Abstract
Because of the use of alternate exons 1, mammals express two distinct forms of Gsα-subunits: the canonical 394-aa Gsα present in all tissues and a 700+-aa extra-long αs (XLαs) expressed in a more restricted manner. Both subunits transduce receptor signals into stimulation of adenylyl cyclase. The XL exon encodes the XL domain of XLαs and, in a parallel ORF, a protein called Alex. Alex interacts with the XL domain of XLαs and inhibits its adenylyl cyclase-stimulating function. In mice, rats, and humans, the XL exon is thought to contribute 422.3, 367.3, and 551.3 codons and to encode Alex proteins of 390, 357, and 561 aa, respectively. We report here that the XL exon is longer than presumed and contributes in mice, rats, and humans, respectively, an additional 364, 430, and 139 codons to XLαs. We called the N-terminally extended XLαs extra-extra-long Gsα, or XXLαs. Alex is likewise longer. Its ORF also remains open in the 5′ direction for ≈2,000 nt, giving rise to Alex-extended, or AlexX. RT-PCR of murine total brain RNA shows that the entire XXL domain is encoded in a single exon. Furthermore, we discovered two truncated forms of XXLαs, XXLb1 and XXLb2, in which, because of alternative splicing, the Gsα domain is replaced by different sequences. XXLb proteins are likely to be found as stable dimers with AlexX. The N-terminally longer proteins may play regulatory roles.
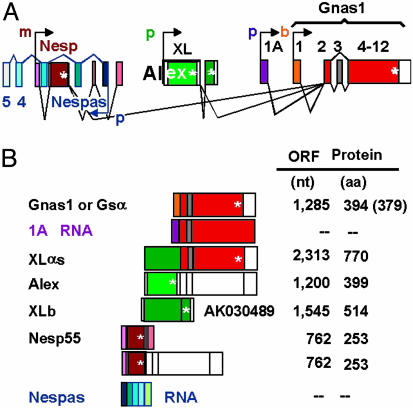
The murine α-subunit of the Gs G protein is encoded in the Gnas complex locus at the distal end of chromosome 2. This locus not only contains the promoter responsible for initiation of transcription at exon 1 of Gnas1 (46.3 codons), but it encodes at least four additional promoters, of which three drive transcription using the sense strand as template beginning at exons (5′ to 3′ direction) Nesp1, XL, and 1A, all upstream of Gnas1 exon 1; the fourth drives transcription using the antisense strand as template, beginning at exon Nespas1. Transcripts beginning with exons 1A and 1 of Nespas do not appear to be translated and code for RNAs of unknown function (reviewed in 1). The Nesp transcript codes for Nesp55 (neuroendocrine secretory protein of 55 kDa), which may play an important role in modulating central nervous system neurotransmission (2). Fully processed transcripts beginning with the XL exon give rise to the extra-long Gsα (XLαs) (3).
By analogy to the human Gsα (4), mouse (and rat) Gsα is presumed to be encoded in 13 exons, and, as happens with human and rat Gsα, mouse Gsα comes in at least two easily distinguishable flavors, long and short (394 and 379 aa, respectively), that depend on the inclusion or not of the 45-nt exon 3, of which each in turn can arise without (default) or with an added Ser, because of use of an alternative splice donor site at exon 4 (4, 5). Processing of the XL transcript has been reported to join its XL exon to exon 2 of Gsα, thus giving rise to XLαs, formed of the XL domain followed by the canonical GTPase and helical domains of the Gsα-subunit, in both its long and short variants. The XL domain is listed at 367 aa for the rat (GenBank accession no. X84047), corrected from an initial report of 498 aa (3, 7), 551 aa for the human (GenBank accession no. NM_080425), and 422 aa for the mouse (GenBank accession no. AF116268).
In addition to the ORF coding for the XL portion of XLαs, the XL exon was found to have a concurrent ORF that runs along the XL ORF and terminates with a stop codon at the exact end of the exon (6). The protein encoded in this ORF was termed Alex. It interacts with the protein encoded in the XL domain of XLαs, whose activity it appears to suppress (8). Alex has been reported as having lengths that vary among species, as did those of the XL domain of XLαs, with human, mouse, and rat Alex having 357, 390, and 561 aa, respectively. The binding of a recombinant human XL domain protein comprising the C-terminal 374 aa (stop codon at -7 from the end of the XL exon; GenBank accession no. NM_080425) to an N-terminal truncated Alex of 323 aa was shown to be lost in humans bearing a particular polymorphism associated with prolonged bleeding times and metabolic and neurologic abnormalities. This finding provided evidence for the physiological relevance of the extra-long forms of Gsα (8). Strong developmental and metabolic phenotypic changes found in mice harboring mutations in the locus encoding Gsα and XLαs have provided further evidence for roles of XLαs and Gnas1 in normal mammalian development (9, 10).
Fig. 1 illustrates the intron-exon structures of the murine Gnas complex locus, the processed mRNAs as obtained from GenBank supercontig NT_039211, region 3831406-3893830, and incorporates some of the features just described. In addition, the figure also incorporates the very interesting findings that transcription off the various promoters is under epigenetic control, being either exclusively from the maternal allele, as in the case of Nesp55, or from the paternal allele, as in the cases of XLαs, the antisense Nespas, and the transcript beginning with exon 1A. Gnas1 (the Gsα-subunit proper) mRNA is transcribed biallelically in most tissues (denoted by “b”). However, it should be noted here that in some tissues, such as proximal renal tubules, white and brown adipose cells, anterior pituitary cells, and thyroid, it is transcribed mostly from the maternal allele (for reviews see refs. 1 and 11; see also refs. 13-15).
Fig. 1.
Intronexon organization of the murine Gnas complex locus at the distal end of chromosome 2, orientation of the transcription units, and allelic expression characteristics as known before the present studies. (A) Intronexon organization. Neither exons (boxes) nor introns (interbox distances) are to scale; however, their relative positions reflect their positions in the genome. Exact parameters can be obtained from GenBank genomic contig NT_039211, region 3831406-3893830. p (paternal), m (maternal), and b (biallelic) next to arrows depicting promoter location and direction indicate a transcribed allele, where p transcripts are maternally silenced (imprinted) and m transcripts are paternally imprinted. (B) Main fully processed transcripts and their translation products. Two, 1A and Nespas, appear to be noncoding. *, Stop codon.
The present report had its origin in that, upon analysis of the mouse genome upstream of the presumed translation initiation site of the XL exon, the first in-frame stop codon is located 2.4 kb from this presumed translation initiating ATG. This raised the possibility that, inadvertently, the so far known XLαs is a truncated form of the real translation product.
PCR analysis of reverse transcripts synthesized by using adult mouse total brain RNA as template revealed that, indeed, the mRNA is longer than it had been thought to be. We propose that the XLαs protein synthesized (henceforth referred to as XXLαs, for extra-extra-long Gsα) is likely to have an N-terminal extension that in the mouse is of 364 aa. We also found that Gsα amino acids 221-280, which in the human gene are encoded in exons 9 and 10, are encoded in a single exon in the mouse.
Materials and Methods
Standard methods of molecular biology and of database searching for homologous and orthologous nucleotide similarities were used (ref. 16; see also www.ncbi.nih.gov). Brain RNA was isolated from adult male FVB/N mice by using RNeasy kits (Qiagen, Valencia, CA). Reverse transcripts were prepared from total RNA by using the SuperScript First-Strand Synthesis System (Invitrogen) with reagents and protocols supplied by the manufacturer. PCR was performed by using High Fidelity polymerase and reagents supplied by Roche Molecular Biochemicals. Primers used for PCR are shown in the figures and/or their legends. Amplifications were carried out as follows: 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 65°C for 1 min, and 72°C for 2 min, followed by 72°C for 7 min. The resulting PCR products were cloned into pCR2.1-TOPO (Invitrogen).
DNA sequencing was performed by using the dideoxynucleotide chain termination method. Templates consisting of plasmid DNA were subjected to the cycle PCR method by using protocols supplied by the National Institute on Environmental Health Sciences DNA sequencing facility. Briefly, ≈100 ng of plasmid DNA was used for each sequence reaction containing 8 μl of BigDye Terminator Ready Reaction mix (Applied Biosystems) and 1 μM of forward or reverse primer in a final volume of 20 μl. Cycling conditions were as follows: 1 cycle of 96°C for 2 min, followed by 25 cycles of 94°C for 10 s, 50°C for 5 s, and 60°C for 4 min. The sequence reactions were then cleaned by using a Qiagen DyeEx purification column. Purified sequence reactions were run on a PE-Applied Biosystem 3100 Automated Capillary DNA Sequencer. Resulting DNA sequences were compared with known database sequences by using both the GCG DNA Analysis Wisconsin package and National Center for Biotechnology Information (NCBI) blast to determine differences between the RNA (i.e., cloned PCR product) sequences and genomic sequences.
Results and Discussion
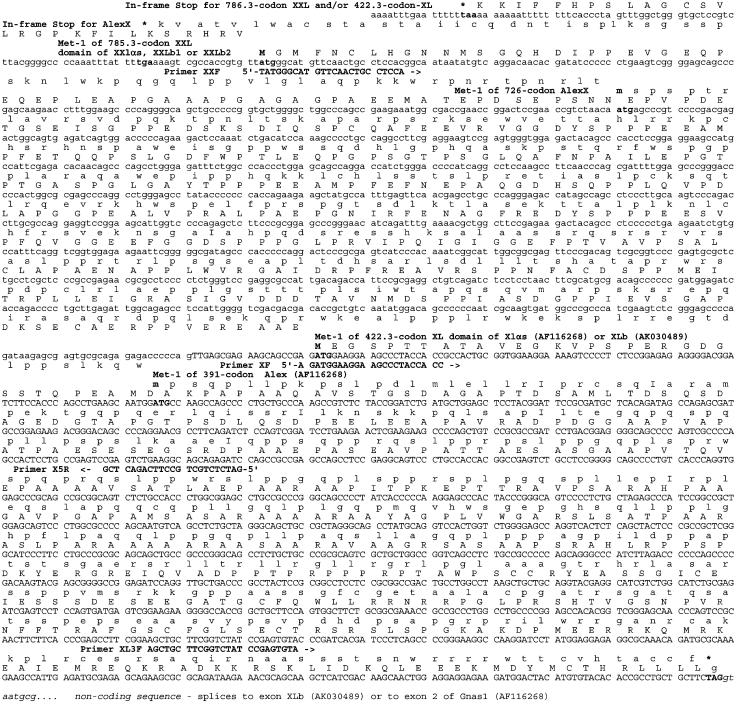
ORFs Associated with the Coding Sequence of the XL Domain and Alex Protein Have the Potential of Encoding a Significantly Longer Protein. As depicted in Fig. 2, translation of the genomic DNA sequence of the murine Gnas complex locus reveals that the ORFs coding for the XL portion of the mouse XLαs, as well as that coding for Alex, remain open in the 5′ direction for an additional 364 and 335 codons, respectively, for a total length of 785.3 and 726 codons. Analysis of the human and rat genome databases of the NCBI reveals a similar situation for rats and humans. Figs. 2 and 3A present the alignments of the amino acid sequences deduced from the mouse, human, and rat genomic DNA sequences of the extended forms of XL and Alex (referred to as XXL, for extra-extra-long, and AlexX, for Alex-extended).
Fig. 2.
The genomic sequence of mouse distal chromosome 2 encodes extended XL and Alex domains, XXL and AlexX, that are potentially much longer than suspected. Analysis of the mouse genomic DNA sequence (GenBank accession no. NT_039211, region 3844806-3847406), which encodes the XL and Alex domains of the Gnas1 complex locus, shows the existence of ORFs of 785.3 and 725 codons that run in parallel and encode in their 3′ end the murine XL and Alex domains of the 422.3-codon XL and 391-codon Alex domains reported in GenBank's AF116268 record. Shown are the nucleotide sequence and, above, the amino acids encoded in the two ORFs. Nucleotides in uppercase are from AF116268 (cDNA), as well as from NT_039211 (genomic sequence); nucleotides in lowercase are from NT_039211 only; nucleotides in italics are from intronic sequences. Amino acids in uppercase indicate the XL domain and its potential N-terminal extension, referred to as XXL; amino acids in lowercase indicate Alex and its potential N-terminal extension, referred to as AlexX (Fig. 4). Also shown, in bold and uppercase characters, are the forward (F) and reverse (R) PCR primers XXF, XF, X3F, and X5R (Fig. 4), which were used to test for the existence of the XXL/AlexX domains.
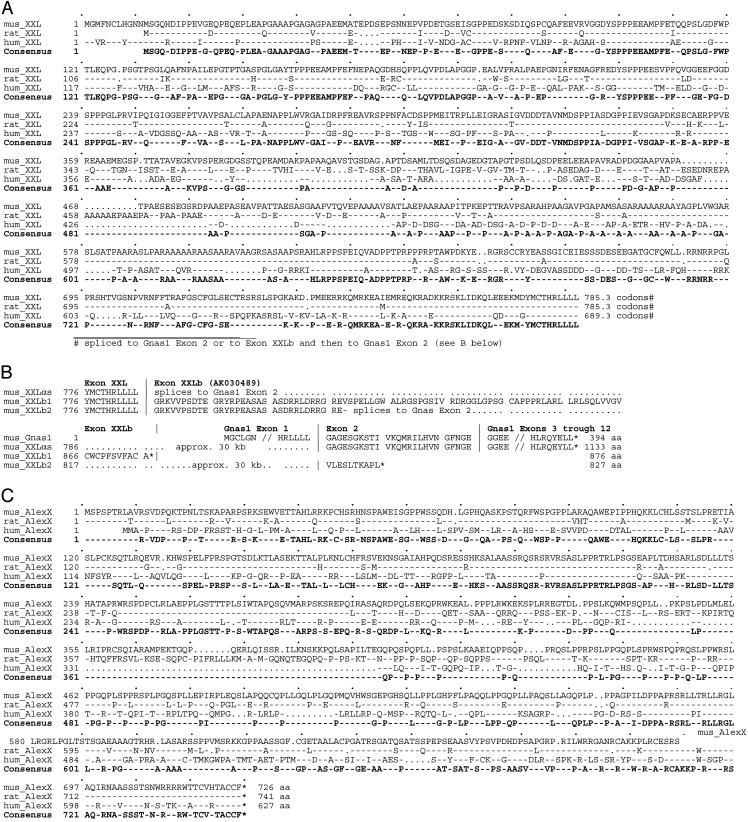
Fig. 3.
Amino acid sequence alignments of the extended mouse, rat, and human XXL (A) and AlexX (B) domains as deduced on the basis of RT-PCR analysis of mouse brain transcripts and mouse, rat, and human genomic DNA sequences accessible at www.ncbi.nih.gov. Amino acid sequences are based on: (i) the RT-PCR analysis reported in this study (Fig. 4); (ii) GenBank cDNA records X84047 (rat XLαs), AF116268 (mouse XLαs), AK030489 (mouse XLb), and NM_080425 (human XLαs); and (iii) orthologous sequences found in genomic contigs NT_039211 (mouse), NT_011362 (human), and NW_047679 (rat). (A) Alignment of XXL amino acid sequences; (B) Amino acid alignment comparing variations in sequence in the region coded for by exons lying between the end of the XXL exon and Gnas1 exon 2; (C) Alignment of AlexX amino acid sequences. The key to alignments follows. First line: master sequence to which the other sequences are compared. Second and third lines: -, identical to the master sequence; uppercase letter, different from the master sequence. Consensus sequence (bold): uppercase letters, amino acids identical in all sequences compared; -, different by at least one from the master sequence. *, Stop codon.
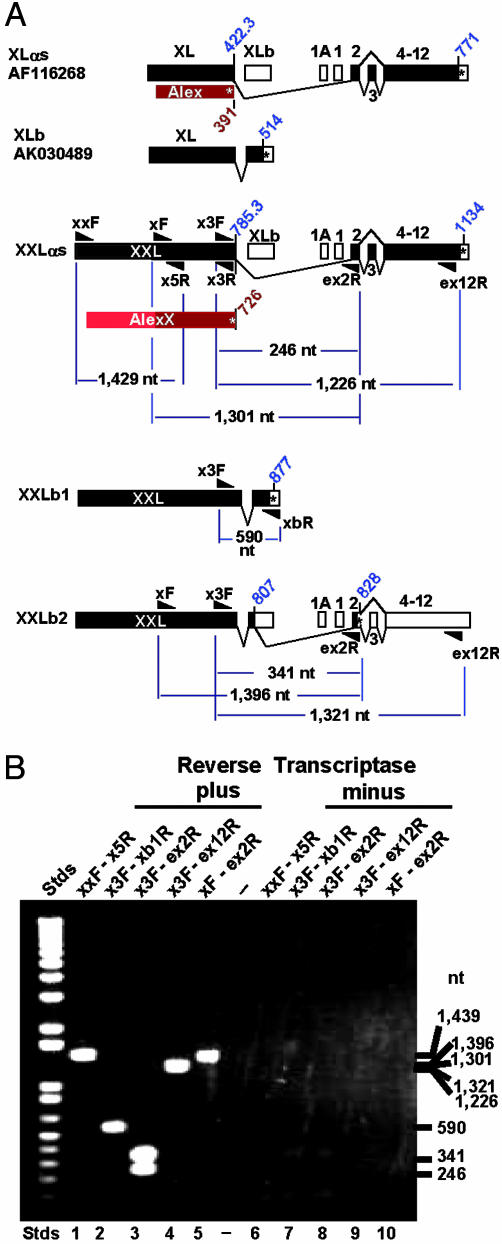
Using mouse brain RNA as template, we tested by RT-PCR for the existence of RNA transcripts of the predicted size and composition required for translation of XXLαs and AlexX, shown in Fig. 3. Fig. 4A presents the PCR strategy used, with predicted product sizes. Fig. 4B shows the PCR products obtained. These products were cloned into pCR2.1-TOPO and sequenced. The results confirmed the existence of an mRNA coding for XXLαs/AlexX. Importantly, lane 1, next to the sizing standards, shows a positive band of ≈1,450 bp that upon sequencing proved to be the expected 5′ 1,092-nt extension of the XL exon plus the first 347 nt of the XL exon (from primer XXF to XL5R, Fig. 2).
Fig. 4.
RT-PCR analysis of total mouse brain RNA shows presence of transcripts derived from the 5′ end of the extended XXL/AlexX ORFs depicted in Fig. 3. (A) Diagrams of proven and potential XL-related transcripts and approximate location of primers used to test for their existence. Primers XXF, XF, X3F, and X5R are defined in Fig. 3. Other primers were as follows: XLb1R, <-3′-CCAAT GGAAC ACTAA ACGCG GTAC-5′; Ex2R, <-3′-CTCAG ACCGT TTTCG TGGTA TGGTA ACAC-5′; Ex12R, <-3′-ACCTG TGACT CTTGT AGGCG GCACA-5′. Blue diagonal numbers indicate codons of ORFs. (B) Agarose gel electrophoresis of PCR products: XXL and AlexX exist, and XXL splices to Gnas1 exon 2 either directly, giving XXLαs, or via an intermediary alternative exon, XLb, giving rise to XXLb2. Note that the PCR analysis shows the presence of a splice variant that gives rise to XXLb2, whose mRNA differs from that of XXLαs in that it incorporates 95 nt of the XXLb exon. Lanes: 1-5, PCR products obtained by using reverse-transcribed single-stranded cDNAs as templates; 6-12, PCR products obtained by using the reaction products of a mock single-stranded cDNA synthesis reaction containing the RNA template but obtained by omitting the reverse transcriptase. Absence of products indicates absence of contaminating DNA that could have been used as template in the reactions shown in lanes 2-6.
The consecutively overlapping PCR products shown in lanes 2-5 (Fig. 4) proved the existence of extra-extra-long forms of XLb cDNA (GenBank accession no. AK030489) coding for XXLb1 and of XLαs coding for XXLαs. The XXLb1 nucleotide sequence predicts a protein of 876 aa [363 longer than that predicted by the AK030489 cDNA, which is part of a collection of 60,770 full-length cDNAs cloned by the RIKEN genome project (17)], which included numerous transcripts derived from the Gnas complex locus (18). Unexpectedly, PCR products derived from primers spanning from the XL domain to Gnas1 exon 2 and beyond (X3F to ex2R and ex12R, and XF to ex2R) comprised two products, each differing in 95 nt, that by sequence analysis constitute the 5′ end of the XLb exon spliced to the splice donor site of Gnas1 exon 2. We refer to the protein encoded in these mRNAs as XXLb2 (Fig. 3C). Exon XLb splices to exon 2 out of frame and leads to a second form of the XXLb protein, XXLb2, encoded in exons XXL, XLb (first 95 nt), and Gnas exons 2-12. XXLb1 in turn is encoded in exons XXL and XLb. The nucleotide sequence predicts the length of XXLb2 to be 827 aa. The nucleotide sequence of XXLαs, XXLb1, XXLb2, and AlexX have been deposited in the GenBank database with accession numbers AY519501, AY519503, AY519504 and AY519502, respectively.
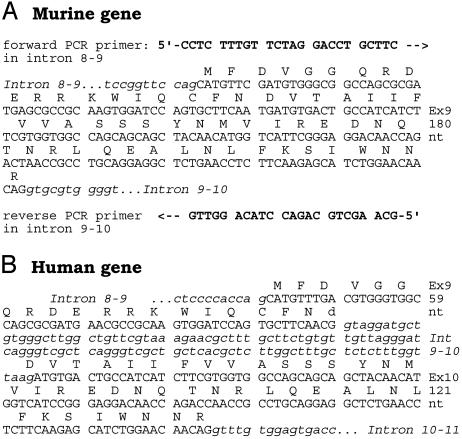
The Human Gsα-Subunit Is Encoded in 13 Exons, but the Mouse Orthologue Is Encoded in only 12. Throughout the text we have referred to the murine Gsα gene as a gene encoded in 12 exons, not 13 as is commonly done in the literature. The reason for this is that upon direct sequencing of the intron-exon boundaries of the mouse Gsα gene, we found that the intron that separates exons 9 and 10 in the human Gsα gene is absent from the murine gene (Fig. 5). As a consequence, the coding sequence of mouse Gsα has only 12 exons. The exon 9 sequences of DNA obtained from 129Sv, FVB/N, BALB/c, and C57BL/6J mice have been deposited in the GenBank database with accession numbers AY518306, AY518305, AY518307, and AY518308.
Fig. 5.
Nucleotide sequence of the region coding for Gsα amino acids Met-221 through Arg-280, which in Homo sapiens are encoded in exons 9 and 10, reveals them to be encoded in a single exon in Mus musculus. (A) The genomic region of interest was amplified by PCR (forward and reverse primers as shown) using genomic DNA from an FVB/N mouse and sequenced by the dideoxynucleotide chain termination method. Shown is the complete sequence of the amplified DNA and the deduced amino acid sequence encoded in the fully spliced RNA. This sequence (only partially shown) spans from the middle of exon 7 to the middle of the intron that follows the exon 9-10 coding region. The sequence shown for the murine gene was identical for DNA from 129SV, FVB/N, BALB/c, and C57BL/6J mice. (B) Human Gnas1 nucleotide and deduced amino acid sequence (ref. 4; GenBank accession no. NT_011362 region). Nucleotide sequences (in blocks of 10): uppercase, coding (exonic); lowercase italic, noncoding (intronic).
It is curious that the full-length sequence of the proteins encoded in the XXL exon has gone undetected for so long. We therefore searched the databases for ESTs and partial cDNAs that had as their origins the genomic region upstream of presumed Met-1 of XLαs. These searches returned one rat cDNA (GenBank accession no. AF093569) encoding a protein of unknown function, but whose N-terminal 70 aa are those of the rat AlexX shown in Fig. 3. Its sequence then diverges with alternating stretches of XXL and AlexX, ending with Alex. In addition, we found four ESTs, BY135711, BY785179, BX387357, and BX444740, that had template RNAs derived from the region immediately surrounding the ATGs of mouse XXL and AlexX. It would seem that the secondary structure of the RNA has prevented the cloning of the full-length cDNAs of the XXL/AlexX domains. The sequence alignments shown in Fig. 3 indicate a remarkable divergence in sequence identity for the XXL and AlexX domains (as was already true for XL and Alex) when the comparison is made among species. This divergence contrasts with the almost absolute conservation of the Gsα domain sequences and raises questions as to the functions (if any) that the XXLb and AlexX proteins may have.
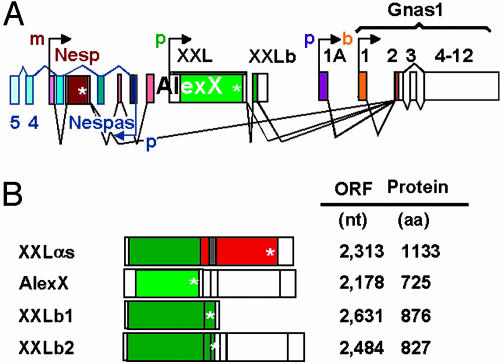
Given that the XL and Alex domains interact (6, 8), the existence of the XXLb proteins raises the possibility that the availability of XXLαs for signal transduction can be regulated by the relative abundance of the message coding for the adenylyl cyclase-stimulating XXLαs protein versus message coding for the Alex-binding, but adenylyl cyclase-indifferent, XXLb proteins. Whether cells at steady state accumulate AlexX in preference over XXLαs or XXLb will depend not only on the levels of mRNA but also on the relative turnover of these proteins. Finally, the data predict the existence of AlexX/XXLb dimers, whose function and subcellular locations also need to be explored. In Fig. 6 we summarize the expression and splice patterns uncovered in this report (Fig. 6A) and present an updated list of mRNAs not previously recognized in the available GenBank database (Fig. 6B). XXLαs, AlexX, XXLb1, and XXLb2 replace the XLαs, Alex, and XLb mRNAs depicted in Fig. 1 and are expressed in addition to the Gnas1 (Gsα), 1A RNA, Nesp55, and Nespas RNAs shown.
Fig. 6.
Update of splice patterns (A) and translation products (B) under control of the XXL promoter.
Abbreviations: XLαs, extra-long Gsα; XXLαs, extra-extra-long Gsα; AlexX, Alex-extended.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY518305-AY518308 and AY519501-AY519504).
References
- 1.Weinstein, L. S. (2001) J. Clin. Endocrinol. Metab. 86, 4622-4626. [DOI] [PubMed] [Google Scholar]
- 2.Ischia, R., Lovisetti-Scamihorn, P., Hogue-Angeletti, R., Wolkersdorfer, M., Winkler, H. & Fischer-Colbrie, R. (1997) J. Biol. Chem. 272, 11657-11662. [DOI] [PubMed] [Google Scholar]
- 3.Kehlenbach, R. H., Matthey, J. & Huttner, W. B. (1994) Nature 372, 804-809. [DOI] [PubMed] [Google Scholar]
- 4.Kozasa, T., Itoh, H., Tsukamoto, T. & Kaziro, Y. (1988) Proc. Natl. Acad. Sci. USA 85, 2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray, P., Carter, A., Simons, C., Guo, V., Puckett, C., Kamholz, J., Spiegel, A. & Nirenberg, M. (1986) Proc. Natl. Acad. Sci. USA 83, 8893-8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klemke, M., Kehlenbach, R. H. & Huttner, W. B. (2001) EMBO J. 16, 3849-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehlenbach, R. H., Matthey, J. & Huttner, W. B. (1995) Nature 375, 253. [DOI] [PubMed] [Google Scholar]
- 8.Freson, K., Jaeken, J., Van Helvoirt, M., de Zegher, F., Wittevrongel, C., Thys, C., Hoylaerts, M. F., Vermylen, J. & Van Geet, C. (2003) Hum. Mol. Genet. 12, 1121-1130. [DOI] [PubMed] [Google Scholar]
- 9.Cattanach, B. M., Peters, J., Ball, S. T. & Rasberry, C. (2000) Hum. Mol. Genetics 9, 2263-2273. [DOI] [PubMed] [Google Scholar]
- 10.Skinner, J. A., Cattanach, B. M. & Peters, J. (2002) Genomics 80, 373-375. [DOI] [PubMed] [Google Scholar]
- 11.Weinstein, L. S., Yu, S., Warner, D. R. & Liu, J. (2001) Endocr. Rev. 22, 675-705. [DOI] [PubMed] [Google Scholar]
- 12.Hayward, B. E., Kamiya, M., Strain, L., Moran, V., Campbell, R., Hayashizaki, Y. & Bonthron, D. T. (1998) Proc. Natl. Acad. Sci. USA 95, 10038-10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayward, B. E., Moran, V., Strain, L. & Bonthron, D. T. (1998) Proc. Natl. Acad. Sci. USA 95, 15475-15480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters, J., Wroe, S. F., Wells, C. A., Miller, H. J., Bodle, D., Beechey, C. V., Williamson, C. M. & Kelsey, G. (1999) Proc. Natl. Acad. Sci. USA 96, 3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, T., Vu, T. H., Zeng, Z. L., Nguyen, B. T., Hayward, B. E., Bonthron, D. T, Hu, J. F. & Hoffman, A. R. (2000) Genomics 69, 295-304. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 17.Fantom Consortium and Riken Genome Exploration Project Phase I and II Teams (2002) Nature 420, 563-573.12466851 [Google Scholar]
- 18.Holmes, R., Williamson, C., Peters, J., Denny, P. & Wells, C., RIKEN GER. Group, and GSL Members (2003) Genome Res. 13, 1410-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]