Abstract
Glutathione peroxidase 3 (GPx3) is a glycosylated member of GPx family and can catalyze the reaction of different types of peroxides with GSH to form their corresponding alcohols in vitro. The active center of GPx3 is selenocysteine (Sec), which is incorporated into proteins by a specific mechanism. In this study, we prepared a recombinant human GPx3 (rhGPx3) mutant with all Cys changed to Ser from a Cys auxotrophic strain of E. coli, BL21(DE3)cys. Although lacking post-translational modification, rhGPx3 mutant still retained the ability to reduce H2O2 and PLPC-OOH. Study on the quaternary structure suggested that rhGPx3 mutant existed as a monomer in solution, which is different from native tetrameric GPx3. Loss of the catalytic activity was considered to be attributed to both the absence of glycosylation and the failure of the tetramer. Further analysis was performed to compare the structures of rhGPx3 and GPx4 mutant, which were quite similar except for oligomerization loop. The differences of amino acid composition and electrostatic potentials on the oligomerization loop may affect the binding of large substrates to rhGPx3 mutant. This research provides an important foundation for biosynthesis of functionally selenium-containing GPx3 mutant in E.coli.
Glutathione peroxidases (GPxs) are a family of enzymes that catalyze the reduction of hydroperoxides to their corresponding alcohols. Thus far eight isoforms of GPx enzymes (GPx1–GPx8) have been found in mammals, of which five are selenoenzymes (GPx1–GPx4 and GPx6). GPx3, also known as plasma glutathione peroxidase or extracellular glutathione peroxidase, is the only known glycosylated extracellular isoform, which makes it unique among the selenium-containing members of GPx family. The human Gpx3 gene is located at region q32 of chromosome 5 and divided into five exons spanning about 10 kb, the first of which encodes the signal peptide for secretion1. The kidney is considered to be the main source for the GPx3 in plasma, which is primarily synthesized in the kidney proximal tubular cells2,3. GPx3 mRNA is also detected in other tissues, such as liver, heart, breast, lung4, skeletal muscle, pancreas, brain5, mature absorptive epithelial cells6 and thyroid7, from where Gpx3 is secreted into the surrounding extracellular environment. Although GPx3 can reduce different types of peroxides using GSH as the reducing agent in vitro8,9, the function of this enzyme is still a mystery due to the low concentration of GSH in plasma10,11.
GPx3 exists as a tetramer in its native state with a subunit molecular weight of approximately 23 kDa12. The crystal structure of human GPx3 revealed that each subunit contained a selenocysteine (Sec) residue and the active Sec-53 was located in a pocket on the protein surface with Gln-87, Trp-161 and Asn-162 nearby13, which formed a catalytic tetrad highly conserved in the selenium-dependent GPx family14. Sec, the 21st amino acid, is encoded by UGA, which is normally regarded as a stop signal. The decoding of UGA as Sec depends on a cis-acting element, known as the selenocysteine insertion sequence, and several trans-acting factors. The mechanism of Sec incorporation is quite different between prokaryotes and eukaryotes, thus very little work has been done in the area of preparation of recombinant mammalian GPx in E. coli for the last decades. But recently our group have prepared a GPx1 and GPx4 mutant with all Cys converted to Ser from an E. coli BL21(DE3)cys, which showed significant activity comparable to the native enzymes15,16.
The aim of this paper is to explore the possibility of preparing an active selenium-containing GPx3 mutant from E. coli. After obtaining this GPx3 mutant, we discussed the effect of glycosylation on enzymatic activity. Furthermore, the reason for the difference of quaternary structure between native GPx3 and its mutant was also explored.
Results
Preparation of rhGPx3 mutant
The hGPx3 mutant gene was cloned into expression vector pCold I for preparation of selenium-containing rhGPx3 mutant in E. coli BL21(DE3)cys using a single protein production system (Takara). The expression plasmid pCGPx3M in fusion with translation enhancing element sequence for improving the expression level, the hexahistidine tag for purification and factor Xa cleavage site was constructed successfully. Sequencing analysis confirmed that the Sec and Cys codon(s) of hGPx3 gene was(were) changed to Cys and Ser codon(s), respectively, and no undesired mutation was introduced during the cloning process. SDS-PAGE analysis of rhGPx3 mutant linked with excess amino acid sequence at N-termini showed a single band with a molecular mass of about 25.6 kDa. After being treated with factor Xa, the recombinant protein showed a mass decrease because of the removal of the excess amino acid sequence. Further analysis by Western blot showed that rhGPx3 mutant with excess amino acid could be recognized by the monoclonal antibody against the hexahistidine tag, but not in the case of the protein treated with factor Xa (Figure 1a and 1b). The results indicated that rhGPx3 mutant was successfully expressed and purified from E.coli BL21(DE3)cys. The yield of the recombinant selenoprotein was approximately 2.9 mg/L of culture. The concentration of selenium was found to be about 2.6 μg per mg protein, indicating that substitution ratio of Sec for Cys residues was approximately 84%.
Figure 1.
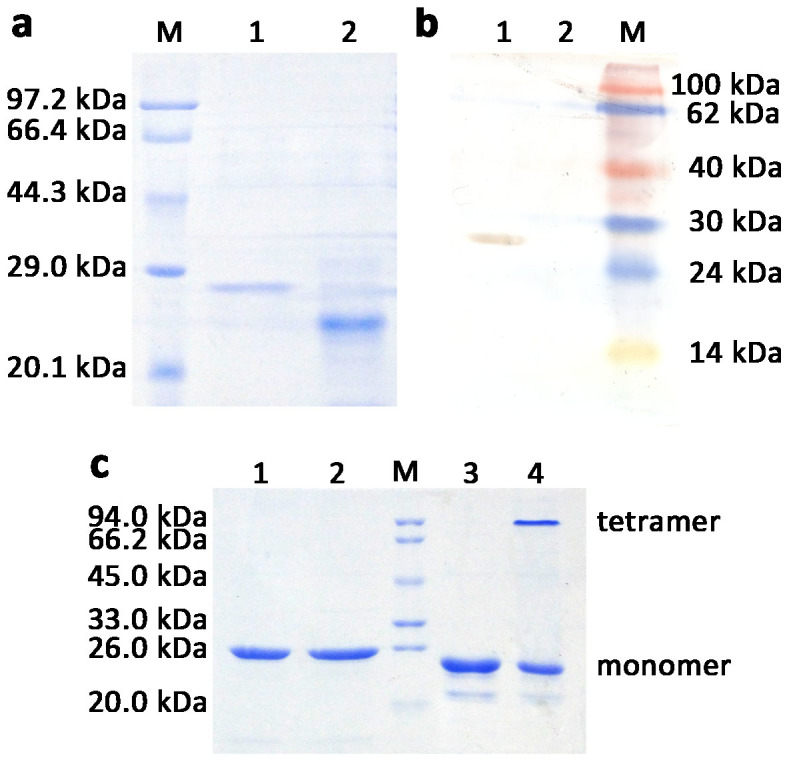
SDS-PAGE (a) and Western Blot (b) analysis of purified rhGPx3 mutant treated with (Lane 1) or without (Lane 2) factor Xa. (c). SDS-PAGE analysis of purified rhGPx3 and rhGPx1 mutants. Lane 1, purified rhGPx3 mutant under reducing condition; Lane 2, purified rhGPx3 mutant under non-reducing condition; Lane 3, purified rhGPx1 mutant under reducing condition; Lane 4, purified rhGPx1 mutant under non-reducing condition. M represents marker.
Analysis of the quaternary structure of rhGPx3 mutant
The purified rhGPx3 mutant was treated with loading buffer with or without β-mercaptoethanol and boiling, and the resulting product was separated by SDS-PAGE. We used a recombinant hGPx1 mutant prepared from E. coli BL21(DE3)cys as a control, which had been determined to exist, at least partially, as a tetramer15. The tetrameric and monomeric forms of rhGPx1 mutant could be detected under non-reducing condition (Figure 1c). Unlike rhGPx1 mutant, the rhGPx3 mutant was found to migrate as a single band under both reducing and non-reducing conditions. The results revealed that the rhGPx3 mutant existed as a monomer, which was different from native human GPx3.
Assay of enzyme activity
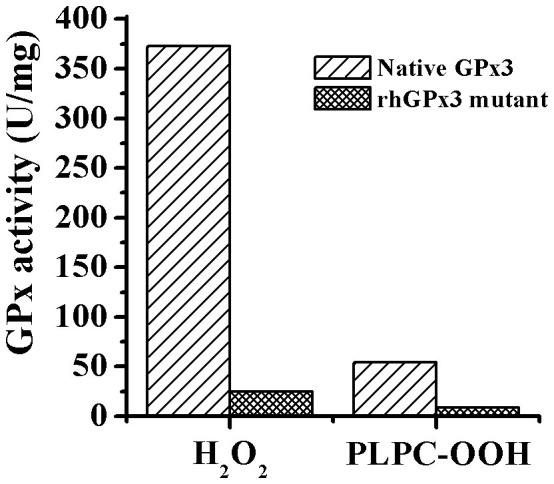
The rhGPx3 mutant showed activity of 25.0 U/mg and 8.5 U/mg using H2O2 and 1-palmitoyl-2-(13-hydroperoxy-cis-9, trans-11-octadecadienoyl)- L-3-phosphatidylcholine (PLPC-OOH) as an oxidizing substrate, respectively (Figure 2). We also determined the activity of the rhGPx3 mutant with the excess amino acid sequence. Equal amounts of proteins were added for spectrophotometric activity assays as described in Methods. And the results revealed that there was no significant difference in the activity between the rhGPx3 mutant with or without the excess amino acid sequence. The results suggested that the additional amino acids had little effect on the enzyme activity.
Figure 2. A comparison of activity between native GPx3 and rhGPx3 mutant by using H2O2 and PLPC-OOH as oxidizing substrate, respectively.
Determination of optimum temperature and pH
The rhGPx3 mutant showed the highest activity at 45°C and pH 8.9 (Figure 3), which was similar to that of native GPx1 (42°C and pH 8.8)17. Enzymatic activity decreased rapidly when temperature was raised above 50°C and the mutant was almost completely inactivated at 55°C. The enzyme was relatively stable over a pH range of 7.4–8.9, but it lost 93% of the original activity if the pH was raised to 9.8.
Figure 3. Effects of temperature and pH on activity of rhGPx3 mutant.
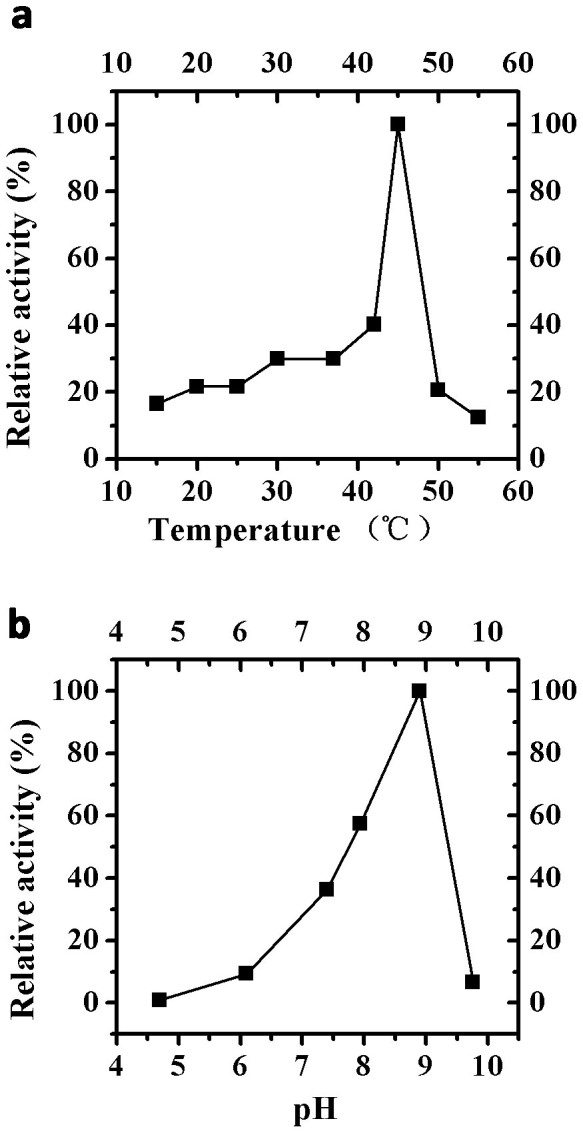
(a) Plot of GPx activity versus temperature. The activities were measured at the temperature range of 15–55°C at pH 7.4. The enzyme activity was assumed as 100% at 45°C. (b) Plot of GPx activity versus pH. The activities were measured in the pH range of 4.7–9.8 at 37°C. The enzyme activity was assumed as 100% at pH 8.9. The concentration of GSH and H2O2 used for assay was 1 mM and 0.5 mM, respectively. The scale of relative activity (%) indicates the percentage of the activity relative to the highest activity (100%) within each experiment.
Molecular modeling
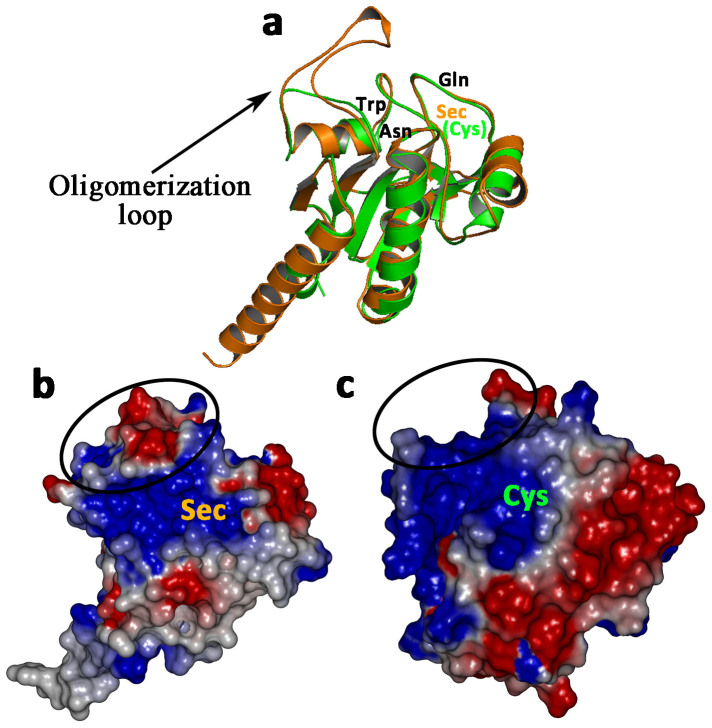
The profile-3D score of rhGPx3 mutant was determined to be 72.2, compared with the expected high score of 86.6 and the expected low score of 39.0. Ramachandran plot analysis showed 88.7% of residues in the core region, 11.3% in the allowed region, and no residues in the disallowed region. These results suggested that the conformation of rhGPx3 mutant was reliable. Figure 4a showed that rhGPx3 mutant was superimposed on U46C GPx4 mutant (PDB entry 2OBI) with a root mean square deviation of 2.87 Å, for 156 C-Alpha atoms.
Figure 4. Comparison between rhGPx3 mutant and U46C GPx4 mutant (PDB entry 2OBI).
(a). The superposition of rhGPx3 mutant (orange) and U46C GPx4 mutant (green). The catalytic tetrad consisting of Sec(Cys), Gln, Trp and Asn is labeled. (b). The electrostatic surface potential of rhGPx3 mutant. (c). The electrostatic surface potential of U46C GPx4 mutant. The red denotes negative electrostatic potential and the blue denotes positive electrostatic potential. Potential isocontours are shown at +3 kT/e (blue) and −3 kT/e (red).The circles in panels B and C indicate oligomerization loop.
Discussion
GPx, one of the most efficient antioxidants, can catalyze the reduction of hydroperoxides and protect cells against oxidative stress. Although GPx is a potential drug target, its therapeutic usage is restricted because of the limited sources. As the catalytic active site of selenium-containing GPx, Sec is irreplaceable with respect to activity. However, it was difficult to prepare mammalian selenium-containing GPx from E. coli due to the unique mechanism of Sec incorporation into protein. Recently, our group found that substitution of Ser for Cys would not cause dramatic loss of GPx activity compared to changing those Cys to Sec15,16. By using this method, we produced a GPx3 mutant, which showed significant activity in reducing both H2O2 and PLPC-OOH. Compared with native human GPx39, less than 10% of the activity was retained when the rhGPx3 mutant catalyzed the reaction of H2O2 to form the corresponding H2O, while 15.6% of the activity was retained using PLPC-OOH as the oxidizing substrate (Figure 2). The number was the lowest compared with the GPx1 mutant (28%) and the GPx4 mutant (40%) produced in our previous work15,16. GPx3 has been reported to be a glycoprotein12, but rhGPx3 mutant produced in E.coli lacked such post-translational glycosylation because of the difference between prokaryotic and eukaryotic cells. Considering the fact that the activity of recombinant GPx3 from mammalian cells was similar to that of partially purified human plasma GPx318, the significant drop in activity should be related to the absence of the post-translational modification. In addition, the substitution ratio of Sec for Cys residues using E. coli BL21(DE3)cys has been proved to be 84% in this study, which would also lower the activity of the mutant.
The tetrameric GPx3 was constituted by two asymmetric dimmers13, but the results above showed that rhGPx3 mutant did not even form such dimmer. Unlike rhGPx3 mutant, the monomeric form of the GPx1 mutant seemed to undergo “self-assembly” to form tetramer, at least partially, although the E.coli cells could not provide the same conditions as mammalian cells. Taking into account that rhGPx3 mutant was prepared using the same method, the failure to form tetramer should be mainly due to the structure factor. Aside from lacking post-translational glycosylation, rhGPx3 mutant have a different amino acid composition compared to native GPx3. In E. coli BL21(DE3)cys, the selenoprotein was produced via tRNACys misloading, suggesting that all Cys in the protein would be inevitably replaced by Sec. And previous studies in our laboratory have shown that conversion of Cys residues into Sec residues in GPx1 and GPx4 could result in dramatic loss of the catalytic activity15,16. To avoid the adverse effects caused by the introduction of multi-Sec residues, all Cys in GPx3 were changed to Ser in this study. However, analysis on native GPx3 showed a different mobility of reduced and nonreduced GPx3 monomer on SDS-PAGE, indicating the possible presence of disulfide bridges in native enzyme19. The results of the crystal structure of human GPx3 suggested the disulfide bridge was probably formed by Cys-12 and Cys-13613. Therefore, it can not be excluded that the existence of the disulfide bridge may contribute to the formation of tetramer, while no such disulfide bridge is present in rhGPx3 mutant. The absence of the disulfide bridge could cause structural change, which is another reason for the failure of formation of tetramer.
It has been reported that native GPx3 has a similar structure to that of the bovine GPx113. Study on bovine GPx1 revealed that only two GSH molecules could bind to each tetramer, indicating that the GSH binding site were formed by residues at least from two subunits20. Therefore, it is tempting to speculate that a similar situation might occur in the case of native GPx3, which would show the highest activity in tetrameric form. In comparison, rhGPx3 mutant seems more likely to catalyze the reaction just like GPx4, which is the only monomeric member among the GPx family, due to the fact that the mutant exists as a monomer in its native state. Actually the structures of U46C GPx4 mutant and rhGPx3 mutant are quite similar (Figure 4a) although their sequence identity is only 34%. The main difference between the two structures is the region opposite the active site, which is termed as oligomerization loop. The type of oligomerization loop in rhGPx3 mutant is mainly lined by hydrophilic residues (Thr-165, Ser-166, Asp-167, Arg-168, Trp-171 and Glu-172) and the surface of oligomerization loop is negatively charged (Figure 4b). The oligomerization loop of rhGPx3 protrudes from an area near the catalytic tetrad. In contrast, this area is a positively charged flat surface in GPx4 (Figure 4c) and has been considered to be involved in the efficient binding of complex lipid molecules at the active site21. The type of oligomerization loop in GPx3 was supposed to be related to the formation of the tetrameric complex22. But in rhGPx3 mutant, the hydrophilic environment and the negatively charged surface of oligomerization loop may limit the access of large, complex lipid substrates to the active site. This could explain why the activity of rhGPx3 mutant using H2O2 as oxidizing substrate was in the same order of magnitude as that of human native GPx4 (34.8 U/mg)9 and GPx4 mutant with all Cys converted to Ser (14.0 U/mg)16, while it was much lower than that of native GPx4 (64.6 U/mg) and the GPx4 mutant (24.6 U/mg) in the case of using PLPC-OOH as the oxidizing substrate.
In conclusion, a rhGPx3 mutant with Cys converted to Ser was obtained from E. coli BL21(DE3)cys. Although the activity of this mutant was about one order of magnitude lower than that of native GPx3, the expression level of rhGPx3 mutant in E. coli was significantly higher than that of recombinant GPx3 in eukaryotic cells. Unlike native enzyme, rhGPx3 mutant existed as a monomer, which was probably because of the differences in amino acid composition and the absence of post translational modifications. To our knowledge, this is the first time that a potential substitute for human GPx3 has been prepared from E. coli. Mass preparation of this enzyme is of great significance to explore the role of GPx3. There is good reason to believe that rhGPx3 mutant would exhibit the same function as native enzyme if the tetramer is formed. Future studies are needed to explore the mechanism of tetramer formation.
Methods
Construction of hexahistidine-tagged recombinant human GPx3 (rhGPx3) mutant expression vector
A GPx3 mutant gene without signal peptide sequence (nucleic acids 1–60) was designed on the basis of the cDNA sequence of human GPx3 (GenBank accession No. NM_002084.3), in which the codons for Cys-12 (TGC), Cys-81 (TGC) and Cys-136 (TGT) were replaced with the codons for Ser (TCG) and the codon for Sec-53 (TGA) was changed to Cys codon (TGC). The GPx3 mutant gene was synthesized and subcloned into the Nde I/Hind III sites of pUC57 by Sangon Biotech (Shanghai, China), yielding pUC57-GPx3M. The expression plasmid pCGPx3M encoding the human GPx3 mutant was constructed by subcloning the Nde I/Hind III fragment from pUC57-GPx3M into the Nde I/Hind III sites of the pCold I vector (TaKaRa). The construct was verified by DNA sequencing.
Overexpression and purification of rhGPx3 mutant
The hGPx3 mutant was expressed in an E. coli BL21(DE3)cys auxotrophic strain23,24,25 following the previously described protocol with slight modifications16,26. In detail, E. coli BL21(DE3)cys cells cotransformed with plasmids pCGPx3M and pMazF (TaKaRa) were grown at 37°C in 400 mL of MM medium (modified M9 minimal medium) supplemented with ampicillin (100 μg/mL), kanamycin (25 μg/mL), chloramphenicol (25 μg/mL) and Cys (50 μg/mL) on a shaker. At OD600 of 0.5, the culture was chilled in an ice-water bath for 5 min and incubated at 15°C for 45 min on a shaker. The cells were collected by centrifugation (5000 g for 5 min at 15°C) and washed twice with 0.9% NaCl at 15°C. The pellet was resuspended in 10 ml of MM medium supplemented with ampicillin (100 μg/mL), kanamycin (25 μg/mL), chloramphenicol (25 μg/mL), isopropyl-β-d-1-thiogalactopyranoside (1 mM) and Sec (600 μM). And then the culture was incubated at 15°C for 16 more hours on a shaker.
The cells resuspended in binding buffer (50 mM sodium phosphate, 300 mM NaCl, and 20 mM imidazole, pH 7.4) were sonicated for 10 min with middle pulse extension on ice. The supernatant was obtained by centrifugation (10000 g for 5 min at 4°C) and filtered using 0.45 μm filter. The supernatant was loaded on a Ni2+-immobilized metal affinity chromatography column previously equilibrated with binding buffer. The rhGPx3 mutant with the hexahistidine tag was bound on the column. The column was washed with 6 bed volumes of wash buffer (50 mM sodium phosphate, 300 mM NaCl, and 100 mM imidazole, pH 7.4) and the target protein was eluted with 6 bed volumes of elution buffer (50 mM sodium phosphate, 300 mM NaCl, and 300 mM imidazole, pH 7.4). Purified rhGPx3 mutant was incubated with factor Xa protease (1 μg/50 μg fusion protein in a total reaction volume of 50 μl) for 48 h at 4°C. Then, the cleavage reaction was dialyzed against 50 mM sodium phosphate (pH 7.4).
SDS-PAGE and Western Blot analysis
Twenty μL of sample containing 5 mg of purified rhGPx3 mutant was mixed with 20 μL of loading buffer (20 mM Tris-HCl, 8% SDS, 10% β-mercaptoethanol, 30% glycerol and 0.004% bromophenol blue at pH 6.8) and heated in boiling water for 5 min. The samples were subjected to standard SDS-PAGE on 12% polyacrylamide gels with a 5% stacking gel27. The gel was stained with Coomassie brilliant blue R-250 to visualize protein. To prepare non-reduced samples, β-mercaptoethanol was omitted from loading buffer and samples were loaded without boiling. Western blot analysis was performed using an anti-hexahistidine monoclonal antibody as described above15. Samples were separated on a 12% SDS-PAGE gel and transferred onto a nitrocellulose membrane. The membrane was incubated for 1 h in blocking buffer composed of Tween-Tris buffered saline (TTBS, 100 mM Tris, pH 7.5, 150 mM NaCl, 0.2% Tween 20) and 5% nonfat dry milk at 37°C. And then the membrane was incubated with mouse monoclonal anti-hexahistidine monoclonal antibody (Sigma) diluted (1:3000) in TTBS at 37°C for 1 h. The membrane was washed three times with TTBS for 5 min each and incubated with horseradish peroxidase conjugated goat anti-mouse IgG (Sigma) diluted (1:3000) in TTBS at 37°C for 1 h. After three washes with TTBS, immunoreactive protein band was identified by DAB staining. Total protein content was estimated by the Bradford method using bovine serum albumin as a standard. The selenium content of rhGPx3 mutant was determined by hydride generation atomic fluorescence spectrometer as described previously28.
Assay of enzyme activity
GPx activities of rhGPx3 mutant were determined using a previously described method29. Sodium phosphate (50 mM pH 7.4), EDTA (1 mM), GSH (2 mM), NADPH (0.25 mM), glutathione reductase (1 U) and samples (2.5 μM) were mixed in a cuvette at 37°C. The reagent mixture was incubated for 3 min at 37°C and the reaction was initiated by addition of (final concentration) 60 μM peroxides, at a total volume of 0.5 mL. The peroxides used in this study were H2O2 and PLPC-OOH, which was prepared by enzymatic hydroperoxidation of 1-palmitoyl-2-linoleoyl-L-3-phosphatidylcholine by lipoxidase as described precisely30. GPx activity was determined by measuring the decrease of NADPH absorption at 340 nm per min. Activity units (U) are defined as the amount of enzyme necessary to oxidize 1 μmol of NADPH per min at 37°C. The specific activity is expressed in U/mg. Samples were run in triplicate, and the values were averaged.
Determination of optimum temperature and pH
The optimum temperature and pH for GPx activity of rhGPx3 mutant was determined by performing enzymatic assays at different temperatures (15 to 55°C) at pH 7.4. The optimum pH was determined by performing enzymatic assays at different pH levels (4.7 to 9.76) at 37°C. GPx activities were measured using H2O2 as an oxidizing substrate with the same method as described above. Samples were run in triplicate, and the values were averaged.
Molecular modeling
Molecular modeling was performed with the Insight II package, version 2000 (Accelrys, San Diego, CA). The X-ray crystal structure of GPx3 with Sec-53 mutated to Gly (Protein Data Bank ID: 2R37) was used for the starting coordinates. The 3D structure of the GPx3 mutant with all Cys converted to Ser was refined by MD simulations and analyzed with Profile-3D and Procheck following the method as described previously31,32. The electrostatic potential was calculated using the DelPhi module.
Author Contributions
J.W., H.S. and D.L. initiated the joint project and J.W. designed all experiments. J.S. and Y.Y. contributed to manuscript preparation. J.S. and R.X. performed the MD simulations. Y.Y. and X.G. performed molecular biology experiments. All authors reviewed and edited the manuscript.
Acknowledgments
The authors thank Prof Marie-Paule Strub and August Böck for providing the E. coli Cys auxotrophic strain, BL21(DE3)cys. This work is supported by the National Natural Science Foundation of China (Nos. 30970633 and 31270851), Doctoral Funding Grants, Norman Bethune Health Science Center of Jilin University (No.470110000006) and Graduate Innovation Fund of Jilin University (No.2014060).
References
- Shinichi Y. et al. The human plasma glutathione peroxidase-encoding gene: organization, sequence and localization to chromosome 5q32. Gene 145, 293–297 (1994). [DOI] [PubMed] [Google Scholar]
- Avissar N. et al. Human kidney proximal tubules are the main source of plasma glutathione peroxidase. Am J Physiol-Cell Ph 266, C367–C375 (1994). [DOI] [PubMed] [Google Scholar]
- Whitin J. C., Bhamre S., Tham D. M. & Cohen H. J. Extracellular glutathione peroxidase is secreted basolaterally by human renal proximal tubule cells. Am J Physiol-Renal Physiol 283, F20–F28 (2002). [DOI] [PubMed] [Google Scholar]
- Chu F.-F., Esworthy R. S., Doroshow J., Doan K. & Liu X.-F. Expression of plasma glutathione peroxidase in human liver in addition to kidney, heart, lung, and breast in humans and rodents. Blood 79, 3233–3238 (1992). [PubMed] [Google Scholar]
- Martín-Alonso J.-M., Ghosh S. & Coca-Prados M. Cloning of the bovine plasma selenium-dependent glutathione peroxidase (GPx) cDNA from the ocular ciliary epithelium: expression of the plasma and cellular forms within the mammalian eye. J Biochem 114, 284–291 (1993). [DOI] [PubMed] [Google Scholar]
- Tham D. M., Whitin J. C., Kim K. K., Zhu S. X. & Cohen H. J. Expression of extracellular glutathione peroxidase in human and mouse gastrointestinal tract. Am J Physiol-Gastr L 275, G1463–G1471 (1998). [DOI] [PubMed] [Google Scholar]
- Howie A., Walker S., Akesson B., Arthur J. & Beckett G. Thyroidal extracellular glutathione peroxidase: a potential regulator of thyroid-hormone synthesis. Biochem J 308, 713 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esworthy R., Chu F., Geiger P., Girotti A. & Doroshow J. Reactivity of plasma glutathione peroxidase with hydroperoxide substrates and glutathione. Arch Biochem Biophys 307, 29–34 (1993). [DOI] [PubMed] [Google Scholar]
- Takebe G. et al. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem 277, 41254–41258 (2002). [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohé R. & Maiorino M. Glutathione peroxidases. BBA-Gen Subjects 1830, 3289–3303 (2013). [DOI] [PubMed] [Google Scholar]
- Wendel A. & Cikryt P. The level and half-life of glutathione in human plasma. FEBS Letts 120, 209–211 (1980). [DOI] [PubMed] [Google Scholar]
- Takahashi K., Avissar N., Whitin J. & Cohen H. Purification and characterization of human plasma glutathione peroxidase: a selenoglycoprotein distinct from the known cellular enzyme. Arch Biochem Biophys 256, 677–686 (1987). [DOI] [PubMed] [Google Scholar]
- Ren B., Huang W., Åkesson B. & Ladenstein R. The crystal structure of seleno-glutathione peroxidase from human plasma at 2.9 Å resolution. J Mol Biol 268, 869–885 (1997). [DOI] [PubMed] [Google Scholar]
- Tosatto S. C. et al. The catalytic site of glutathione peroxidases. Antioxid Redox Sign 10, 1515–1526 (2008). [DOI] [PubMed] [Google Scholar]
- Guo X., Song J., Yu Y. & Wei J. Y. Can recombinant human glutathione peroxidase 1 with high activity be efficiently produced in Escherichia coli? Antioxid Redox Sign 20, 1524–1230 (2013). [DOI] [PubMed] [Google Scholar]
- Yu Y. et al. Characterization and structural analysis of human selenium-dependent glutathione peroxidase 4 mutant expressed in Escherichia coli. Free Radical Bio Med 71, 332–338 (2014). [DOI] [PubMed] [Google Scholar]
- Wendel A. Glutathione peroxidase. Method Enzymol 77, 325–333 (1981). [DOI] [PubMed] [Google Scholar]
- Ottaviano F. G., Tang S.-S., Handy D. E. & Loscalzo J. Regulation of the extracellular antioxidant selenoprotein plasma glutathione peroxidase (GPx-3) in mammalian cells. Mol Cell Biochem 327, 111–126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avissar N. et al. Plasma selenium-dependent glutathione peroxidase. Cell of origin and secretion. J Biol Chem 264, 15850–15855 (1989). [PubMed] [Google Scholar]
- EPP O., LADENSTEIN R. & WENDEL A. The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur J Biochem 133, 51–69 (1983). [DOI] [PubMed] [Google Scholar]
- Scheerer P. et al. Structural Basis for Catalytic Activity and Enzyme Polymerization of Phospholipid Hydroperoxide Glutathione Peroxidase-4 (GPx4) §. Biochemistry-US 46, 9041–9049 (2007). [DOI] [PubMed] [Google Scholar]
- Toppo S., Vanin S., Bosello V. & Tosatto S. C. Evolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid Redox Sign 10, 1501–1514 (2008). [DOI] [PubMed] [Google Scholar]
- Mueller S. et al. The formation of diselenide bridges in proteins by incorporation of selenocysteine residues: biosynthesis and characterization of (Se) 2-thioredoxin. Biochemistry-US 33, 3404–3412 (1994). [DOI] [PubMed] [Google Scholar]
- Sanchez J.-F., Hoh F., Strub M.-P., Aumelas A. & Dumas C. Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein. Structure 10, 1363–1370 (2002). [DOI] [PubMed] [Google Scholar]
- Strub M.-P. et al. Selenomethionine and selenocysteine double labeling strategy for crystallographic phasing. Structure 11, 1359–1367 (2003). [DOI] [PubMed] [Google Scholar]
- Suzuki M., Mao L. & Inouye M. Single protein production (SPP) system in Escherichia coli. Nat Protoc 2, 1802–1810 (2007). [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- Cava-Montesinos P., Cervera M. L., Pastor A. & de la Guardia M. Hydride generation atomic fluorescence spectrometric determination of ultratraces of selenium and tellurium in cow milk. Anal Chim Acta 481, 291–300 (2003). [Google Scholar]
- Wilson S. R., Zucker P. A., Huang R. R. C. & Spector A. Development of synthetic compounds with glutathione peroxidase activity. J Am Chem Soc 111, 5936–5939 (1989). [Google Scholar]
- Maiorino M., Gregolin C. & Ursini F. [47] Phospholipid hydroperoxide glutathione peroxidase. Method Enzymol 186, 448–457 (1990). [DOI] [PubMed] [Google Scholar]
- Yu Y. et al. Characterization of catalytic activity and structure of selenocysteine-containing hGSTZ1c-1c based on site-directed mutagenesis and computational analysis. IUBMB Life 65, 163–170 (2013). [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., MacArthur M. W., Moss D. S. & Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26, 283–291 (1993). [Google Scholar]