Abstract
Studies using low-resolution fiber diffraction, electron microscopy, and atomic force microscopy on various amyloid fibrils indicate that the misfolded conformers must be modular, compact, and adopt a cross-β structure. In an earlier study, we used electron crystallography to delineate molecular models of the N-terminally truncated, disease-causing isoform (PrPSc) of the prion protein, designated PrP 27–30, which polymerizes into amyloid fibrils, but we were unable to choose between a trimeric or hexameric arrangement of right- or left-handed β-helical models. From a study of 119 all-β folds observed in globular proteins, we have now determined that, if PrPSc follows a known protein fold, it adopts either a β-sandwich or parallel β-helical architecture. With increasing evidence arguing for a parallel β-sheet organization in amyloids, we contend that the sequence of PrP is compatible with a parallel left-handed β-helical fold. Left-handed β-helices readily form trimers, providing a natural template for a trimeric model of PrPSc. This trimeric model accommodates the PrP sequence from residues 89–175 in a β-helical conformation with the C terminus (residues 176–227), retaining the disulfide-linked α-helical conformation observed in the normal cellular isoform. In addition, the proposed model matches the structural constraints of the PrP 27–30 crystals, positioning residues 141–176 and the N-linked sugars appropriately. Our parallel left-handed β-helical model provides a coherent framework that is consistent with many structural, biochemical, immunological, and propagation features of prions. Moreover, the parallel left-handed β-helical model for PrPSc may provide important clues to the structure of filaments found in some other neurodegenerative diseases.
In >20 human disorders, including Alzheimer's disease and the prion diseases, the misfolding of an endogenous protein and the subsequent aggregation of the aberrant conformers are essential steps in the pathogenic process (1–3). Although the native proteins are evolutionarily or structurally unrelated, the structural characteristics of the misfolded forms are highly similar and can be mimicked in vitro with a variety of sequences (3, 4), suggesting that the tendency of misfolded proteins to form ordered aggregates, known as amyloid, reflects a predisposition of the polypeptide backbone. The insolubility of the misfolded conformers has hampered their structural analysis by classical techniques, such as x-ray diffraction and NMR spectroscopy. Optical spectroscopy reveals that these aggregates are dominated by β-sheet structures (for review, see ref. 5). In addition, fiber diffraction studies have shown that β-sheets are organized in a cross-β fashion, in which the strands run perpendicular to the axis of the fibril (3, 4). Electron microscopy (EM), cryo-EM, and atomic force microscopy studies show that the protofibrillar units are compact, with typical sizes ranging between 15 and 40 Å (6–8).
In the prion diseases, such as Creutzfeldt–Jakob disease, Gerstmann–Sträussler–Scheinker disease, and fatal insomnia, the cellular prion protein (PrPC) folds into an aberrant, infectious isoform (PrPSc) (9), which accumulates and causes disease. Infectious fibrils, known as prion rods, are composed of PrP 27–30, a fragment of PrPSc resulting from proteinase K digestion of the first ≈88 residues. Prion rods possess the tinctorial properties of amyloid fibers (10, 11) and resemble amyloid fibrils found in vivo (12). Structural models of PrP 27–30 have been developed that incorporate low-resolution optical spectroscopy and x-ray data. However, the insufficient quality of these models limits our understanding of the molecular basis of prion replication. The discovery of 2D crystals of PrP 27–30 and of a fibrillogenic miniprion, PrPSc106 [with residues 141–176 deleted (13)], provided additional structural constraints (14) for modeling PrPSc. Additionally, analysis of these crystals by electron crystallography led to low-resolution projection maps of PrPSc.
Using these data, we surveyed a database of known protein structures (15) to identify protein folds that fulfill the basic structural requirements for amyloid fibrils and identified parallel β-helices as the most likely candidates. After analysis of the position-specific amino acid preferences for these periodic structures, we found that the PrP sequence in the conformationally plastic region (residues 89–175) is compatible with a left-handed, β-helical fold. With threading analysis (16), we created a model that features an oligomeric disc structure for a segment of PrP 27–30. Stacking these discs enables the modeling of the PrP 27–30 assembly into prion rods.
In parallel, we extended our electron crystallographic analysis of 2D crystals of PrP 27–30 and PrPSc106 to a larger number of samples, yielding maps with data to ≈12-Å resolution. These maps provide structural constraints for the models and allowed us to position the α-helical structures in PrP 27–30. We observed concordance between the trimeric β-helical model of PrP 27–30 and PrPSc106 using the refined electron density maps.
Materials and Methods
Survey of Known Protein Structures. The SCOP database (15) was used to survey known protein structures. We inspected the 119 known all-β folds for the suitability of fiber formation by using the following criteria: diameter of <50 Å, cross-β architecture, and accessible β-sheet edges. Of 12 folds identified, 10 represented subtypes of β-sandwiches (Table 1, which is published as supporting information on the PNAS web site) and two were parallel β-helices: single-stranded, right-handed β-helices and single-stranded, left-handed β-helices. The β-roll fold was not considered because it requires calcium ions to stabilize each turn.
Analysis of Left- and Right-Handed β-Helices. Known parallel left- and right-handed β-helices were analyzed to understand their position-specific residue preferences. For the right-handed β-helix, we only considered the family of pectin lyase-like folds, because the four other families only have one or two members. We analyzed 18 right-handed β-helices [Protein Data Bank (PDB) ID codes: 1AIR, 1BHE, 1BN8, 1CZF, 1DAB, 1DB0, 1DGB, 1EE6, 1GQ8, 1H80, 1HG8, 1IA5, 1IDK, 1JTA, 1K5C, 1QCX, 1QJV, and 1TSP] and nine left-handed β-helices (1G97, 1HV9, 1KK6, 1KRR, 1L0S, 1LXA, 1QRE, 1XAT, and 2TDT).
The volume of interior residues was calculated as the sum of side-chain volumes of the conserved interior residues for each complete rung. Interior residues for a complete rung include positions R1, R3, R9, R11, R14, R16, and R23 for the right-handed β-helix and L3, L5, L3′, L5′, L3″, and L5″ for the left-handed β-helix. Side-chain volumes were defined as the Van der Waals volume of individual amino acids (17) minus the volume of glycine.
Molecular Modeling. The model of residues 89–175 of PrP 27–30 was built on the scaffold of N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain (PDB ID code 1G97). Different parts of the same structure were copied and reassembled onto the scaffold so as to follow the selected threading (see Results). The side chains of the PrP sequence were subsequently placed by using scwrl 3.0 (18, 19). Because the NMR structure of the C terminus of mouse (Mo) PrP is poorly defined (20, 21), we used the coordinates of the NMR structure of human PrP (PDB ID code 1QM0) to incorporate the C-terminal α-helices (residues 178–228). When necessary, side chains from the human sequence were replaced with the Mo residues by using scwrl 3.0. The loops and the linker between the β-helical and the α-helical region were built by using rapper (22). In brief, 100 conformations of each of the loops were built independently. To identify conformations that facilitate trimerization and fibrillization, the loops were constructed in the context of a hexamer built by superimposing six β-helical models onto the template of bacterial carbonic anhydrase (PDB ID code 1THJ). The final loop models were selected to give the best fit to the EM maps. The model was optimized through two rounds of energy minimization by using the amber 7 package (23). First, 2,000 steps were computed in which the Cα'softhe β-helical and α-helical domains were kept fixed. The Cα's of the loops and the regions joining the different domains were allowed to move. A second run of 2,000 minimization steps was then performed with no regional constraints.
Contact Order Calculation. Relative contact order (RCO) was computed as defined by Plaxco et al. (24). We calculated the RCO for all-α and all-β folds of the nonredundant set of protein structures (sequences with <95% identity) from the ASTRAL database (25).
EM. Samples were stained on carbon-coated copper grids that were glow-discharged before staining. Samples were adsorbed for up to 5 min, briefly washed, and stained with a freshly filtered 2% uranyl acetate solution. After drying, the samples were viewed in a FEI Tecnai F20 electron microscope at a 200-kV acceleration voltage. Images were recorded on a 1,024 × 1,024 pixel Gatan 694 Slow-Scan charge-coupled device camera at a nominal magnification of ≈111,000 (Gatan, Pleasanton, CA). The effective pixel size was ≈2.2 Å per pixel. A more detailed description of the EM and subsequent image-processing methods will be published elsewhere (H.W., C.G., D. A. Agard, F.E.C., and S.B.P., unpublished work).
Results
Survey of Known β-Folds. Amyloid fibers are β-rich, with extensive cross-β structure capable of adopting a domain architecture that is 20–50 Å in diameter. Analyzing the SCOP database to survey all known protein folds (15), we identified only a few architectures from among the 119 all-β folds that fulfilled our basic structural constraints. These include the parallel β-helical folds and some subtypes of the β-sandwich, a well known antiparallel motif (26).
Increasing evidence indicates that several amyloid fibrils are likely to adopt parallel β-structures. Parallel in-register architecture has been seen for Aβ1–40 by solid-state NMR (27) and recently proposed for α-synuclein after EPR labeling (28). In contrast, the recurring absence of the meridional 9.6-Å reflection in fiber diffraction patterns of most amyloids suggests that antiparallel arrangements are less likely. We have therefore focused on parallel folds and thus analyzed the structures of the nine left-handed and 18 pectin lyase-like, right-handed β-helical folds to identify their features and sequence-dependent conformational tendencies.
Left-handed β-helices are stacks of triangular rungs that typically incorporate 18 residues per rung. Each side of the triangle spans 20–25 Å and is made of six residues, five residues in an extended conformation joined by sharp turns with one residue in an αL conformation (Fig. 1). An analysis of the residues at these six positions indicates that positions pointing to the helix interior (Fig. 1, L3 and L5) are restricted to small hydrophobic residues and Thr and Ser residues that can form side-chain hydrogen bonds with the main chain backbone carbonyl of position L6. This interaction appears to stabilize the tight turn. The outside-facing positions L1, L2, L4, and L6 are less constrained. Amino acids with charged and bulky side chains are observed routinely. Proline residues are often found at position L1 and to a lesser extent at position L2 (Fig. 5, which is published as supporting information on the PNAS web site). All known structures accommodate stretches of bulky and charged residues in loops protruding from the turns. In ≈17% of the cases, the residue at position L1 is absent, creating a shorter turn that provides additional structural variability.
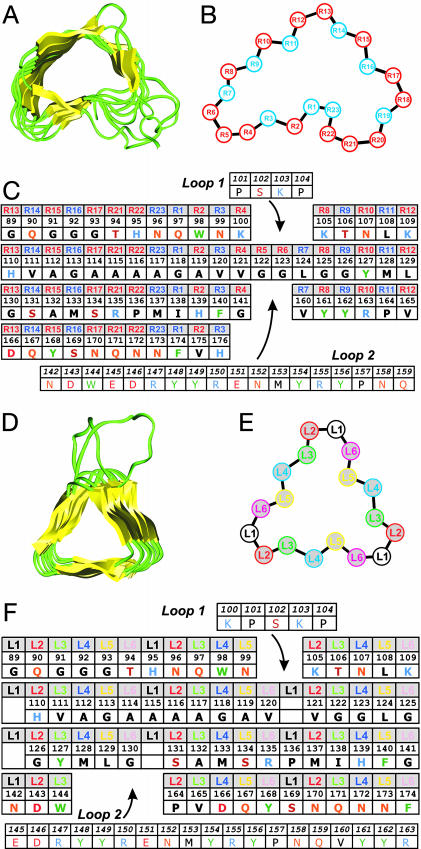
Fig. 1.
Threading the PrP sequence onto β-helical architectures. (A) The structure of pectin lyase from Aspergillus niger (PDB ID code 1QCX) is shown as an example of a parallel right-handed β-helix. (B) Schematic diagram of the right-handed β-helical repeat. Positions in blue face the interior. Regions R4–R8 and R18–R21 are not conserved and may vary in length and conformation. (C) Threading of PrP residues 89–176 onto a right-handed β-helical fold. Each row represents a helical rung, the positions are labeled as in B. The template used here does not include residues at positions R18, R19, and R20, like the example shown in A. Inserted loops are indicated. Amino acids are indicated by their one-letter codes. (D) The structure of UDP N-acetylglucosamine O-acyltransferase from Escherichia coli (PDB ID code 1LXA) is shown as an example of a parallel left-handed β-helix. (E) Schematic diagram of the left-handed β-helical repeat. Each rung is made of six different positions repeated three times. Positions L3 and L5 face the interior. (F) Threading of PrP residues 89–175 onto a left-handed β-helical fold. Each row represents a helical rung; the positions are labeled as in E. Inserted loops are indicated. The absence of residues at L1 indicates short turns.
The kidney-shaped, right-handed β-helices are less regular and slightly larger in diameter (20–30 Å) and can accommodate a wider variety of side chains (Fig. 1 and Fig. 6, which is published as supporting information on the PNAS web site). Each rung, often extended by protruding loops, is composed of 22 to 25 residues. Significant variations in the repeats, in particular, in the turn regions, are seen within each protein and throughout the fold family. A survey of the position-specific residue distribution from 18 nonredundant protein structures indicates that no charged residues are found inside the helix, although polar side chains such as Thr, Ser, Asn, and Gln are common. Notably, very few prolines are found in right-handed β-helices, with <0.5% occurrence and almost exclusively at position R1.
Our finding on the residue distributions in left-handed and right-handed β-helices are in agreement with published studies (29, 30).
Threading the Sequence of PrP onto a Parallel β-Helical Fold. Eisenberg and coworkers (16) introduced the concept of threading a protein sequence onto the scaffold of another protein structure to recognize evolutionarily distant relationships; this can also be applied to relate a sequence to a canonical protein fold. Having defined the position-specific preferences for amino acids in the left- and right-handed β-helical folds, we adapted the threading concept to explore the structural preferences for PrPSc. Fourier transform IR spectroscopy indicates that PrP 27–30 includes 60 to 70 residues in extended conformations and 30 to 40 residues in α-helical conformations (31). Immunological data show that the major conformational changes that differentiate PrPC and PrPSc occur in the region 90–112 and are likely to extend beyond residue 138 (32). By contrast, two antibodies targeting the C-terminal helix of PrPC also recognize PrPSc (32), suggesting that at least one and likely both C-terminal α-helices of PrPC are retained in PrPSc. The preservation of these α-helices is supported by the existence of a disulfide bridge in PrPSc (33), a bond that joins these helices. Furthermore, replacing the cysteines with alanines abolishes the ability of PrP to form infectious prions (34). Therefore, in PrP 27–30, the main conformational change should occur between residues 89 and 170, leading us to probe the sequence of this region for its compatibility with left- and right-handed β-helices. Because 2D crystals of PrP 27–30 and PrPSc106 appear to be isomorphous (14), plausible threading solutions for these forms should be isomorphous as well. This finding implies that residues 141 and 176 must be relatively near one another to accommodate the deletion in PrP106. MoPrP also includes Pro at positions 101, 104, 136, 157, and 164, which must be accommodated in any structural model.
Right-Handed Threading. First, we threaded residues 89–176 onto the three right-handed β-helical subtypes to place residue 141 spatially close to residue 176 and placed the charged residues in exterior positions. Some right-handed β-helices (such as the P22 tailspike protein) are known to trimerize, and 2D crystals of PrPSc show a 3-fold symmetry. This observation provides an additional structural constraint because the C-terminal α-helices cannot fit within the oligomerization interface (positions R8 to R16). Only threading onto the shortest subtype (20 residues per rung, as found in the structure of the P.69 virulence factor of Bordetella pertussis) could fulfill the trimeric arrangement (Fig. 1C). A loop was introduced to accommodate the KPSKPK sequence at residues 100–105, whereas another loop was required to accommodate the bulky and charged residues from the sequence 142–159. This threading (and all other alignments explored) does not place all the prolines at sites found in known structures. For example, P137 and P111 must be inserted into positions that prefer extended conformations. Notably, the Ala- and Gly-rich segment (residues 112–131) creates a cavity in the interior of the β-helix. On average, the inside core of right-handed β-helices occupies 418 ± 18 Å3. In the optimal threading of PrP 27–30 onto the right-handed β-helix, this segment would produce a rung occupying only 209 Å3. In theory, such a cavity could be stabilized by the presence of large side chains in the next rung. However, in the case of PrPSc106, the last rung is absent and the cavity would be exposed before aggregation, which could significantly destabilize the monomer. Finally, we observed that known right-handed β-helices contain at least eight successive rungs, whereas a model following this threading includes only two rungs for PrPSc106 and three for PrP 27–30. Taken together, these elements indicate that the sequence of PrP 27–30 between residues 89–176 is unlikely to adopt a right-handed β-helical fold.
Left-Handed Threading. As with the right-handed threading, threading MoPrP onto a left-handed β-helix introduced two loops (Fig. 1F). The first short loop accommodates the KPSKPK sequence at a corner. P → L mutations at residues 101 and 104 cause Gerstmann–Sträussler–Scheinker disease. L101 could allow other registers and would not require the presence of the short loop. A longer loop would be necessary to position residues 145–163, a region rich in charged and bulky residues, outside the β-helical repeat. This type of loop is observed in all known β-helices in which protrusions of the main chain allow for the insertion of series of charged side chains. This threading includes four short turns (absence of position L1) and six long turns, with a total of four rungs. Short left-handed β-helices including only four or five rungs have been observed in the truncated form of the C-terminal domain of N-acetylglucosamine 1-phosphate uridyltransferase from E. coli (PDB ID code 1FXJ). The Ala/Gly-rich stretch (residues 112–131) also introduces a packing defect (35) in one of the helical rungs (core volume of the corresponding rung is 171 Å3 vs. an average of 335 ± 48 Å3 per rung in known structures), but it can be stabilized by side chains from the next rung that are present in both PrP 27–30 and PrPSc106 (for example, Y127, L129, or I138). Overall, this threading indicates a structural compatibility between the sequence of MoPrP and the left-handed β-helical fold.
This threading can be readily extended to the sequences of human, bovine, ovine, and hamster PrP. All polymorphic residues can be accommodated in the parallel, left-handed β-helix with the exception of residue Q167 at the interior position L5 in the last rung (Fig. 7, which is published as supporting information on the PNAS web site). This position is Glu in human PrP and Arg in sheep PrP. We note that similar charged side chains are observed occasionally inside the first or last rung of β-helical proteins of known structure. The presence of a positive or negative charge in the last rung may create a problem for certain heteromultimeric interactions. For example, these charge differences could create a species barrier that would prevent sheep scrapie transmission to humans and impair human Creutzfeldt–Jakob disease transmission to mice. Polymorphisms and mutations at codon 167 have been associated with relative resistance to prion infection (36).
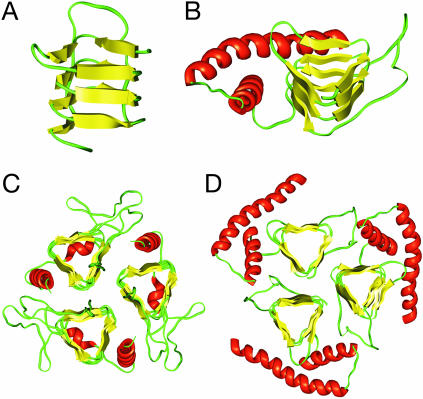
A Molecular Model of PrP 27–30. Using the left-handed β-helical solution, we built a molecular model of residues 89–174 (Fig. 2A). The sequence was first threaded onto the structure of the β-helical part of uridyltransferase (GlmU) of Streptococcus pneumoniae. Where necessary, the turns were built by using a spare-parts approach (37), in which suitable models were taken from similar regions of GlmU. For the longer loops, conformational ensembles were built with the rapper program (22), and the final model was selected as to best fit the EM data. The side chains of the relevant MoPrP sequence were placed by using a backbone-dependent rotamer library with the scwrl program (18, 19). Energy minimization was performed to optimize the geometries of the backbone and side chains. The β-helical region was connected to the C-terminal helices from the NMR structure of PrP, producing a monomeric model of PrP 27–30 (Fig. 2B).
Fig. 2.
Modeling PrP residues 89–174 onto a left-handed β-helical fold. (A) The β-helical model of the N-terminal part of PrP 27–30 corresponding to the threading of Fig. 1F. (B) Model of the monomer of PrP 27–30. The α-helical region (residues 177–227) as determined by NMR spectroscopy (PDB ID code 1QM0) was linked to the β-helical model shown in A.(C) The crystal structure of the trimeric carbonic anhydrase from Methanosarcina thermophila (PDB ID code 1THJ). (D) Trimeric model of PrP 27–30 built by superimposing three monomeric models onto the coordinates of the Cα's of the 1THJ structure.
The α-helices are packed onto the β-helix in an arrangement reminiscent of known β-helical structures. The exact position of the α-helices was optimized to fit the densities observed in the projection maps of the 2D crystals. Electron crystallographic analysis indicates a P3 symmetry, consistent with a trimeric assembly in the unit cell (ref. 14 and see below). Several parallel left-handed β-helices are known to trimerize at the level of their β-helix, providing a natural template to assemble three monomers. Super-imposing the β-helical regions onto the trimeric structure of bacterial carbonic anhydrase (Fig. 2C), we created a trimeric model of PrP 27–30. The Cα's of the residues in the β-strands of the model were superimposed onto the atomic coordinates of corresponding Cα's in the crystal structure. In the resulting model (Fig. 2D), the three monomers interact at the level of the β-helices, whereas the C-terminal α-helices are located on the outside of the trimer, packed closely against the β-sheets, with the glycosylated asparagines pointing away from the center.
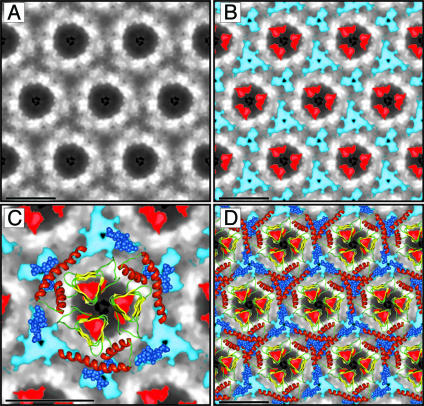
Improved Electron Crystallographic Data. Here we report significant improvements in the detail visible in projection and difference maps of the 2D crystals of PrP 27–30 and PrPSc106, which were analyzed in an earlier study (14). Images of individual 2D crystals were processed by averaging small subsets of images with the single-particle approach (14). After several rounds of correlation-mapping and averaging, the algorithm converged. The final correlation average was then used for crystallographic analysis and averaging, thereby taking advantage of the intrinsic crystal lattice symmetry. To improve the signal-to-noise ratio, we combined the results of three independent 2D crystals into one final average (Fig. 3A). A similar analysis was performed on 2D crystals of PrPSc106 (data not shown).
Fig. 3.
Projection map of PrP 27–30 and statistically significant differences from PrPSc106. (A) Projection map of PrP 27–30 obtained by processing and averaging three independent 2D crystals of PrP 27–30. (B) Statistically significant differences between PrP 27–30 and PrPSc106 overlaid onto the projection map of PrP 27–30. The differences attributed to the internal deletion of PrPSc106 (residues 141–176) are shown in red; the differences in glycosylation between PrP 27–30 and PrPSc106 are shown in blue. (C) Superimposition of the trimeric left-handed model onto the EM maps. The trimeric left-handed β-helical model of PrP 27–30 is superimposed on a 1:1 scale (bar = 50 Å) with the electron crystallographic maps of PrP 27–30. For the sugars linked to N180 and N196 shown as blue space-filling spheres, only the conserved core region (two N-acetylglucosamine and three mannose molecules) is depicted. Sensible side chain dihedral angles for the asparagines and oligosaccharides were selected to optimize the fit with the EM maps. (D) The scaled trimeric model was copied onto the neighboring units of the crystals (bar = 50 Å) to show the crystallographic packing suggested by the model.
As before, we calculated the statistically significant differences between the projection maps of PrP 27–30 and PrPSc106 and overlaid the results onto the projection map of PrP 27–30 (Fig. 3B). The difference density attributed to the internal deletion of PrPSc106 is shown in red, and the difference in glycosylation between the two forms is depicted in blue. A more detailed description and analysis of the 2D crystals and image processing is needed (H.W., C.G., D. A. Agard, F.E.C., and S.B.P., unpublished work). At present, the quality of the 2D crystals seems to be the limiting factor in obtaining higher resolution data.
The β-Helical Model Fits and Rationalizes Structural Data. Because PrP 27–30 and PrPSc106 assemble into isomorphous crystals, it is very likely that their molecular arrangements are comparable. This similarity implies that the boundaries of the Δ141–176 deletion must be spatially close in PrP 27–30. Otherwise, the C-terminal helices would have to adopt different orientations in the crystals of the two forms. In our model, residues 141 and 176 are 15 Å apart and on the same face of the β-helix. A model of PrPSc106 can be constructed by simply removing one β-helical turn and the loop spanning residues 145–164, then reconnecting the β-helix to α-helix B. This assembly requires minor relocation of the C-terminal helices and produces a very similar image in a 2D projection (Fig. 8, which is published as supporting information on the PNAS web site).
A to-scale superimposition of the trimeric model of PrP 27–30 onto the EM projection map depicting the location of the sugars and the Δ141–176 deletion (Fig. 3C) shows that the overall size of the model fits remarkably well with that of the crystal unit cell. Note that the overall geometry of the trimeric model is dictated by the intrinsic tendency of some β-helices to trimerize and was not modified to fit the EM data. The α-helices overlap with the stain-excluding regions, consistent with the high protein density observed in this area. With the selection of appropriate rotamers for N180 and N196, the N-linked sugars can be aligned with the observed location of the sugar moieties. Finally, the estimated location of the Δ141–176 deletion agrees well with the position of the β-helical region and the protruding loop. In the PrP 27–30 model, the 141–176 segment covers exactly one β-helical turn and the long loop. We constructed a molecular model of the 2D crystal by replicating the prototypic trimer, following the crystallographic symmetry. Superimposing this model on the EM image reveals that the projection of the model on the densities is sensible (Fig. 3D). In addition, owing to the spacing along the z axis, sufficient room exists for helices from neighboring molecules to interdigitate.
Although uranyl acetate is generally used as a negative stain for EM samples, it was surprising to observe that 2D crystals of prions specifically bind these ions near the center of the unit cell. This unexpected behavior provided evidence for a trimeric arrangement of PrP 27–30 monomers. We examined our model for possible uranyl-binding sites. Uranyl acetate is known to bind negatively charged side chains, but with only one Asp present in the β-helical region of the PrP 27–30 model and none in the PrPSc106 model, uranyl acetate binding must occur through a different mechanism if it binds to the conformationally plastic region of PrP. Left-handed β-helices expose backbone carbonyl moieties on each turn. In the center of the trimers, these moieties are often involved in the multimerization interface through H bonds with polar side chains of the neighboring monomer. Analysis of the β-helical trimeric model shows that the distances between the carbonyls near the center and their interacting side chains provide suitable functionality to coordinate uranyl ions, either by replacing coordinated waters or by competing with the acetate counterion (38). In our model of PrP 27–30, Q90, H110, and D143 could be used to coordinate three to six uranyl acetate ions per trimer. This binding mode would place electron-dense uranyl ions in the center of the image, colocating with the densities observed in the EM maps.
Discussion
Although it is conceivable that a single structural motif could describe all amyloids at a molecular level, it seems more likely that these conformers share some common underlying architecture (3, 5). The availability of 2D crystals of PrP 27–30 and PrPSc106 offers an opportunity to explore this question.
β-Helical Fibers. Our survey of known folds indicates that parallel β-helices provide a solution to the monomeric structural features observed in amyloid fibers. In addition, because these structures can dimerize or trimerize, they also offer a mechanism to understand how monomers can assemble into oligomeric discs and how discs can assemble into protofibrils (39). Our experimental and computational results point to a trimer of parallel left-handed β-helices as the fundamental unit of PrPSc structure. This finding is consistent with ionization radiation experiments, which identified the target size of the proteinaceous part of the smallest infectious unit as being 55 ± 9 kDa (40, 41). For comparison, the molecular masses of trimers of PrPSc and PrP 27–30 would be 69 and 48 kDa, respectively.
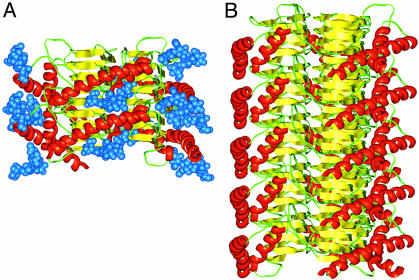
To initiate a fibril, two trimers could assemble by hydrogen bonds linking the top of one β-helix and the bottom of the next layer of PrP 27–30 (Fig. 4A). As shown, the C-terminal α-helices can easily stack in a fashion that allows the sugar moieties to point away from the trimer. The result of this head-to-tail arrangement is an oriented fibril (Fig. 4B). As expected, the β-strands run perpendicular to the fiber axis, producing a cross-β architecture. In this structure, the β-helical residues would be largely inaccessible to antibody binding or proteolytic cleavage. The C-terminal α-helical structure and the N-linked sugars would form an external coating that would be reminiscent of one face of PrPC, providing plausible explanations of why PrPSc binds PrPC (42), of why antibodies to α-helix B bind both PrPC and PrPSc (32), and of the nonimmunogenic nature of PrPSc (43, 44). Of course, full-length PrPSc does not polymerize into fibrils in vitro until N-terminal residues 23–88 are removed and detergent is added (10, 11). We suggest that the retention of the N terminus is compatible with formation of the β-helical trimer but would sterically inhibit stacking of the discs to form fibrils. This hindrance is akin to capping of β-helices by N- or C-terminal extensions of the chain that are thought to block polymerization of these proteins (45). In PrPSc, aggregation without ordered polymerization is expected and observed.
Fig. 4.
Fibrillization of the trimeric left-handed β-helical discs. (A) Two discs of PrP 27–30 can assemble through polar backbone interactions between the lower β-helical rung of the top disk and the upper rung of the bottom disk. This assembly provides enough room for the α-helices to stack and the N-linked sugars to extend away from the center of the structure. (B) A model for the PrP 27–30 fiber was constructed by assembling five trimeric discs. For clarity, the sugars were omitted.
Recently, the crystal structure of a Choristoneura fumiferana antifreeze protein revealed that left-handed β-helices can also dimerize (46), indicating that, depending on the sequence of the misfolded proteins and the specific conditions, β-helical protofilaments may assemble into different oligomeric states. This could lead to variability in the morphology of amyloid fibers and provide one route to differentiate prion strains.
Low Contact Order and Extended Conformation. Several studies have shown that protein misfolding in the prion diseases and disorders featuring amyloid formation is under kinetic control (1, 47, 48). Glockshuber and colleagues (49) demonstrated that PrP folds rapidly to the α-helical isoform. Under conditions that destabilize the helical state, a β-rich oligomeric isoform is formed (50). From a computational perspective, Baker and colleagues (24) have correlated the folding rate with a measurement of contact order. Proteins that have large contact order (i.e., average sequence spacing between interacting residues is large) tend to fold more slowly than structures that are characterized by local contacts. For the proteins of known structure, we calculated the RCO for all-α and all-β folds and compared these with the RCO of left-handed and right-handed β-helices. On average, all-α and all-β folds have an RCO of 11% and 19%, respectively, consistent with the rapid folding observed for α-helical proteins like myoglobin (51), cytochrome c (52), and PrPC. Left-handed and right-handed parallel β-helices have an RCO of 8% and 7%, respectively, indicating that β-helices are dominated by a greater extent of local contacts than most all-α folds. This finding is consistent with the observed structure in the early folding intermediates of β-helical proteins (53, 54). Such studies have also shown that the folding of isolated β-helical domains is very cooperative under dilute conditions. The isolated β-helices have a high tendency to form amyloid-like aggregates, and the early folding intermediates are present at the transition point between either aggregation or folding (54).
Extension to Other Amyloids. Multimeric assemblies of parallel β-helices have an intrinsic cross-β-structure, a simple aggregation mechanism and a plausible propagation mechanism. Thus, this structural motif is an attractive architecture for misfolded aggregates and amyloid fibers. The position-specific amino acid preferences for the left-handed, parallel β-helix structure are compatible with the conformationally plastic region of PrP. It will be interesting to investigate whether other amyloid-forming sequences can be threaded sensibly onto a β-helical structure. Huntington's disease occurs when at least 36 consecutive glutamines are present in the polyglutamine region of huntingtin (55), which corresponds exactly to the number of residues needed to build two complete rungs of a left-handed β-helix. Asn stacks have been observed in parallel β-helices (30).
Although a highly repetitive sequence is not a sensible fit to a sequence profile of a globular protein, we assessed whether poly-glutamines could be threaded onto a left-handed β-helix (Fig. 9, which is published as supporting information on the PNAS web site) and found that all the outside-facing positions can accept glutamines. L5, an interior position, can accommodate a Gln side chain, and polar residues (Cys, Tyr, and Ser) are occasionally found. Glutamines from one side of the β-helix can easily interact with glutamines from the two other sides and build a network of hydrogen bonds. Although Gln can be accommodated in various ways at L3, the other interior position, we note that the amide moiety of this side chain could form hydrogen bonds with the backbone carbonyl of L1, a juxtaposition reminiscent of the stabilizing interactions achieved by Ser, Thr, and Cys at L3 in known structures. Based on fiber diffraction studies, Perutz et al. (56) proposed a helical topology for polyglutamine fibers that is cylindrically symmetric but reminiscent of the known β-helical architectures.
With shorter sequences or sequence fragments, it is conceivable that each peptide forms 1, 4/3, or 5/3 of a β-helix rung and subsequent monomers continue the motif. For example, solid-state NMR studies on the Aβ1–40 peptide indicate that the β-strands are arranged in a parallel fashion with a sharp bend around residues 25–29 (see ref. 27). As a result, the observed conformation for this misfolded peptide could be compatible with a β-helical architecture. Recently, using proline scanning analysis, Wetzel and coworkers (57) concluded that a β-helix is a likely architecture for Aβ fibrils. For β2-microglobulin amyloid, a β-helical arrangement was also suggested (58) based on hydrogen/deuterium exchange protection data (59). Further studies will be necessary to probe the structural compatibility between the various sequences of proteins prone to misfolding and the different types of known β-helices.
Acknowledgments
We thank Dr. Ingmar Grenthe for helpful discussions on uranyl chemistry and Mark DePristo for providing a stand-alone version of rapper. C.G. was a fellow of the Belgian American Educational Foundation and is now a fellow of the John Douglas French Alzheimer's Foundation. This work was supported by National Institutes of Health Grants AG02132, AG10770, and AG021601 and by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations: PrP, prion protein; PrPC, normal cellular isoform; PrPSc, disease-causing isoform; PrP 27–30, N-terminally truncated PrPSc; PrPSc106, prion of 106 residues; RCO, relative contact order; EM, electron microscopy.
References
- 1.Prusiner, S. B., Scott, M. R., DeArmond, S. J. & Cohen, F. E. (1998) Cell 93, 337-348. [DOI] [PubMed] [Google Scholar]
- 2.Kelly, J. W. (1998) Curr. Opin. Struct. Biol. 8, 101-106. [DOI] [PubMed] [Google Scholar]
- 3.Dobson, C. M. (2001) Philos. Trans. R. Soc. London B 356, 133-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunde, M., Serpell, L. C., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. (1997) J. Mol. Biol. 273, 729-739. [DOI] [PubMed] [Google Scholar]
- 5.Sunde, M. & Blake, C. C. F. (1998) Q. Rev. Biophys. 31, 1-39. [DOI] [PubMed] [Google Scholar]
- 6.Khurana, R., Ionescu-Zanetti, C., Pope, M., Li, J., Nielson, L., Ramirez-Alvarado, M., Regan, L., Fink, A. L. & Carter, S. A. (2003) Biophys. J. 85, 1135-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jimenez, J. L., Guijarro, J. I., Orlova, E., Zurdo, J., Dobson, C. M., Sunde, M. & Saibil, H. R. (1999) EMBO J. 18, 815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiménez, J. L., Nettleton, E. J., Bouchard, M., Robinson, C. V., Dobson, C. M. & Saibil, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9196-9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prusiner, S. B. (2001) N. Engl. J. Med. 344, 1516-1526. [DOI] [PubMed] [Google Scholar]
- 10.Prusiner, S. B., McKinley, M. P., Bowman, K. A., Bolton, D. C., Bendheim, P. E., Groth, D. F. & Glenner, G. G. (1983) Cell 35, 349-358. [DOI] [PubMed] [Google Scholar]
- 11.McKinley, M. P., Meyer, R. K., Kenaga, L., Rahbar, F., Cotter, R., Serban, A. & Prusiner, S. B. (1991) J. Virol. 65, 1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeArmond, S. J., McKinley, M. P., Barry, R. A., Braunfeld, M. B., McColloch, J. R. & Prusiner, S. B. (1985) Cell 41, 221-235. [DOI] [PubMed] [Google Scholar]
- 13.Supattapone, S., Bosque, P., Muramoto, T., Wille, H., Aagaard, C., Peretz, D., Nguyen, H.-O. B., Heinrich, C., Torchia, M., Safar, J., et al. (1999) Cell 96, 869-878. [DOI] [PubMed] [Google Scholar]
- 14.Wille, H., Michelitsch, M. D., Guénebaut, V., Supattapone, S., Serban, A., Cohen, F. E., Agard, D. A. & Prusiner, S. B. (2002) Proc. Natl. Acad. Sci. USA 99, 3563-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreeva, A., Howorth, D., Brenner, S. E., Hubbard, T. J., Chothia, C. & Murzin, A. G. (2004) Nucleic Acids Res. 32, D226-D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowie, J. U., Luthy, R. & Eisenberg, D. (1991) Science 253, 164-170. [DOI] [PubMed] [Google Scholar]
- 17.Creighton, T. E. (1993) Proteins: Structures and Molecular Properties (Freeman, New York).
- 18.Bower, M. J., Cohen, F. E. & Dunbrack, R. L. J. (1997) J. Mol. Biol. 267, 1268-1282. [DOI] [PubMed] [Google Scholar]
- 19.Canutescu, A. A., Shelenkov, A. A. & Dunbrack, R. L., Jr. (2003) Protein Sci. 12, 2001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riek, R., Hornemann, S., Wider, G., Billeter, M., Glockshuber, R. & Wüthrich, K. (1996) Nature 382, 180-182. [DOI] [PubMed] [Google Scholar]
- 21.Calzolai, L., Lysek, D. A., Güntert, P., von Schroetter, C., Riek, R., Zahn, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8340-8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo, M. A., de Bakker, P. I., Lovell, S. C. & Blundell, T. L. (2003) Proteins 51, 41-55. [DOI] [PubMed] [Google Scholar]
- 23.Case, D. A., Pearlman, D. A., Caldwell, J. W., Cheatham, T. E., Wang, J., Ross, W. S., Simmerling, C. L., Darden, T. A., Merz, K. M., Stanton, R. V., et al. (2002) amber 7 (Univ. of California, San Francisco).
- 24.Plaxco, K. W., Simons, K. T. & Baker, D. (1998) J. Mol. Biol. 277, 985-994. [DOI] [PubMed] [Google Scholar]
- 25.Chandonia, J. M., Hon, G., Walker, N. S., Lo Conte, L., Koehl, P., Levitt, M. & Brenner, S. E. (2004) Nucleic Acids Res. 32, D189-D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen, F. E., Sternberg, M. J. & Taylor, W. R. (1980) Nature 285, 378-382. [DOI] [PubMed] [Google Scholar]
- 27.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742-16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Der-Sarkissian, A., Jao, C. C., Chen, J. & Langen, R. (2003) J. Biol. Chem. 278, 37530-37535. [DOI] [PubMed] [Google Scholar]
- 29.Heffron, S., Moe, G. R., Sieber, V., Mengaud, J., Cossart, P., Vitali, J. & Jurnak, F. (1998) J. Struct. Biol. 122, 223-235. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins, J. & Pickersgill, R. (2001) Prog. Biophys. Mol. Biol. 77, 111-175. [DOI] [PubMed] [Google Scholar]
- 31.Wille, H., Zhang, G.-F., Baldwin, M. A., Cohen, F. E. & Prusiner, S. B. (1996) J. Mol. Biol. 259, 608-621. [DOI] [PubMed] [Google Scholar]
- 32.Peretz, D., Williamson, R. A., Matsunaga, Y., Serban, H., Pinilla, C., Bastidas, R. B., Rozenshteyn, R., James, T. L., Houghten, R. A., Cohen, F. E., et al. (1997) J. Mol. Biol. 273, 614-622. [DOI] [PubMed] [Google Scholar]
- 33.Turk, E., Teplow, D. B., Hood, L. E. & Prusiner, S. B. (1988) Eur. J. Biochem. 176, 21-30. [DOI] [PubMed] [Google Scholar]
- 34.Muramoto, T., Scott, M., Cohen, F. E. & Prusiner, S. B. (1996) Proc. Natl. Acad. Sci. USA 93, 15457-15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards, F. M. (1977) Annu. Rev. Biophys. Bioeng. 6, 151-176. [DOI] [PubMed] [Google Scholar]
- 36.Perrier, V., Kaneko, K., Safar, J., Vergara, J., Tremblay, P., DeArmond, S. J., Cohen, F. E., Prusiner, S. B. & Wallace, A. C. (2002) Proc. Natl. Acad. Sci. USA 99, 13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, T. A. & Thirup, S. (1986) EMBO J. 5, 819-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craw, J. S., Vincent, M. A., Hillier, I. H. & Wallwork, A. L. (1995) J. Phys. Chem. 99, 10181-10185. [Google Scholar]
- 39.Rochet, J. C. & Lansbury, P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60-68. [DOI] [PubMed] [Google Scholar]
- 40.Bellinger-Kawahara, C. G., Kempner, E., Groth, D. F., Gabizon, R. & Prusiner, S. B. (1988) Virology 164, 537-541. [DOI] [PubMed] [Google Scholar]
- 41.Kempner, E. S., Miller, J. H. & McCreery, M. J. (1986) Anal. Biochem. 156, 140-146. [DOI] [PubMed] [Google Scholar]
- 42.Meier, P., Genoud, N., Prinz, M., Maissen, M., Rülicke, T., Zurbriggen, A., Raeber, A. J. & Aguzzi, A. (2003) Cell 113, 49-60. [DOI] [PubMed] [Google Scholar]
- 43.Tsukamoto, T., Diringer, H. & Ludwig, M. (1985) Tohoku J. Exp. Med. 146, 483-484. [DOI] [PubMed] [Google Scholar]
- 44.Prusiner, S. B., Groth, D., Serban, A., Koehler, R., Foster, D., Torchia, M., Burton, D., Yang, S.-L. & DeArmond, S. J. (1993) Proc. Natl. Acad. Sci. USA 90, 10608-10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson, J. S. & Richardson, D. C. (2002) Proc. Natl. Acad. Sci. USA 99, 2754-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leinala, E. K., Davies, P. L. & Jia, Z. (2002) Structure (London) 10, 619-627. [DOI] [PubMed] [Google Scholar]
- 47.Baskakov, I. V., Legname, G., Prusiner, S. B. & Cohen, F. E. (2001) J. Biol. Chem. 276, 19687-19690. [DOI] [PubMed] [Google Scholar]
- 48.Fandrich, M. & Dobson, C. M. (2002) EMBO J. 21, 5682-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wildegger, G., Liemann, S. & Glockshuber, R. (1999) Nat. Struct. Biol. 6, 550-553. [DOI] [PubMed] [Google Scholar]
- 50.Baskakov, I. V., Legname, G., Baldwin, M. A., Prusiner, S. B. & Cohen, F. E. (2002) J. Biol. Chem. 277, 21140-21148. [DOI] [PubMed] [Google Scholar]
- 51.Jennings, P. A. & Wright, P. E. (1993) Science 262, 892-896. [DOI] [PubMed] [Google Scholar]
- 52.Roder, H. & Colon, W. (1997) Curr. Opin. Struct. Biol. 7, 15-28. [DOI] [PubMed] [Google Scholar]
- 53.Schuler, B. & Seckler, R. (1998) J. Mol. Biol. 281, 227-234. [DOI] [PubMed] [Google Scholar]
- 54.Schuler, B., Rachel, R. & Seckler, R. (1999) J. Biol. Chem. 274, 18589-18596. [DOI] [PubMed] [Google Scholar]
- 55.The Huntington's Disease Collaborative Research Group. (1993) Cell 72, 971-983. [DOI] [PubMed] [Google Scholar]
- 56.Perutz, M. F., Finch, J. T., Berriman, J. & Lesk, A. (2002) Proc. Natl. Acad. Sci. USA 99, 5591-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams, A. D., Portelius, E., Kheterpal, I., Guo, J. T., Cook, K. D., Xu, Y. & Wetzel, R. (2004) J. Mol. Biol. 335, 833-842. [DOI] [PubMed] [Google Scholar]
- 58.Wetzel, R. (2002) Structure (London) 10, 1031-1036. [DOI] [PubMed] [Google Scholar]
- 59.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol 9, 332-336. [DOI] [PubMed] [Google Scholar]