Abstract
Cell lines have many advantages: they can be manipulated genetically, expanded, and stockpiled for organ transplantation. Freshly isolated hepatocytes, oval cells, pancreatic cells, and hematopoietic stem cells have been shown to repopulate the damaged liver. Here we show that bipotential mouse embryonic liver (BMEL) stem cell lines participate in liver regeneration in albumin–urokinase plasminogen activator/severe combined immunodeficiency disease (Alb-uPA/SCID) transgenic mice. In the liver, BMEL-GFP cells proliferate and differentiate into both hepatocytes and bile ducts, forming small to large clusters detected throughout the 3–8 weeks analyzed after transplantation. Moreover, they respond like host cells to signals for growth, differentiation, and even zonal expression of metabolic enzymes, showing regulated expression of cytokeratins and liver-enriched transcription factors. Immunostaining for MHC class I molecules revealed that cells do not coexpress donor and recipient H-2 haplotypes, as would be the case had cell fusion occurred. This report shows that immortalized stem cell lines not only are competent to participate in the repair of a damaged tissue but also can differentiate into the two major epithelial cell types of a complex organ, hepatocytes and bile ducts.
Regeneration of some organs necessitates the reconstitution of more than one cell type, which will require the utilization of stem cells for tissue repair. In the adult, however, in the absence of stimuli for proliferation, stem cells remain quiescent. To stimulate proliferation of transplanted cells, models of specific organ or tissue deficiencies are essential. Success has been achieved with freshly isolated hematopoietic stem cells for lethally irradiated recipients and hepatocytes in models of liver degeneration (1–4). In only a few cases have long-term cultivable cells been demonstrated to effect tissue repair, including keratinocytes for skin grafts (5) and mesangioblasts or muscle-derived stem cells for muscle repair of dystrophic mice (6, 7).
Freshly isolated adult mouse hepatocytes can regenerate a damaged liver through up to seven serial transplants (1–3). Several types of hepatocytes can participate in liver repopulation: diploid, tetraploid, octaploid, cells isolated from old or young mice, and cells of different sizes (8, 9). However, only freshly isolated fetal hepatoblasts have been shown to be bipotential in vivo, differentiating as hepatocytes and bile duct cells (10–12). While there are reports that human or primate cultured liver cells can home to the mouse liver, the data do not clearly demonstrate growth or cluster formation (13, 14).
Here we report that bipotential mouse embryonic liver (BMEL) cell lines participate in liver repair in the albumin–urokinase plasminogen activator/severe combined immunodeficiency disease (Alb-uPA/SCID) mouse. The nontransformed BMEL cell lines can be readily isolated from wild-type mice of many genotypes, and we have demonstrated for three lines that the potential to differentiate into hepatocytes or cholangiocytes (bile duct epithelial cells) in culture is heritable in daughter clones (15). BMEL cell lines are nontransformed because they do not grow in soft agar and do not form tumors in nude mice (see Materials and Methods for details). The cells are grown in basal culture conditions, where they proliferate without undergoing differentiation. Modifying the culture conditions induces differentiation of the cells, which express a number of markers. However, no functional test of the differentiation capacity of the cells has been available. Indeed, for stem cells, the ability to contribute to tissue repair in the organism is the most stringent test. Because BMEL cells are nontransformed and bipotential, they seemed excellent candidates for carrying out liver repair, which requires not only that the cells home to the liver but also that they respond to in vivo stimuli for proliferation and differentiation.
Recent reports have used genetic markers to demonstrate that incorporation and differentiation in solid tissues of cells of hematopoietic origin are due to the fusion of donor cells with host cells (16, 17). Because it has been shown, both in vivo and in cultured cells, that parental H-2 antigens are consistently coexpressed in fused cells (18, 19), we use differences between the H-2 haplotypes of the BMEL cells and recipient mice to demonstrate that cell fusion is not involved in liver repopulation by BMEL cells.
Materials and Methods
BMEL Cell Line Transduction with a TRIP Lentiviral Vector. BMEL cell culture has been described (15). Cell lines 9A1 and 14B3, at early passages after isolation, were incubated overnight with the equivalent of 500 ng/ml p24 TRIP-CMV-GFP vector and 5 μg/ml DEAE-dextran in RPMI medium 1640 according to ref. 20. Cells were expanded and fluorescence-activated cell sorter (FACS) analysis was performed: 9A1-GFP was 86% GFP-positive and 14B3-GFP was 77% GFP-positive. Twelve generations later 9A1-GFP was 61% and 14B3-GFP was 53% GFP-positive by FACS analysis.
Test for Tumor Formation. Five nude mice were s.c. injected at several sites with 4–6 × 106 BMEL cells. Each experimental mouse was injected with one of the two cell lines, 9A1 or 14B3, before and after transduction with the lentiviral vector. As a positive control, transformed BW1J hepatoma cells were injected. No tumors were detected after 11 months in the mice injected with BMEL cells, whereas tumors were apparent 1 week after injection for the control.
BMEL-GFP Cell Injection into Alb-uPA/SCID Mice. Alb-uPA transgenic mice and SCID mice were purchased from The Jackson Laboratory and Iffa Credo, respectively. Mice were crossed, and animals homozygous for the SCID trait and hemizygous for the Alb-uPA transgene were used for transplantation experiments. BMEL-GFP cells were thawed and expanded for 2 days before injection. Single-cell suspensions were resuspended in Williams medium (Invitrogen) at 5 × 106 cells per ml. Alb-uPA/SCID mice 14–16 days after birth were anesthetized, and 0.5 × 106 cells were injected into the spleen (21). Each week the mice were subjected, by intraperitoneal injection, to an antimacrophage treatment of liposome-encapsulated dichloromethylene diphosphonate (250 μg of clodronate) (22). No repopulation was observed when BMEL-GFP cells were injected without this treatment (eight mice analyzed). Animal care was in accordance with institutional guidelines.
Immunohistochemical Analysis of Liver Sections and Quantification. Mouse livers were frozen in OCT compound (Sakura) and 10-μm serial cryostat sections were fixed in 4% paraformaldehyde (Merck) for 15 min at 20°C (used throughout). Sections were permeabilized in 0.1% Triton X-100 (Sigma) for 10 min, then incubated in 0.3% H2O2 for 5 min, then for 30 min in 10% serum. Primary antibodies were incubated for 2 h: anti-GFP (Molecular Probes), anti-H-2Kk (Pharmingen), anti-H-2Dd (Pharmingen), anti-Ki67 (Pharmingen), anti-hepatocyte nuclear factor (HNF)4α (Santa Cruz Biotechnology, and a gift from F. Sladek, University of California, Riverside), anti-HNF1α and anti-HNF1β (kind gifts from M. Yaniv, Pasteur Institute, Paris), anti-dipeptidyl peptidase IV (DPPIV) (CD26 Pharmingen), anti-glutamine synthetase (Santa Cruz Biotechnology), anticarbamoyl-phosphate synthetase I (CPSI) (Santa Cruz Biotechnology), anti-cytokeratin (CK)7 (Progen, Heidelberg), anti-CK19 (Troma 3, a gift from R. Kemler, Max Planck Institute of Immunobiology, Freiburg, Germany), anti-CD45 (Pharmingen), anti-F4/80 (Serotec), anti-Nimp-R14 (a gift from G. Milon, Pasteur Institute, Paris). Secondary antibodies coupled to peroxidase (DAKO and Caltag) were added for 30 min and the reaction was developed with diaminobenzidine (DAKO), and tissue was counterstained with Mayer's hematoxylin (Merck).
Quantification of BMEL-GFP cell participation in liver repopulation was performed on GFP- or H-2Kk-stained sections by using photoshop 5.5 (Adobe Systems, Mountain View, CA). Areas were calculated from the number of pixels in total liver tissue fields (excluding gaps) compared with the number of pixels in selected GFP-positive or H-2Kk-positive fields.
Results
BMEL Cells Home to the Liver, Proliferate, and Integrate into the Parenchymal Tissue. To identify the transplanted BMEL cells, early passage cultures were transduced with a lentiviral vector expressing GFP under control of the cytomegalovirus (CMV) promoter, to genetically mark the cells and their progeny. This vector transduces and stably integrates into mitotic and nonmitotic cells (20). Fluorescence-activated cell sorter (FACS) analysis showed that 70–90% of cells expressed GFP at the time of the first transplantations, and 12 generations later this number had dropped to 50–60%. All experiments were performed with cells within this interval.
The Alb-uPA model for liver repopulation is a transgenic mouse over-expressing urokinase plasminogen activator (uPA) under control of the albumin (Alb) promoter (23). These mice die of liver failure unless rare hepatocytes excise the transgene. These wild-type hepatocytes then proliferate, and they regenerate the entire liver within 8 weeks. During this interval, the Alb-uPA mouse suffers chronic liver damage and long-term regeneration (1, 9). SCID alleles were bred into the Alb-uPA mice to encourage engraftment and survival of nonsyngeneic cells.
Half a million 9A1-GFP and 14B3-GFP cells were injected intrasplenically into 14- to 16-day-old Alb-uPA/SCID pups. Recipient mice were killed 3, 5, and 8 weeks after injection, serial sections of the livers were prepared, and adjacent sections were used whenever two or more markers were analyzed. At all times examined, immunohistochemical staining of GFP revealed clusters of BMEL-GFP cells distributed throughout the liver, indicating that the cells were able to home to the liver, cross the hepatic sinusoidal barrier, and integrate into the tissue. Moreover, the GFP-expressing cells not only were present in parenchymal tissue but were organized into bile ducts as well. Fig. 1A shows a cluster of BMEL-GFP parenchymal cells displaying apparently normal architecture, and Fig. 1B shows a BMEL-GFP bile duct next to host-derived bile ducts.
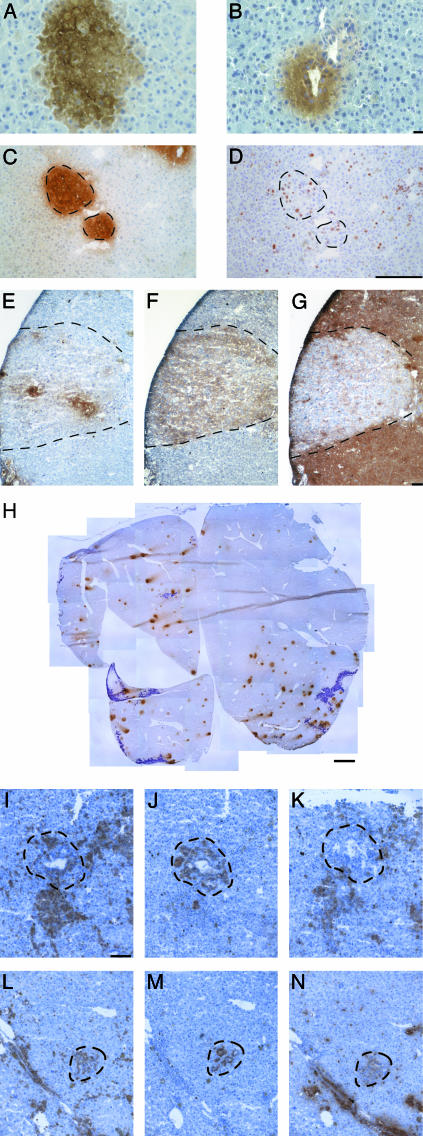
Fig. 1.
BMEL cells integrate and proliferate in the liver of Alb-uPA/SCID mice without having undergone cell fusion and elicit an immune reaction. (A and B) Immunohistochemistry revealing BMEL-GFP cells (brown) in the hepatic parenchyma (A) and as a bile duct-like structure (B). (C and D) Clusters of BMEL-GFP cells (C) are composed of proliferating cells as shown by the nuclear localization of the Ki67 antigen (D). Broken lines in this and the following figures delimit the fields of interest. GFP (E), H-2Kk (F), and H-2Dd (G) immunohistochemical analyses of adjacent serial sections are shown. Transduced BMEL cells do not all express GFP (E). H-2Kk reveals all BMEL cells injected (F) and H-2Dd identifies the host cells (G). The H-2Kk-positive field (F) is H-2Dd-negative (G), demonstrating that cell fusion, which would result in double-positive cells, is not involved. (H) A complete liver section after GFP staining reconstituted from 31 photographic images and illustrating the material used for quantitation detailed in Table 1. BMEL cell clusters are visible as brown spots, whereas the purple-stained regions are necrotic. (I–N) Infiltrations of immune cells are seen around BMEL cells after transplantation. BMEL H-2Kk-positive bile ducts (J and M) are surrounded by CD45+ cells (I and L), of which many are macrophages (antibody F/480) (K) and neutrophils (antibody Nimp-R14) (N). (Scale bars: A and B,10 μm; C–G, 100 μm; H, 1 mm; I–N,40 μm.)
The round clonal appearance as well as the size of the clusters suggested that the BMEL-GFP cells had proliferated in vivo. An antibody directed against Ki67, which is present in all cycling cells, demonstrates that within clusters of GFP-positive cells (Fig. 1C), many nuclei stain for Ki67 (Fig. 1D), indicating that the transplanted cells, similarly to the host cells, are proliferating.
Cell Fusion Is Not Implicated in the Formation of BMEL Cell Clusters in the Liver. It has been demonstrated that purified hematopoietic stem cells (HSC) are able to contribute to liver repair and rescue FAH-/- mice, which are mutant for an essential liver-specific gene encoding fumarylacetoacetate hydrolase (24). However, recent studies have revealed that the rescue by HSC is due to their fusion with resident liver cells (16, 17). As was previously observed in hepatoma–lymphoid cell hybrids (25), expression of the silent wild-type FAH allele would be activated by fusion of the HSC with a resident FAH-/- hepatocyte, conferring the required selective advantage in the mutant mouse.
We made use of differences in H-2 haplotypes between CBA/C57Bl6-derived BMEL cells and the BALB/c/C57Bl6 host livers: BMEL cells are H-2Kk and host livers are H-2Dd. Fig. 1 E, F, and G show, respectively, immunostaining of GFP and H-2Kk for the transplanted cells and H-2Dd for the host liver cells. The mirror images reveal two crucial points. First, the transplanted cells express donor H-2Kk (Fig. 1F) but not host H-2Dd (Fig. 1G). Therefore they are not hybrid cells. In none of the numerous sections examined have clusters of cells expressing both H-2 alleles been observed. Second, in some repopulated areas, more cells express H-2Kk than GFP, implying either that the expression of GFP is extinguished or that some clusters are from a mixture of GFP-positive and -negative cells. We conclude that H-2Kk is a more reliable marker of donor cells.
In addition to the experimental demonstration that cell fusion is not involved in BMEL cluster formation, it is relevant to recall that a hybrid cell would continue to express the Alb-uPA transgene, and thereby would be destined for elimination, not for selection.
Quantitation of BMEL Cell Contribution to the Liver. To determine the extent of BMEL cell participation in liver repopulation, selected fields were quantitated, using either GFP or H-2Kk staining. In addition, for objective evaluation, entire liver sections were photographed, the tissue was reconstructed, and donor cell occupation was determined (Fig. 1H). The analysis of selected fields (Table 1) indicated that BMEL cells constituted 1.1–6.1% of the liver when analyzed by GFP, and in each case 2- to 5-fold more after staining for H-2Kk, attaining values up to 22%. However, the more objective analysis achieved by quantitation of entire liver sections gave lower numbers. For example, in one liver BMEL cells repopulated at 4.5% judging from H-2Kk staining, whereas GFP gave lower values. The differences in measured levels of repopulation can be explained by the facts that up to 45% of BMEL cells do not express GFP, and that cytomegalovirus promoter-directed transgene expression may be extinguished with time in vivo as described (12). Counts of the total numbers of clusters per liver section (Fig. 1H and Table 1) revealed unexplained variability in the efficiency with which the cells homed to and engrafted individual livers. Finally, despite the proliferation of BMEL cells in vivo, we have not seen a clear increase of BMEL cell participation in liver regeneration with time. This lack of increase is most probably due to elimination of the nonsyngeneic BMEL cells by the residual immune system of the recipient SCID mice. Indeed, CD45+ immune cells (Fig. 1 I and L), including macrophages (Fig. 1K) and neutrophils (Fig. 1N), are seen throughout the livers and surrounding BMEL cells (Fig. 1 J and M).
Table 1. Quantification of BMEL-GFP cell participation in regeneration of Alb-uPA/SCID mouse livers.
| Percent repopulation
|
||||||
|---|---|---|---|---|---|---|
| Chosen fields
|
Total section
|
|||||
| Donor cells | Time in vivo, wk | GFP | H-2Kk | GFP | H-2Kk | Clusters per section |
| 9A1-GFP | 3 | 6.1 ± 2.3 | 2.1 ± 0.7 | 141 (18) | ||
| n = 9 | n = 5 | |||||
| 14B3-GFP | 3 | 1.7 ± 0.9 | 3.1 | 0.8 | 19 (2) | |
| n = 4 | n = 1 | |||||
| 9A1-GFP | 5 | 4.5 ± 5.2 | 5.3 | 0.8 | 8 (4) | |
| n = 4 | n = 1 | |||||
| 14B3-GFP | 5 | 6.0 ± 3.1 | 11.5 ± 3.1 | 2.0 | 68 (16) | |
| n = 6 | n = 3 | |||||
| 9A1-GFP | 8 | 4.7 ± 3.2 | 22.3 ± 3.7 | 1.6 | 4.5 | 15 (7) |
| n = 6 | n = 3 | |||||
| 14B3-GFP | 8 | 1.1 ± 1.0 | 0.3 | 7 (4) | ||
| n = 2 | ||||||
Total liver sections are the composite of ≈30 images through a ×2.5 objective. Although only one field is presented for five of the six livers, each represents 30 chosen fields. Results are presented as mean ± SD; n = number of fields analyzed. Numbers in parentheses represent the number of bile duct areas per liver section.
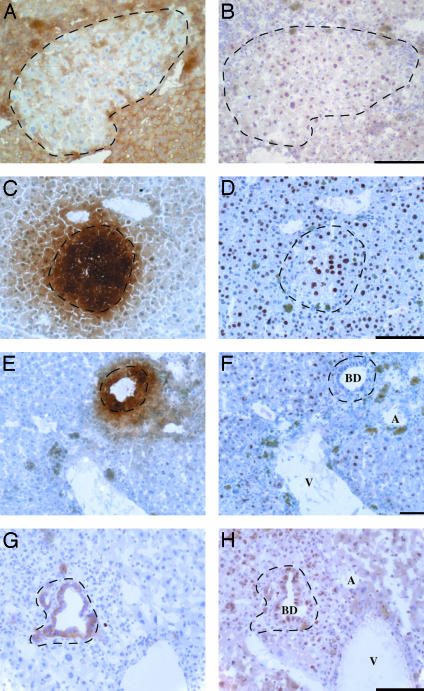
The Fine Regulation of Transcription Factor Expression Is Respected in BMEL Cells in Vivo. An important criterion to evaluate the differentiation of the transplanted BMEL cells is expression of transcription factors in hepatocytes and bile duct cells. HNF1α and HNF4α are among the most characteristic markers of differentiated hepatocytes, whereas HNF1β is expressed more abundantly in bile duct cells than in hepatocytes, and HNF4α is absent from bile ducts (26–28). HNF1α (Fig. 2B) and HNF4α (Fig. 2D) are expressed in parenchymal areas that are occupied by donor BMEL cells (Fig. 2 A and C) as well as in the surrounding host hepatocytes. Immunostaining for both hepatocyte-enriched transcription factors shows variability in positivity of the large round hepatocyte nuclei, whether of host or donor origin. As is true for host cells, bile ducts formed by BMEL cells (Fig. 2 E and G) do not express HNF4α (Fig. 2F) and strongly express HNF1β (Fig. 2H). The transcription factors and hepatic functions expressed in bile ducts and hepatocytes in vivo, as well as in BMEL cells in basal culture, or engrafted in the livers described here, are summarized in Table 2.
Fig. 2.
BMEL-derived hepatocytes and bile ducts localized within a portal triad show appropriate nuclear and cell-type-specific expression of liver-enriched transcription factors. BMEL-GFP cells are recognized as H-2Dd-negative (A), GFP-positive (C and E), or H-2Kk-positive (G). (B and D) HNF1α and HN4α, respectively, revealing nuclear staining of cells within the BMEL cluster of hepatocytes. (F) HNF4α is expressed in hepatocytes but not in bile ducts, and is absent from the BMEL-derived bile duct shown. (H) HNF1β is strongly expressed in bile ducts, including the duct formed by BMEL cells, whereas it is weakly expressed in hepatocytes. In B, D, F, and H, immune cells with small nuclei are visible surrounding the BMEL cell islands. BD, bile duct; A, artery; V, vein. (Scale bars: 100 μm.)
Table 2. BMEL cells differentiate in vivo with the same gene expression pattern as host cells.
| BMEL cells in vivo
|
Host liver cells
|
||||
|---|---|---|---|---|---|
| Markers | BMEL in vitro | Hepatocytes | Bile ducts | Hepatocytes | Bile ducts |
| HNF1α, β | + | + | + | + | + |
| HNF4α | + | + | — | + | — |
| DPPIV | ND | + | — | + | — |
| CPSI, GS | — | + | — | + | — |
| CK7, CK19 | + | — | + | — | + |
HNF1α, HNF1β, and HNF4α are liver-enriched transcription factors. DPPIV, CPSI, and GS (glutamine synthetase) are hepatocyte enzymes. CK7 and CK19 are expressed only by bile duct cells. ND, not determined.
BMEL Cells Differentiate as Mature Hepatocytes. A similar analysis was performed to determine whether the hepatocyte-like BMEL-GFP clusters express the appropriate hepatocyte functions in synchrony with the host hepatocytes. Because the cells were derived from embryonic day 14 liver, when mainly hepatoblasts are present, it was important to verify that the cells expressed the functions characteristic of adult hepatocytes. Therefore, we choose to examine genes that are expressed late during development: DPPIV, which shows apical expression in the bile canaliculi (29), and two enzymes involved in ammonia metabolism characterized by mutually exclusive and zonal expression along the hepatic acinus: glutamine synthetase and CPSI (30). The hepatocytes that span the region between the portal and central veins are designated as an acinus. Nutrientrich blood flows from the portal to the central veins, traversing the acinus and establishing a physiological gradient, reflected by the zonal expression of metabolic enzymes. CPSI is expressed by all hepatocytes except those nearest the central vein, which express glutamine synthetase instead.
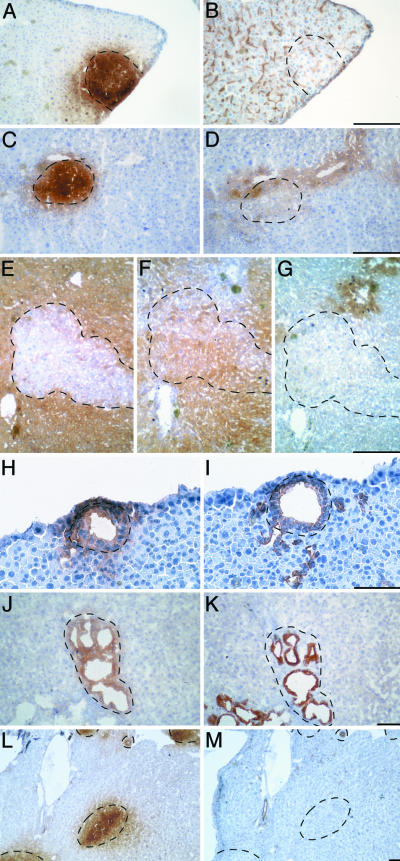
Fig. 3 A and B shows adjacent serial sections stained for GFP and DPPIV, for which the canalicular staining is visible as channels running between hepatocytes. The homogenous staining for DPPIV shows that BMEL cells are appropriately polarized and suggests that neighboring host- and donor-derived hepatocytes communicate through their bile canaliculi. While the region corresponding to GFP-positive cells does show canalicular staining, the staining appears somewhat weaker in the center. In Fig. 3 C and D are depicted GFP and glutamine synthetase: the GFP-positive area covers a region extending to a vessel identified as a central vein where glutamine synthetase is accumulated. The enzyme-rich regions contain both host and donor hepatocytes that show indistinguishable staining. BMEL cells, recognized as H-2Dd-negative (Fig. 3E), located in the hepatic acinus express CPSI (Fig. 3F) and not glutamine synthetase (Fig. 3G). The anticipated mirror image staining pattern is obtained. This series of results demonstrates that donor BMEL cells not only express the appropriate functions characteristic of adult liver but also respond to the signals controlling hepatocyte polarity and to the physiological stimuli responsible for zonal expression along the liver acini.
Fig. 3.
BMEL cells differentiate as hepatocytes and as bile ducts in the regenerating liver. (A–G) Analysis of serial sections with antibodies specificto hepatocyte markers showing that BMEL cells differentiate as hepatocytes in the liver, displaying the same characteristics as host hepatocytes. BMEL cells are recognized in A, C (GFP staining), and E (H-2Dd staining). All host and donor cells (A) in the section localize the bile canalicular marker DPPIV (B) to the apical pole. Note that clusters of large hepatocytes are seen in B for transplanted cells, and in C and D for host cells. Glutamine synthetase (D) is expressed by hepatocytes close to the central vein and by BMEL-GFP cells (C) located in this region. CPSI (F) is produced by hepatocytes throughout the liver parenchyma except those located in the central vein region, where glutamine synthetase (G) is expressed instead. The BMEL-derived hepatocytes (E) express these adult functions exactly as the neighboring host-derived hepatocytes. (H–M) BMEL-GFP cells differentiate as bile ducts in regenerating liver and express bile duct-specific CK7 and CK19. A bile duct formed by BMEL-GFP cells (H) expresses CK19 (I). A BMEL cell-derived bile duct recognized as H-2Kk-positive (J) expresses CK7 (K) as host-derived bile ducts. Regions of proliferating bile ducts are intrinsic to the Alb-uPA/SCID model because they are also observed in mice that have not been injected with BMEL cells. CK19 staining is restricted to bile ducts (L, GFP; M CK19). (Scale bar: 100 μm.)
BMEL Cells Form Bile Ducts in Vivo. Even though the BMEL cells formed structurally normal bile ducts, we investigated whether the appropriate genes were expressed. A characteristic marker of cholangiocytes is their pattern of CK expression, in particular CK7 and CK19, which are bile duct specific (31). Fig. 3 H and I demonstrates that a BMEL-GFP-positive bile duct (Fig. 3H) expresses CK19 (Fig. 3I). Fig. 3J shows a cluster of bile ducts, only some of which are BMEL derived as seen by the presence of H-2Kk. CK7 staining (Fig. 3K) reveals that donor and host bile ducts show indistinguishable staining. Finally, Fig. 3L shows areas of BMEL-GFP hepatocytes as well as a bile duct; the CK19 staining (Fig. 3M) is restricted to the bile ducts. While BMEL cells express CK7 and CK19 in vitro, this expression is retained only in the bile ducts and is lost when the cells are in hepatocyte clusters (Table 2). Thus, BMEL cells respond to their environment within the liver by extinguishing or activating the expression of genes in a cell type-specific manner.
Discussion
Taken together, these results demonstrate that permanent lines of nontransformed BMEL cells home to the liver, where they participate in regeneration of hepatocytes and bile ducts. They respond to signals from the host for regulated proliferation and differentiation, expressing just like the host cells a series of markers for bile ducts and hepatocytes, including transcription factors, cytoskeletal proteins, and enzymes. Furthermore, the donor-derived hepatocytes are polarized and show zonal expression of liver-specific metabolic enzymes. The repopulation percentages of the two BMEL lines appear at first sight to be modest (Table 1). However, this occurs despite a demonstrable and significant immune response against the donor cells (Fig. 1 I–N). In future experiments two parameters can be addressed to improve BMEL cell participation in liver regeneration. First, a significant enhancement of repopulation efficiency can be obtained with homozygous rather than heterozygous Alb-uPA mice (32). Second, the cases in the literature reporting majority or total repopulation of the liver all involve congenic systems (2, 12). To meet future challenges of cell-based therapy for liver regeneration it will be essential to elaborate effective strategies for overcoming immune rejection.
It can legitimately be questioned whether there is a future for cell lines in the repair of damaged tissues, in particular because cells of lines can usually generate transformed progeny. A first point is to recall the behavior of teratoma cell lines and embryonic stem (ES) cells: both are cell types that develop into tumors when injected s.c. in mice, but when placed in the environment of a developing embryo they participate in the formation of numerous tissues, without causing tumors (33–35). Second, the development of cell lines appears to be a discipline where much remains to be explored, including the effects of environmental factors (substratum, diffusible factors, cell density) on the normal or abnormal behavior of the cells (36). The feasibility of shuttling between cell culture and an organ in vivo will provide a valuable and stringent experimental means of characterizing stem cell potential and defining factors implicated in commitment and differentiation, where the possible emergence of an aggressive variant is not an issue. Finally, should projects to use cell lines for experimental gene therapy be developed, the integration of an inducible suicide gene should be considered.
The results reported here indicate the existence of a previously unrecognized class of permanent cell lines that can be derived from essentially any mouse embryo: cells that are bipotential and immortalized, yet nontransformed and capable of contributing to tissue repair. They are even able to give rise to two distinct cell types within a tissue. It may prove possible, by using similar strategies (15), to isolate such lines from other embryonic tissues, and other species including human, although immortalizing genes could be necessary (37, 38). The use of cell lines for tissue repair would have many advantages in stem cell-based gene therapy. Before transplantation such cells could be manipulated to harbor genes whose regulated expression would stimulate or limit their growth in vivo, deliver into the blood stream a product deficient in the host, or permit ablation of the cells by activation of a suicide gene.
Acknowledgments
We thank J. di Santo, P. Sansonetti, M. Yaniv, G. Milon, and L. Fiette for counsel, Céline Mulet for cryostat sections, and members of the laboratory for discussions. The Weiss laboratory is supported by grants from the Association pour la Recherche contre le Cancer, the Human Frontiers Science Program RG0303/2000-M, an agreement with Baylor College of Medicine/Institut Pasteur, which in turn was supported by Grant 1 U01 DK63588-01 from the National Institutes of Health (coordinator G. Darlington), and the Institut Pasteur Grand Program Horizontal 07 on Stem Cells. H.S.M. was supported by fellowships from the Fondation pour la Recherche Médicale and the Grand Program Horizontal on Stem Cells.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Alb-uPA, albumin–urokinase plasminogen activator; BMEL, bipotential mouse embryonic liver; CPSI, carbamoyl-phosphate synthetase I; CK, cytokeratin; DPPIV, dipeptidyl peptidase IV; HNF, hepatocyte nuclear factor; SCID, severe combined immuno-deficiency disease.
References
- 1.Rhim, J. A., Sandgren, E. P., Degen, J. L., Palmiter, R. D. & Brinster, R. L. (1994) Science 263, 1149-1152. [DOI] [PubMed] [Google Scholar]
- 2.Overturf, K., Al-Dhalimy, M., Tanguay, R., Brantly, M., Ou, C. N., Finegold, M. & Grompe, M. (1996) Nat. Genet. 12, 266-273. [DOI] [PubMed] [Google Scholar]
- 3.Overturf, K., Al-Dhalimy, M., Ou, C. N., Finegold, M. & Grompe, M. (1997) Am. J. Pathol. 151, 1273-1280. [PMC free article] [PubMed] [Google Scholar]
- 4.Laconi, E., Oren, R., Mukhopadhyay, D. K., Hurston, E., Laconi, S., Pani, P., Dabeva, M. D. & Shafritz, D. A. (1998) Am. J. Pathol. 153, 319-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallico, G. G., O'Connor, N. E., Compton, C. C., Kehinde, O. & Green, H. (1984) N. Engl. J. Med. 311, 448-451. [DOI] [PubMed] [Google Scholar]
- 6.Qu-Petersen, Z., Deasy, B., Jankowski, R., Ikezawa, M., Cummins, J., Pruchnic, R., Mytinger, J., Cao, B., Gates, C., Wernig, A. & Huard, J. (2002) J. Cell Biol. 157, 851-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampaolesi, M., Torrente, Y., Innocenzi, A., Tonlorenzi, R., D'Antona, G., Pellegrino, M. A., Barresi, R., Bresolin, N., Cusella De Angelis, M. G., Campbell, K. P., et al. (2003) Science 301, 487-492. [DOI] [PubMed] [Google Scholar]
- 8.Overturf, K., Al-Dhalimy, M., Finegold, M. & Grompe, M. (1999) Am. J. Pathol. 155, 2135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weglarz, T. C., Degen, J. L. & Sandgren, E. P. (2000) Am. J. Pathol. 157, 1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabeva, M. D., Petkov, P. M., Sandhu, J., Oren, R., Laconi, E., Hurston, E. & Shafritz, D. A. (2000) Am. J. Pathol. 156, 2017-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandhu, J., Petkov, P. M., Dabeva, M. D. & Shafritz, D. A. (2001) Am. J. Pathol. 159, 1323-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oertel, M., Rosencrantz, R., Chen, Y. Q., Thota, P. N., Sandhu, J. S., Dabeva, M. D., Pacchia, A. L., Adelson, M. E., Dougherty, J. P. & Shafritz, D. A. (2003) Hepatology 37, 994-1005. [DOI] [PubMed] [Google Scholar]
- 13.Malhi, H., Irani, A. N., Gagandeep, S. & Gupta, S. (2002) J. Cell Sci. 115, 2679-2688. [DOI] [PubMed] [Google Scholar]
- 14.Allain, J. E., Dagher, I., Mahieu-Caputo, D., Loux, N., Andreoletti, M., Westerman, K., Briand, P., Franco, D., Leboulch, P. & Weber, A. (2002) Proc. Natl. Acad. Sci. USA 99, 3639-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strick-Marchand, H. & Weiss, M. C. (2002) Hepatology 36, 794-805. [DOI] [PubMed] [Google Scholar]
- 16.Wang, X., Willenbring, H., Akkari, Y., Torimaru, Y., Foster, M., Al-Dhalimy, M., Lagasse, E., Finegold, M., Olson, S. & Grompe, M. (2003) Nature 422, 897-901. [DOI] [PubMed] [Google Scholar]
- 17.Vassilopoulos, G., Wang, P. R. & Russell, D. W. (2003) Nature 422, 901-904. [DOI] [PubMed] [Google Scholar]
- 18.Spencer, R. A., Hauschka, T. S., Amos, D. B. & Ephrussi, B. (1964) J. Natl. Cancer Inst. 33, 893-903. [DOI] [PubMed] [Google Scholar]
- 19.Wiener, F., Fenyö, E. M. & Klein, G. (1972) Nat. New Biol. 238, 155-159. [DOI] [PubMed] [Google Scholar]
- 20.Zennou, V., Petit, C., Guetard, D., Nerhbass, U., Montagnier, L. & Charneau, P. (2000) Cell 101, 173-185. [DOI] [PubMed] [Google Scholar]
- 21.Giannini, C., Morosan, S., Tralhao, G., Guidotti, J. E., Battaglia, S., Mollier, K., Hannoun, L., Kremsdorf, D., Gilgenkrantz, H. & Charneau, P. (2003) Hepatology 38, 114-122. [DOI] [PubMed] [Google Scholar]
- 22.Zito, M. A., Koennecke, L. A., McAuliffe, M. J., McNally, B., van Rooijen, N. & Heyes, M. P. (2001) Brain Res. 892, 13-26. [DOI] [PubMed] [Google Scholar]
- 23.Sandgren, E. P., Palmiter, R. D., Heckel, J. L., Daugherty, C. C., Brinster, R. L. & Degen, J. L. (1991) Cell 66, 245-256. [DOI] [PubMed] [Google Scholar]
- 24.Lagasse, E., Connors, H., Al-Dhalimy, M., Reitsma, M., Dohse, M., Osborne, L., Wang, X., Finegold, M., Weissman, I. L. & Grompe, M. (2000) Nat. Med. 6, 1229-1234. [DOI] [PubMed] [Google Scholar]
- 25.Brown, J. E. & Weiss, M. C. (1975) Cell 6, 481-494. [Google Scholar]
- 26.Ott, M. O., Rey-Campos, J., Cereghini, S. & Yaniv, M. (1991) Mech. Dev. 36, 47-58. [DOI] [PubMed] [Google Scholar]
- 27.Nagy, P., Bisgaard, H. C. & Thorgeirsson, S. S. (1994) J. Cell Biol. 126, 223-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sladek, F. M. & Seidel, S. D. (2001) in Nuclear Receptors and Genetic Disease, eds. Burris, T. P. & McCabe, E. R. B. (Academic, London), pp. 309-361.
- 29.Feracci, H., Connolly, T. P., Margolis, R. N. & Hubbard, A. L. (1987) Dev. Biol. 123, 73-84. [DOI] [PubMed] [Google Scholar]
- 30.Moorman, A. F., de Boer, P. A., Geerts, W. J., van den Zande, L., Lamers, W. H. & Charles, R. (1988) J. Histochem. Cytochem. 36, 751-755. [DOI] [PubMed] [Google Scholar]
- 31.Germain, L., Blouin, M. J. & Marceau, N. (1988) Cancer Res. 48, 4909-4918. [PubMed] [Google Scholar]
- 32.Mercer, D. F., Schiller, D. E., Elliot, J. F., Douglas, D. N., Hao, C., Rinfret, A., Addison, W. R., Fischer, K. P., Churchill, T. A., Lakey, J. R. T., et al. (2001) Nat. Med. 7, 927-933. [DOI] [PubMed] [Google Scholar]
- 33.Evans, E. P. & Kaufman, M. H. (1981) Nature 292, 154-156. [DOI] [PubMed] [Google Scholar]
- 34.Martin, G. R. (1981) Proc. Natl. Acad. Sci. USA 78, 7634-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradley, A., Evans, M., Kaufman, M. H. & Robertson, E. (1984) Nature 309, 255-256. [DOI] [PubMed] [Google Scholar]
- 36.Weiss, M. C. & Strick-Marchand, H. (2003) Semin. Liver Dis. 23, 313-324. [DOI] [PubMed] [Google Scholar]
- 37.Strick-Marchand, H. & Weiss, M. C. (2003) Mech. Dev. 120, 89-99. [DOI] [PubMed] [Google Scholar]
- 38.Wege, H., Hai, T. L., Chui, L. L., Wu, J. C., Giri, R., Malhi, H., Sappal, B. S., Kumaran, V., Gupta, S. & Zern, M. A. (2003) Gastroenterology 124, 432-444. [DOI] [PubMed] [Google Scholar]