Abstract
FK506-binding protein 52 (FKBP52), which binds FK506 and possesses peptidylprolyl isomerase activity, is an important immunophilin involved in the heterocomplex of steroid receptors with heat-shock protein 90. Here we report the crystal structures of two overlapped fragments [N(1–260) and C(145–459)] of FKBP52 and the complex with a C-terminal pentapeptide from heat-shock protein 90. Based on the structures of these two overlapped fragments, the complete putative structure of FKBP52 can be defined. The structure of FKBP52 is composed of two consecutive FKBP domains, a tetratricopeptide repeat domain and a short helical domain beyond the final tetratricopeptide repeat motif. Key structural differences between FKBP52 and FKBP51, including the relative orientations of the four domains and some important residue substitutions, could account for the differential functions of FKBPs.
Immunophilins are proteins possessing peptidylprolyl isomerase (PPIase) domains that bind immunosuppressant drugs. According to their binding affinity for different drugs, immunophilins have been divided into two families: FK506-binding proteins (FKBPs), which bind FK506 and rapamycin, and cyclophilins, which bind cyclosporin A (1, 2).
FKBP52 is an immunophilin belonging to the FKBP family, with a molecular mass of 52 kDa, and was first discovered as a component of an inactive steroid receptor/heat-shock protein 90 (Hsp90) complex (3). The assembly pathway of steroid receptors involves multiple chaperone and cochaperone proteins such as Hsp90, Hsp40, Hsp70, Hsp70/90 organizing protein (Hop), and P23 (4–6). Matured steroid receptor complexes contained Hsp90 and at least one immunophilin: FKBP52, another FKBP (FKBP51), the cyclosporin A-binding protein cyclophilin 40 (Cyp40), or the protein phosphatase 5 (PP5). These cochaperones all possess tetratricopeptide repeat (TPR) domains, which form the binding sites for Hsp90 (7). The C-terminal MEEVD sequence motif of Hsp90 has been shown to be critical for the binding of Hsp90 to various TPR-containing proteins (8).
Immunophilins are not required for steroid receptors to bind hormones in vitro (5, 9, 10), but they are known to influence steroid signaling pathways. Receptor-associated FKBP52 and PP5 are suggested to play a role in the shuttling of steroid receptors between cytoplasm and nuclear compartments (7, 11–13). Overexpression of FKBP51 may be responsible for the low hormone-binding affinity of glucocorticoid receptor in both human and squirrel monkey (14–16). Although FKBP52 and FKBP51 share ≈75% sequence similarity (Fig. 1a), they affect hormone binding by glucocorticoid receptor in opposing manners and have different Hsp90-binding characteristics. It is not clear what structural features are responsible for these functional differences between FKBP52 and FKBP51. The crystal structure of FKBP51 has been solved recently, as well as that of the PPIase domain of FKBP52 (16, 17). FKBP52 is difficult to crystallize because of its instability in solution. To overcome this problem, we have determined the structures of two overlapping fragments of FKBP52: N(1–260) [(1–260)a (space group P21) and N(1–260)b (space group P212121)], and C(145–459). Based on these two structures, we have defined the whole putative structure of FKBP52, which is architecturally similar to FKBP51 except that the orientations between different domains are diverse. The structural alterations give the clues for the differential effects of FKBP52 and FKBP51. We have also determined the structure of C(145–459) in complex with the C-terminal pentapeptide (MEEVD) from Hsp90. The complex structure maps out the binding pocket in the TPR domain and reveals several key residues involved in Hsp90 binding.
Fig. 1.
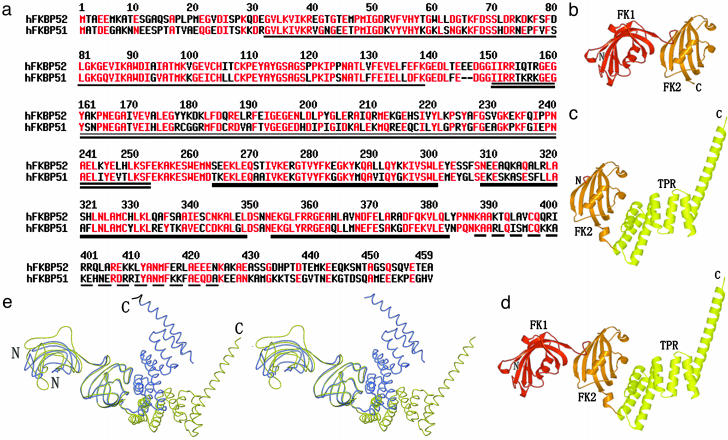
(a) Sequence alignment of human FKBP52 (hFKBP52) and human FKBP51 (hFKBP51). Amino acids with high consensus are shown in red. Human FKBP52 shares 60% amino acid sequence identity and 75% similarity with human FKBP51. Four domains are indicated by different underlines: single underline, FK1 domain; double underline, FK2 domain; thick underline, TPR domain; dashed underline, calmodulin-binding domain. (b) The final N(1–260) model contains residues 21–257. (c) The final C(145–459) model contains residues 145–427. (d) The overall structure of FKBP52 has been defined based on the superposition of overlapped regions of N(1–260) and C(145–459). (e) Stereo view of the structural comparison between FKBP51 (blue) and FKBP52 (yellow) shows their similar structural architectures but the different orientations of their corresponding domains.
Methods
Protein Expression and Purification. Codons 1–260 [N(1–260)] and 145–459 [C(145–459)] of human FKBP52 were cloned into the His-6-tag expression plasmid pET28a(+) (Novagen). N(1–260) was overexpressed in Escherichia coli strain BL21 (DE3), and the selenomethionine-labeled C(145–459) was produced by expression in methionine-deficient E. coli strain B834 (DE3). The soluble proteins were purified by using Ni2+-nitrilotriacetic acid agarose (Qiagen, Valencia, CA) and chromatography on Superdex 75 and Resource Q (Pharmacia).
Crystallization. The protein solution of N(1–260) used for crystallization contained 20 mM Tris (pH 8.0), 150 mM NaCl, and 20 mg/ml protein. Crystals were obtained by using the hangingdrop vapor-diffusion technique with reservoir solutions containing 28–31% polyethylene glycol 6000, 3–5% DMSO, and 100 mM Tris [pH 8.0 for N(1–260)a] and 200 mM calcium chloride, 20% polyethylene glycol 3350 [for N(1–260)b] in a drop formed by mixing 1 μl of protein solution and 1 μl of reservoir solution at 291 K. Crystals of C(145–459) were grown by using a reservoir solution of 100 mM Tris, pH 8.0/2.2–2.4 M ammonium sulfate/2–4% (vol/vol) ethanol. Protein concentration was 10 mg/ml in 20 mM Tris, pH 8.0/100 mM NaCl/5 mM DTT. Crystals of the C(145–459)/MEEVD complex (protein/peptide, 1:1.5) were grown by using a reservoir solution of 1.2 M sodium citrate (pH 6.7).
Data Collection and Processing. X-ray data were collected from a single C(145–459) crystal to 2.7 Å at 90 K. Multiwavelength anomalous diffraction data were collected on a charge-coupled device detector at the BL41XU beamline of SPring-8 (Hyogo, Japan). Data sets were indexed and scaled by using hkl2000 (18). X-ray data of N(1–260)a were collected at the 13th beamline at Beijing Synchrotron Radiation Facility (Beijing, China) to 1.9 Å. Data of N(1–260)b were collected on a Rigaku (Tokyo) RU2000 rotating Cu Kα anode source to 2.0 Å. Data sets were processed by using denzo and scalepack (18). The data for the C(145–459)/MEEVD complex were collected to 3.0 Å at the Advanced Photon Source (Argonne, IL) and processed by using hkl2000.
Structure Determination and Refinement. All 12 possible selenium sites of C(145–459) were found and refined at a 2.8-Å resolution by using solve (19), which produced a mean figure of merit of 0.56. After density modification with resolve (20), ≈50% of all the residues of C(145–459) were automatically traced into the experimental density map; the remaining residues were traced manually by using o (21). cns (22) was used for refinement and addition of solvent molecules. Structures of N(1–260)a and N(1–260)b were solved by molecular replacement using cns. The structures of the FKBP52 N-terminal domain (residues 24–140) (PDB ID code 1N1A) and the N-terminal fragment of C(145–459) (residues 149–255) were used as starting models. This model was subjected to rigid-body refinement, and manual adjustments were made to the model in o. Water molecules were added by using cns. The structure of the C(145–459)/MEEVD complex was solved by molecular replacement with cns, with C(145–459) as the search model. Data collection, processing, and refinement statistic are given in Table 1.
Table 1. Data collection and refinement statistics.
| C(145—459) | N(1—260)a | N(1—260)b | C(145—459)—MEEVD | |||
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | C2221 | P21 | P212121 | C2221 | ||
| Unit cell | ||||||
| a/b/c, Å | 114.4/143.1/171.2 | 48.8/42.2/79.1 | 45.1/59.5/103.2 | 111.6/144.4/170.8 | ||
| α/β/γ, ° | 90.0/90.0/90.0 | 90.0/102.3/90.0 | 90.0/90.0/90.0 | 90.0/90.0/90.0 | ||
| Wavelength, Å | (peak) 0.9798 | (edge) 0.9800 | (remote) 0.900 | 1.2800 | 1.5418 | 0.9793 |
| Resolution, Å | 50.0—2.7 | 50.0—2.7 | 50.0—2.8 | 20.0—1.9 | 50.0—2.4 | 50.0—3.0 |
| Completeness, % | 100.0 (99.8) | 100.0 (99.9) | 100.0 (100.0) | 99.2 (98.3) | 99.9 (99.9) | 98.7 (98.8) |
| Reflections | ||||||
| Total | 291,352 | 291,742 | 258,271 | 72,917 | 78,753 | 169,154 |
| Unique | 38,801 | 38,872 | 34,752 | 24,881 | 11,441 | 27,894 |
| Redundancy | 7.5 | 7.5 | 7.4 | 2.9 | 6.9 | 6.1 |
| Rmerge, %* | 6.4 (32.9) | 5.0 (34.4) | 6.2 (42.2) | 10.1 (40.1) | 9.3 (31.4) | 9.7 (58.9) |
| I/σ(I) | 11.9 (4.2) | 13.8 (4.7) | 11.5 (4.0) | 8.3 (2.7) | 8.9 (5.4) | 6.5 (1.9) |
| Refinement statistics | ||||||
| Resolution, Å | 50.0—2.8 | 20.0—1.9 | 50.0—2.4 | 50.0—3.0 | ||
| R factor, %† | ||||||
| Working set | 21.4 | 20.8 | 21.6 | 23.0 | ||
| Test set | 28.4 | 24.7 | 27.3 | 28.7 | ||
| Rms deviation | ||||||
| Bonds, Å | 0.013 | 0.019 | 0.007 | 0.008 | ||
| Angles, ° | 1.7 | 1.9 | 1.5 | 1.4 | ||
| Ramachandran plot, %‡ | ||||||
| Most favored | 82.0 | 89.9 | 85.2 | 78.7 | ||
| Allowed | 16.9 | 9.1 | 12.3 | 16.9 | ||
| Generously allowed | 0.7 | 0.5 | 2.5 | 3.3 | ||
| Disallowed | 0.4 | 0.5 | 0.0 | 1.0 |
Rmerge = ΣhΣl | Iih — 〈Ih〉 |/ΣhΣl 〈Ih〉, where 〈Ih〉 is the mean of the observations Iih of reflection h
Rwork = Σ (∥Fobs| — Fcalc∥)/Σ|Fobs|; Rfree is the R factor for a subset (10%) of reflections that was selected before refinement calculations and not included in the refinement
Ramachandran plots were generated by using procheck (14)
Results and Discussion
N(1–260), C(145–459), and Putative FKBP52 Structures. FKBP52 can be divided into four domains according to sequence analysis (17). Two overlapped segments of FKBP52 [designated N(1–260) and C(145–459)] have been cloned, expressed, purified, and crystallized. N(1–260) includes the first two FKBP domains [named FK1 (first FKBP domain of FKBPs) and FK2 (second FKBP domain of FKBPs)], whereas C(145–459) is composed of the second FKBP domain, the TPR domain, and a calmodulin-binding domain. The final refined N(1–260) model contains residues 21–257 of FKBP52 (Fig. 1b), whereas the C(145–459) model contains residues 145–427 (Fig. 1c). Based on the superposition of overlapped regions of N(1–260) and C(145–459), the overall structure of FKBP52 has been clearly defined (Fig. 1d).
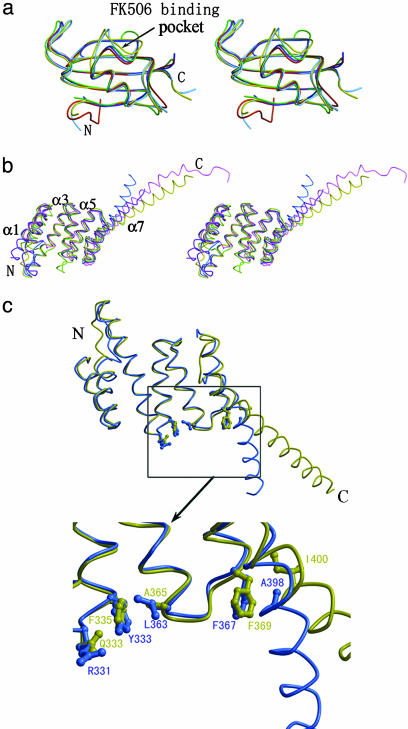
The FKBP domains of FKBP52 both consist of a five- to six-stranded antiparallel β-sheet, which is wrapped around a short α-helix with a right-handed twist, and are similar to those of FKBP51, FKBP12, and macrophage infectivity potentiator protein (16, 23, 24) (Fig. 2a). The TPR domain of FKBP52 is all helical and consists of three units of a consensus 34-aa motif. Each single unit consists of two consecutive α-helices containing 12–15 residues (except α1 and α3, containing 21 and 23 residues, respectively) that cross at an angle of ≈20° to each other. The organizational pattern of the FKBP52 TPR domain is similar to those of FKBP51, Cyp40, PP5, and Hop (8, 16, 25, 26). There is an additional α-helix (α7) in the C terminus beyond the final TPR motif that contains the calmodulin-binding site (Fig. 2b). The overall structure of FKBP52 is very similar to that of FKBP51 except for their relative domain orientations (16) (Fig. 1e). The rms deviations between corresponding FK1, FK2, and TPR domains are 0.6, 0.9, and 1.0 Å, respectively, for all Cα atoms.
Fig. 2.
Stereo view of the superposition of FK and TPR domains. (a) Two FKBP domains of FKBP51 and FKBP52 were superimposed onto FKBP12. FKBP12 (green), 51-FK1 (blue), and 52-FK1 (red) are similar. The structures of 51-FK2 (cyan) and 52-FK2 (yellow) are more closed than the others. (b) TPR domains are superimposed onto the TPR domains of FKBP52. FKBP52 is shown in yellow, FKBP51 is shown in cyan, Hop is shown in green, Cyp40 is shown in purple, and PP5 is shown in pink. The conformations of all the TPR domains are similar, containing six α-helices (α1–α6). The orientations of the extra α-helix (α7) are different. (c) Superposition of the structures of TPR domains and the α7-helixes of FKBP51 (blue) and FKBP52 (yellow). Gln-333, Phe-335, and Ala-365 of FKBP52 are replaced by Arg-331, Tyr-333, and Leu-363 in FKBP51, which may be responsible for the differential binding pattern of FKBPs to Hsp90. The side chain of Ile-400 of FKBP52, corresponding to Ala-398 of FKBP51, will clash with Phe-369, and this may cause the different orientations of the α7-helix.
The FK1 and FK2 domains of FKBP52 are linked by a highly hydrophilic hinge (residues 139–148), and the FK506-binding pocket of FK1 is oriented ≈180° from the putative FK506-binding pocket of FK2. Residues 140–142 form an antiparallel β-strand that interacts with residues 150–152 in the β1 strand of FK2. This loop is stabilized by the formation of a hydrogen-bond network with the side chains of neighboring residues (Fig. 3a). The extensive interactions in the hinge region suggest that the flexibility of the hinge is restrained. The side chains of several residues in the interface (Met-48, Ile-49, Ile-154, and Arg-157) form a hydrophobic core, together with those of Asp-141, Leu-142, Ile-150, Ile-151, and Arg-210. Although a small difference is found in the orientation between FK1 and FK2 of N(1–260)a and N(1–260)b, which have different crystal packing, the conformations of the loop are mostly the same, as well as the hydrogen-bond interactions and the hydrophobic cores. These tight interactions and the rigid hinge stabilize the relative orientation of FK1 and FK2 and restrict the great change of this orientation.
Fig. 3.
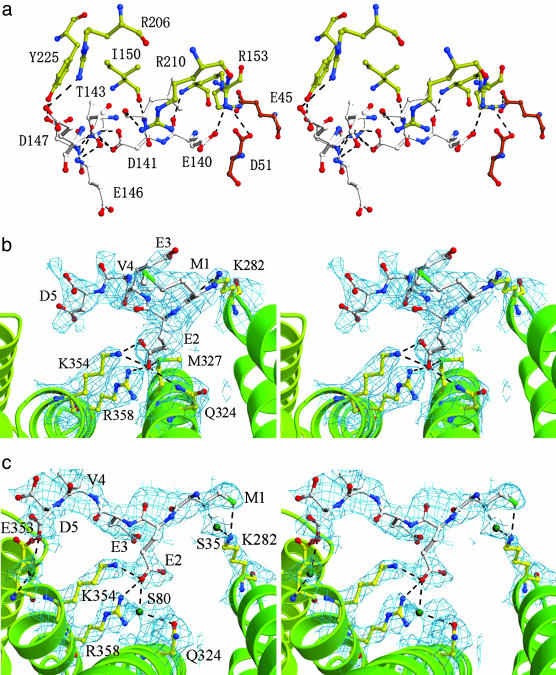
(a) Stereo view of the hydrogen bonds between FK1 and FK2 of FKBP52. Hydrogen bonds at the interface of FK1 and FK2 form a complicated network, which stabilizes the conformation. Residues in FK1 are shown in red, residues in FK2 are shown in yellow, and residues in the loop are shown in white. (b and c) Stereo view of the MEEVD peptide bound to molecules A (b)and B(c) of C(145–459). The omit electron-density map is contoured at 0.7 σ above the mean. Residues of the peptide are shown in white, and residues of the TPR domain are shown in yellow. Residues involved in important interactions are shown in ball-and-stick representation. Hydrogen bonds are shown as dotted lines.
The FK1 domain is responsible for the PPIase activity of FKBP52, whereas the FK2 domain shows no PPIase activity (27, 28). Detailed structural information for FK1 has been reported (17). FK2 is architecturally similar to FK1 despite sharing only 32% sequence identity but lacks the large bulge-splitting strand β4 of FK1 (Fig. 2a). FK1 contains 14 residues directly related to substrate binding, yet only five residues are conserved in FK2. In FK1, Ile-87 and Tyr-113 form highly conserved hydrogen bonds with substrates, and Trp-90 provides a platform for substrate binding (17), whereas in FK2 the corresponding residues are substituted by Pro-201, Phe-232, and Leu-204, respectively. The insertion of one residue (Lys-234) in the 221–242 loop and five residues (Gly-195, Glu-196, Met-197, Leu-198, and Asp-199) in the 192–199 loop, which both flank the binding pocket of FK2, pushes the loops into the binding pocket. The large and extended side chains of Lys-232, Glu-233, and Phe-235 (corresponding to Ser-118, Pro-119, and Lys-121 of FK1, respectively) block the opening of the pocket and change the electrostatic state, in addition to the insertions mentioned above. We conclude that alterations in loops surrounding the binding pocket and substitution of residues important for the substrate binding account for the loss of the PPIase activity of FK2. Previously, a deletion mutant (deletion of Asp-195, His-196, and Asp-197) of FKBP51 has shown that the FK2 domain relates to progesterone receptor preference and target protein interaction (16). Presumably, FK2 domains of FKBP proteins may result from a duplication event. During evolution, FK2 domains lost their PPIase activity but act as organizers of different domains and provide interaction interfaces with other target proteins such as Hsp90 and steroid receptors.
Structure of the C(145–459)/MEEVD Complex. The overall structure of the peptide-bound C(145–459) is similar to that of the free protein. The peptide binds in a cavity formed by the α1, α3, and α5 helices of the TPR domain. Of the three molecules in the asymmetric unit, two (molecules A and B) bind the MEEVD peptide, whereas the third (molecule C) does not. The peptidebinding pockets in molecules A and B are composed of exactly the same residues, namely Lys-282, Asn-324, Met-327, Lys-354, and Arg-358 (Fig. 3 b and c). Some interactions are important to stabilize the binding of the peptide and are conserved in both molecules. In particular, there is a hydrogen bond formed between Lys-282 and Met-1, and two hydrogen bonds are formed between Lys-354, Arg-358, and Glu-2. Nevertheless, the orientations of the peptides in these two molecules are different. Additional amino acids N-terminal to this peptide sequence in Hsp90 also contribute to the binding and render specificity to the interaction between the TPR and Hsp90 (8). As a consequence, the binding between the MEEVD peptide and C(145–459) is not strong, which may account for the different observed binding modes. In the unit cell, the pocket of molecule A faces the pocket of molecule B, and there is enough space for the peptides to fill in, but the binding pocket of molecule C is blocked by the FK2 domain of another molecule C, which explains why this molecule does not bind the peptide. Previous mutation analysis and crystal structures of TPR domains have identified residues in Hop, Cyp40, and PP5 that are essential for Hsp90 binding (8, 25, 26, 29, 30). Comparing these proteins with FKBP52, the key residues for binding Hsp90 are conserved, which suggests a functional similarity between these proteins.
Comparison with the Structure of FKBP51. Although the overall architectures of FKBP52 and FKBP51 are very similar, the relative orientations of their four domains are distinctly different (Fig. 1e). Superimposing the structures of FKBP52 and FKBP51 by using the FK2 domain as the reference, we observe that domain FK1 of FKBP51 is rotated at an angle (θ = 24°, ω = 33°) relative to FK1 of FKBP52. The hinge linking FK1 and FK2 is stabilized by extensive hydrogen bonds and hydrophobic interactions in both FKBP proteins. The different hinge conformations of these two proteins and specific interactions in the interface between FK1 and FK2, together with a different hydrophobic core, may account for their different orientations. Thr-143 and Arg-206 of FKBP52 form four pairs of hydrogen bonds with Asp-141, Glu-144, and Glu-146. In FKBP51, the corresponding residues are Phe-143 and Lys-204, which do not form hydrogen bonds. Additionally, the side chains of Ile-154 and Arg-157 in FKBP52 (corresponding to Thr-152 and Lys-155 of FKBP51, respectively) provide tighter hydrophobic interactions. Available evidence suggests that the hinge may be involved in the interaction between FKBP52 and Hsp90. Thr-143 of FKBP52 has been identified as the major phosphorylation site for casein kinase II, and it is known that casein kinase II-phosphorylated FKBP52 does not bind to Hsp90 (31). Furthermore, Thr-143 is changed to Phe-143 in FKBP51, which may have a differential effect on the Hsp90 binding.
The TPR domain orientation of FKBP52 is considerably different from that of FKBP51. There are fewer hydrogen bonds and hydrophobic interactions in the interface between FK2 and TPR domains, as well as the loop linking FK2 and TPR domains. Considering the flexibility of this area, it is likely that the different orientations of TPR domains are due to different crystal packing.
The TPR domain chimera and truncation experiments have shown that the TPR domain of FKBP51 requires appropriate downstream sequences for Hsp90 binding, but the TPR domain of FKBP52 does not. Additionally, the C-terminal region of FKBP51, which functionally interacts with the TPR domain to permit Hsp90 binding, confers preferential association with progesterone receptor (32, 33). Comparing the TPR domain structures of FKBP52 and FKBP51 (Fig. 2), most residues in the binding interface between FKBP52 and Hsp90 are strictly conserved, with the exception of Gln-333, Phe-335, and Ala-365 (corresponding to Arg-331, Tyr-333, and Leu-363 of FKBP51, respectively). The substitutions of Gln to Arg and Ala to Leu results in changes to the electronic state and steric relationship of the interaction interface, which would account for the differential binding pattern of FKBPs to Hsp90. Superimposing the TPR domains and the extra seventh α-helix (α7) of FKBPs, it can be observed that structural architectures of TPR domain are very similar. However, the α7-helix of FKBP52 shifts away by >30° from the Hsp90-binding interface relative to FKBP51 (Fig. 2). The different orientations of the α7-helix in FKBPs can be explained by the side chain of Ile-400 (corresponding to Ala-398 in FKBP51) in the α7-helix of FKBP52, which would otherwise clash with the phenyl group of Phe-369 in the FKBP51 conformation. The more compact orientation of the C terminus of FKBP51 may result in the proximity and direct interaction with Hsp90, which is consistent with previous studies.
Conclusions
A detailed structural analysis of FKBP52 and the complex with the MEEVD peptide has revealed the essential basis for the loss of PPIase activity of the FK2 domain and the key residues for Hsp90 binding. We observe a number of interesting structural variances between FKBP52 and FKBP51, including the relative orientations of corresponding domains and some important residue substitutions. The very real differences in orientation between the FK1 and FK2 domains in FKBP52 and FKBP51 could explain their differential effects on Hsp90 binding.
Acknowledgments
We thank Xuemei Li, Yuna Sun, and Wei Zheng (Z.R. group) for technical assistance; Min Yao (Hokkaido University, Sapporo, Japan), Rongguang Zhang, Andrzej Joachimiak (Advanced Photon Source, Argonne, IL), Fei Sun, Feng Xu, and Feng Gao (Z.R. group) for assistance with data collection; and Dr. R. B. Sim for critical reading of the manuscript. The peptide MEEVD was a gift from Yuzhang Wu (Third Military Medical University, Chongging, China). This work was supported by Project 863 Grant 2002BA711A12, Project 973 Grant G1999075600, and National Natural Science Foundation of China Grant 30221003.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PPIase, peptidylprolyl isomerase; FKBP, FK506-binding protein; Hsp, heat-shock protein; Hop, Hsp70/90 organizing protein; Cyp40, cyclophilin 40; PP5, protein phosphatase 5; TPR, tetratricopeptide repeat; FK1, first FKBP domain of FKBPs; FK2, second FKBP domain of FKBPs.
Data deposition: The atomic coordinates and structure factors for N(1–260) (space group P21), C(145–459), and C(145–459)/MEEVD have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1Q1C, 1P5Q, and 1QZ2, respectively).
References
- 1.Dolinski, K., Muir, S., Cardenas, M. & Heitman, J. (1997) Proc. Natl. Acad. Sci. USA 94, 13093-13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galat, A. (1993) Eur. J. Biochem. 216, 689-707. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez, E. R. (1990) J. Biol. Chem. 265, 22067-22070. [PubMed] [Google Scholar]
- 4.Pratt, W. B. & Toft, D. O. (1997) Endocr. Rev. 18, 306-360. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, J. & Smith, D. F. (2000) Mol. Endocrinol. 14, 939-946. [DOI] [PubMed] [Google Scholar]
- 6.Morishima, Y., Kanelakis, K. C., Silverstein, A. M., Dittmar, K. D., Estrada, L. & Pratt, W. B. (2000) J. Biol. Chem. 275, 6894-6900. [DOI] [PubMed] [Google Scholar]
- 7.Riggs, D. L., Roberts, P. J., Chirillo, S. C., Cheung-Flynn, J., Prapapanich, V., Ratajczak, T., Gaber, R., Picard, D. & Smith, D. F. (2003) EMBO J. 22, 1158-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheufler, C., Brinker, A., Bourenkov, G., Pegoraro, S., Moroder, L., Bartunik, H., Hartl, F. U. & Moarefi, I. (2000) Cell 101, 199-210. [DOI] [PubMed] [Google Scholar]
- 9.Kosano, H., Stensgard, B., Charlesworth, M. C., McMahon, N. & Toft, D. (1998) J. Biol. Chem. 273, 32973-32979. [DOI] [PubMed] [Google Scholar]
- 10.Dittmar, K. D. & Pratt, W. B. (1997) J. Biol. Chem. 272, 13047-13054. [DOI] [PubMed] [Google Scholar]
- 11.Czar, M. J., Lyons, R. H., Welsh, M. J., Renoir, J. M. & Pratt, W. B. (1995) Mol. Endocrinol. 9, 1549-1560. [DOI] [PubMed] [Google Scholar]
- 12.Galigniana, M. D., Radanyi, C., Renoir, J. M., Housley, P. R. & Pratt, W. B. (2001) J. Biol. Chem. 276, 14884-14889. [DOI] [PubMed] [Google Scholar]
- 13.Dean, D. A., Urban, G., Aragon, I. V., Swingle, M., Miller, B., Rusconi, S., Bueno, M., Dean, N. M. & Honkanen, R. E. (2001) BMC Cell Biol. 2, www.biomedcentral.com/1471-2121/2/6. [DOI] [PMC free article] [PubMed]
- 14.Reynolds, P. D., Ruan, Y., Smith, D. F. & Scammell, J. G. (1999) J. Clin. Endocrinol. Metab. 84, 663-669. [DOI] [PubMed] [Google Scholar]
- 15.Denny, W. B., Valentine, D. L., Reynolds, P. D., Smith, D. F. & Scammell, J. G. (2000) Endocrinology 141, 4107-4113. [DOI] [PubMed] [Google Scholar]
- 16.Sinars, C. R., Cheung-Flynn, J., Rimerman, R. A., Scammell, J. G., Smith, D. F. & Clardy, J. (2003) Proc. Natl. Acad. Sci. USA 100, 868-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, P., Ding, Y., Wu, B., Shu, C., Shen, B. & Rao, Z. (2003) Acta Crystallogr. D 59, 16-22. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 22.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 23.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1991) Science 252, 839-842. [DOI] [PubMed] [Google Scholar]
- 24.Riboldi-Tunnicliffe, A., Konig, B., Jessen, S., Weiss, M. S., Rahfeld, J., Hacker, J., Fischer, G. & Hilgenfeld, R. (2001) Nat. Struct. Biol. 8, 779-783. [DOI] [PubMed] [Google Scholar]
- 25.Das, A. K., Cohen, P. W. & Barford, D. (1998) EMBO J. 17, 1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor, P., Dornan, J., Carrello, A., Minchin, R. F., Ratajczak, T. & Walkinshaw, M. D. (2001) Structure (London) 9, 431-438. [DOI] [PubMed] [Google Scholar]
- 27.Chambraud, B., Rouviere-Fourmy, N., Radanyi, C., Hsiao, K., Peattie, D. A., Livingston, D. J. & Baulieu, E. E. (1993) Biochem. Biophys. Res. Commun. 196, 160-166. [DOI] [PubMed] [Google Scholar]
- 28.Pirkl, F., Fischer, E., Modrow, S. & Buchner, J. (2001) J. Biol. Chem. 276, 37034-37041. [DOI] [PubMed] [Google Scholar]
- 29.Ward, B. K., Allan, R. K., Mok, D., Temple, S. E., Taylor, P., Dornan, J., Mark, P. J., Shaw, D. J., Kumar, P., Walkinshaw, M. D., et al. (2002) J. Biol. Chem. 277, 40799-40809. [DOI] [PubMed] [Google Scholar]
- 30.Russell, L. C., Whitt, S. R., Chen, M. S. & Chinkers, M. (1999) J. Biol. Chem. 274, 20060-20063. [DOI] [PubMed] [Google Scholar]
- 31.Miyata, Y., Chambraud, B., Radanyi, C., Leclerc, J., Lebeau, M. C., Renoir, J. M., Shirai, R., Catelli, M. G., Yahara, I. & Baulieu, E. E. (1997) Proc. Natl. Acad. Sci. USA 94, 14500-14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barent, R. L., Nair, S. C., Carr, D. C., Ruan, Y., Rimerman, R. A., Fulton, J., Zhang, Y. & Smith, D. F. (1998) Mol. Endocrinol. 12, 342-354. [DOI] [PubMed] [Google Scholar]
- 33.Cheung-Flynn, J., Roberts, P. J., Riggs, D. L. & Smith, D. F. (2003) J. Biol. Chem. 278, 17388-17394 [DOI] [PubMed] [Google Scholar]