.1074/jbc.M606542200 [DOI] [PubMed] [Google Scholar]
- Schoenen, H., Bodendorfer B., Hitchens K., Manzanero S., Werninghaus K., Nimmerjahn F., Agger E.M., Stenger S., Andersen P., Ruland J., et al. 2010. Cutting edge: Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J. Immunol. 184:2756–2760. 10.4049/jimmunol.0904013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla, M.J., and Odds F.C.. 1986. Development of Candida albicans hyphae in different growth media—variations in growth rates, cell dimensions and timing of morphogenetic events. J. Gen. Microbiol. 132:3083–3088 [DOI] [PubMed] [Google Scholar]
- Shou, C., Farnsworth C.L., Neel B.G., and Feig L.A.. 1992. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature. 358:351–354. 10.1038/358351a0 [DOI] [PubMed] [Google Scholar]
- Slack, E.C., Robinson M.J., Hernanz-Falcón P., Brown G.D., Williams D.L., Schweighoffer E., Tybulewicz V.L., and Reis e Sousa C.. 2007. Syk-dependent ERK activation regulates IL-2 and IL-10 production by DC stimulated with zymosan. Eur. J. Immunol. 37:1600–1612. 10.1002/eji.200636830 [DOI] [PubMed] [Google Scholar]
- Steele, C., Rapaka R.R., Metz A., Pop S.M., Williams D.L., Gordon S., Kolls J.K., and Brown G.D.. 2005. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 1:e42. 10.1371/journal.ppat.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara, K., Tokieda S., Nagaoka K., Takeda T., Kimura Y., and Inaba K.. 2011. C-type lectin SIGNR1 enhances cellular oxidative burst response against C. albicans in cooperation with Dectin-1. Eur. J. Immunol. 41:1435–1444. 10.1002/eji.200940188 [DOI] [PubMed] [Google Scholar]
- Takai, Y., Sasaki T., and Matozaki T.. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153–208 [DOI] [PubMed] [Google Scholar]
- Taylor, P.R., Tsoni S.V., Willment J.A., Dennehy K.M., Rosas M., Findon H., Haynes K., Steele C., Botto M., Gordon S., and Brown G.D.. 2007. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 8:31–38. 10.1038/ni1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu, S., and Karin M.. 2009. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- van der Velden, W.J., Plantinga T.S., Feuth T., Donnelly J.P., Netea M.G., and Blijlevens N.M.. 2010. The incidence of acute graft-versus-host disease increases with Candida colonization depending the dectin-1 gene status. Clin. Immunol. 136:302–306. 10.1016/j.clim.2010.04.007 [DOI] [PubMed] [Google Scholar]
- Vonk, A.G., Netea M.G., van Krieken J.H., Iwakura Y., van der Meer J.W., and Kullberg B.J.. 2006. Endogenous interleukin (IL)-1 α and IL-1 β are crucial for host defense against disseminated candidiasis. J. Infect. Dis. 193:1419–1426. 10.1086/503363 [DOI] [PubMed] [Google Scholar]
- Wang, Q., Siminovitch K.A., Downey G.P., and McCulloch C.A.. 2013. Ras-guanine-nucleotide-releasing factors 1 and 2 interact with PLCγ at focal adhesions to enable IL-1-induced Ca(2+) signalling, ERK activation and MMP-3 expression. Biochem. J. 449:771–782. 10.1042/BJ20121170 [DOI] [PubMed] [Google Scholar]
- Wang, X., Wang W., Lin Z., Wang X., Li T., Yu J., Liu W., Tong Z., Xu Y., Zhang J., et al. 2014. CARD9 mutations linked to subcutaneous phaeohyphomycosis and TH17 cell deficiencies. J. Allergy Clin. Immunol. 133:905–908: e3. 10.1016/j.jaci.2013.09.033 [DOI] [PubMed] [Google Scholar]
- Wheeler, R.T., and Fink G.R.. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2:e35. 10.1371/journal.ppat.0020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, R.T., Kombe D., Agarwala S.D., and Fink G.R.. 2008. Dynamic, morphotype-specific Candida albicans β-glucan exposure during infection and drug treatment. PLoS Pathog. 4:e1000227. 10.1371/journal.ppat.1000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniok, I., Hornbach A., Schmitt C., Frosch M., Einsele H., Hube B., Löffler J., and Kurzai O.. 2008. Induction of ERK-kinase signalling triggers morphotype-specific killing of Candida albicans filaments by human neutrophils. Cell. Microbiol. 10:807–820. 10.1111/j.1462-5822.2007.01086.x [DOI] [PubMed] [Google Scholar]
- Wu, W., Hsu Y.-M.S., Bi L., Songyang Z., and Lin X.. 2009. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat. Immunol. 10:1208–1214. 10.1038/ni.1788 [DOI] [PubMed] [Google Scholar]
- Zhu, T.-N., He H.-J., Kole S., D’Souza T., Agarwal R., Morin P.J., and Bernier M.. 2007. Filamin A-mediated down-regulation of the exchange factor Ras-GRF1 correlates with decreased matrix metalloproteinase-9 expression in human melanoma cells. J. Biol. Chem. 282:14816–14826. 10.1074/jbc.M611430200 [DOI] [PubMed] [Google Scholar]
- Zhu, L.-L., Zhao X.-Q., Jiang C., You Y., Chen X.-P., Jiang Y.-Y., Jia X.-M., and Lin X.. 2013. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity. 39:324–334. 10.1016/j.immuni.2013.05.017 [DOI] [PubMed] [Google Scholar]