Abstract
The metatranscriptomic recharacterization in the present study captured microbial enzymes at the unprecedented scale of 40,000 active genes belonged to 2,269 KEGG functions were identified. The novel information obtained herein revealed interesting patterns and provides an initial transcriptional insight into the thermophilic cellulose methanization process. Synergistic beta-sugar consumption by Thermotogales is crucial for cellulose hydrolysis in the thermophilic cellulose-degrading consortium because the primary cellulose degraders Clostridiales showed metabolic incompetence in subsequent beta-sugar pathways. Additionally, comparable transcription of putative Sus-like polysaccharide utilization loci (PULs) was observed in an unclassified order of Bacteroidetes suggesting the importance of PULs mechanism for polysaccharides breakdown in thermophilic systems. Despite the abundance of acetate as a fermentation product, the acetate-utilizing Methanosarcinales were less prevalent by 60% than the hydrogenotrophic Methanobacteriales. Whereas the aceticlastic methanogenesis pathway was markedly more active in terms of transcriptional activities in key genes, indicating that the less dominant Methanosarcinales are more active than their hydrogenotrophic counterparts in methane metabolism. These findings suggest that the minority of aceticlastic methanogens are not necessarily associated with repressed metabolism, in a pattern that was commonly observed in the cellulose-based methanization consortium, and thus challenge the causal likelihood proposed by previous studies.
Cellulose is Earth's most abundant biomass, and it is gaining worldwide attention as a renewable resource for bioenergy production1. In both natural and engineered systems, the bioconversion of lignocellulosic biomass benefits from synergistic reactions among microorganisms within a microbiome2. Our understanding of community dynamics and the ecological roles of microorganisms living in various cellulose-degrading communities has been revolutionized by the application of whole genome shotgun sequencing based on Next Generation Sequencing (NGS), which is also referred to as metagenomic sequencing. The NGS-based metagenomes of cellulose deconstruction microbiomes have revealed an unexpectedly high diversity of genes related to polysaccharide hydrolysis3,4 and were later shown to be an extensive resource for the discovery of novel glycoside hydrolases5. However, these excellent frontier explorations of genes embedded in genomes, which outline the broad genomic potential of a cellulolytic system, lack the ability to identify the pathways that are actually expressed or the key genes that are differentially transcribed.
Metaproteomics and metatranscriptomics (also known as RNA-seq) are both capable of providing useful insight into this expression issue. Nevertheless, the relatively low protein separation throughput of metaproteomics ranges from hundreds to thousands of identified proteins6,7,8 and, in some cases, fails to favor sufficient gene/protein coverage for reliable data interpretation. Some contradictory results were reported in these metaproteomic studies; for instance, Hanreich, A. et al. identified an unusually high expression of enzymes related to the methanogenesis process6, and other studies suggested that genes involved in carbohydrate metabolism are much more active8.
Relative to proteomic-based approaches, NGS-based metatranscriptomic sequencing has the advantage of providing massive gene identification, and the expression of these genes enables a comprehensive observation of functional microbes and genes involved in microbial processes. Thus far, this method has been applied most extensively in human-related medical disciplines to analyze the transcription profiles of genes related to certain symptoms9,10 and, to a lesser extent, to study gene expression in environmental samples, especially polysaccharide-active microbiota11. The aim of the present study was to gain insight into the transcriptional activities of the key genes and microbial populations involved in thermophilic cellulose deconstruction and to evaluate metatranscriptomics as a potentially suitable technology for this purpose. To this end, total RNA and DNA extracts were subjected to metatranscriptomic and metagenomic profiling, respectively, with an Illumina platform. The technical reproducibility of the nucleotide isolation method (DNA or RNA), Illumina library preparation and subsequent sequencing were quantified.
This first metatranscriptomic attempt to disclose the expression activity of genes that are involved in thermophilic cellulose decomposition may provide novel insights into the following subjects: (1) the differences between transcriptional activities and the genetic potential of carbohydrate-active genes (CAGs) in thermophilic cellulose deconstruction and (2) the active roles of various microbial populations in thermophilic cellulose deconstruction and methanogenesis. Additionally, the analytic procedure established here will serve as a reference for fundamental issues in the transcriptional quantification of gene activity in a metatranscriptome with unevenly distributed microbial populations.
Results
Reproducibility of RNA and DNA libraries
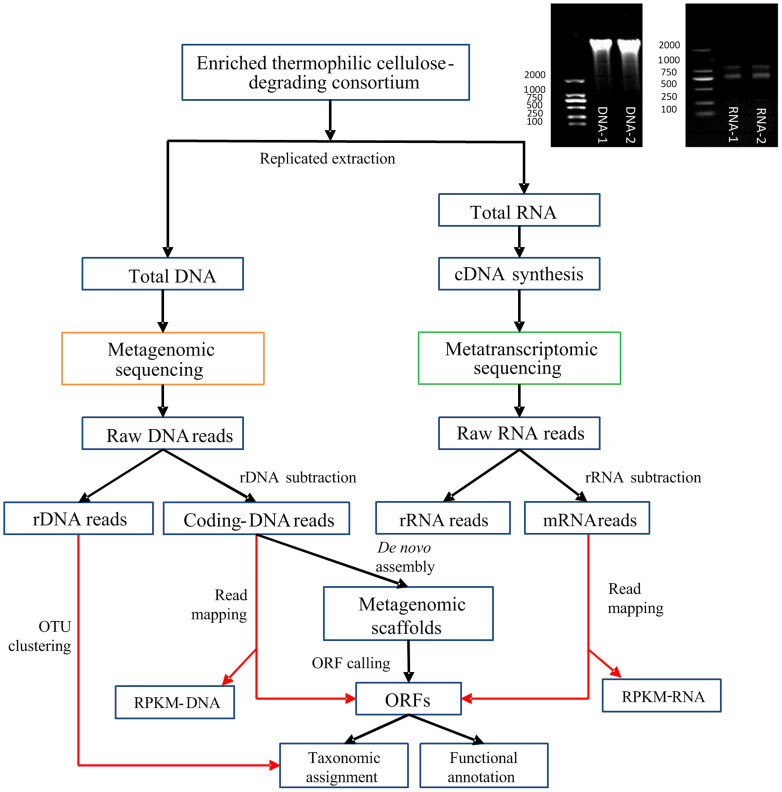
The analysis workflow combining replicated metatranscriptomic and metagenomic datasets is illustrated in Figure 1. First, the technical reproducibility of metagenomic and metatranscriptomic sample preparation (including RNA/DNA extraction, library construction and Illumina sequencing) was investigated using DNA and RNA library replicates, respectively. The reliable reproducibility of the metagenome preparation was confirmed by the strong consistency (R2 > 0.9, Table S1) of both the taxa composition and functional annotation between metagenomic replicates. However, the mRNA proportion of metatranscriptomics showed noticeable variations (with an R2 value of approximately 0.7, Table S1) when only two RNA samples from a single environmental sample were analyzed.
Figure 1. Schematic of the experimental and analytic workflow.
An electrophoresis gel of two replicates of extracted DNA (left) and total RNA (right) are shown.
Community structure
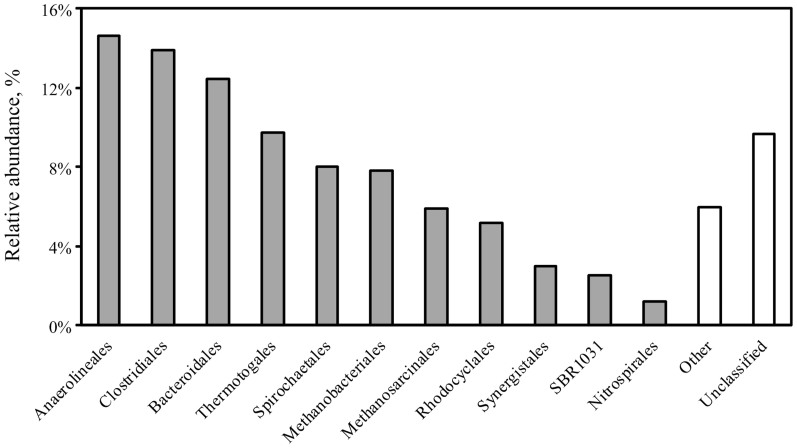
Approximately 0.06% of total DNA reads were designated as rDNA sequences in the DNA dataset (Table S2). Of the total rDNA reads, 87% could be aligned to the reference 16S sequences in the Greengenes database12. Based on clustering analysis, the thermophilic cellulose-degrading consortium is dominated by four bacterial orders, Anaerolineales (14.6%), Clostridiales (13.9%), Bacteroidales (12.4%) and Thermotogales (9.7%), in addition to the two archaeal populations, Methanobacteriales (7.8%) and Methanosarcinales (5.9%) (Figure 2).
Figure 2. Distribution of dominating orders (with a relative abundance larger than 1%) within the thermophilic cellulolytic community.
Metagenomic gene recovery and annotation
The rarefaction curve indicated that the sequencing depth was sufficient for de novo assembly (Figure S1). Ninety percent of the 50 million total DNA sequences were included in the de novo assembly by IDBA_UD. Assembled scaffolds longer than 300 bp exhibited an N50 of 10,963 bp (see Table S3 for detailed information of the metagenomic assembly). From the assembled scaffolds, 155,454 ORFs were predicted. Among these ORFs, 26.3% showed transcriptional activity with at least one aligned mRNA sequence. Among the 40,589 ORFs with transcriptional activity, 65.4% could be assigned to known phyla in the NCBI nr database, and 42.7% could be functionally annotated by SEED subsystems.
Transcriptional activities of major populations in the community
A comparison of transcriptional activities across different populations was initially based on the ratio of mRNA reads to coding DNA reads as proposed by Yu, K. et al.13 As shown in Figure S2, a significant annotation bias towards Proteobacteria was observed when using this approach in which the short reads were used directly as search queries. This bias mirrored the uneven representation of different taxa in the NCBI nr database, the effect of which was significantly enlarged by the low classification ratio of short reads (less than 40%, Table S4). Given the inherent difficulty of finding homologies for short NGS reads, the reliability of results built on this quantification approach is questionable14.
To circumvent this limitation, the transcriptional activities of protein-coding genes were investigated in an alternative manner based on the MRPKM value (the ratio of RPKM-RNA to RPKM-DNA as defined in Equations S1, S2, and S3) of the ORFs, which tended to have a much higher classification ratios (63.4% in present study, Table S4). As shown in Figure S3, species in Synergistetes and Nitrospirae showed the highest average MRPKM values of 1.5 and 1.2, respectively, compared to the other microbial groups in the community. Genes encoding enzymes related to cell mobility via bacterial flagella showed extraordinarily high expression (with MRPKM values larger than 100) in both of these phyla, suggesting their superior motility within the community.
Global function of the community
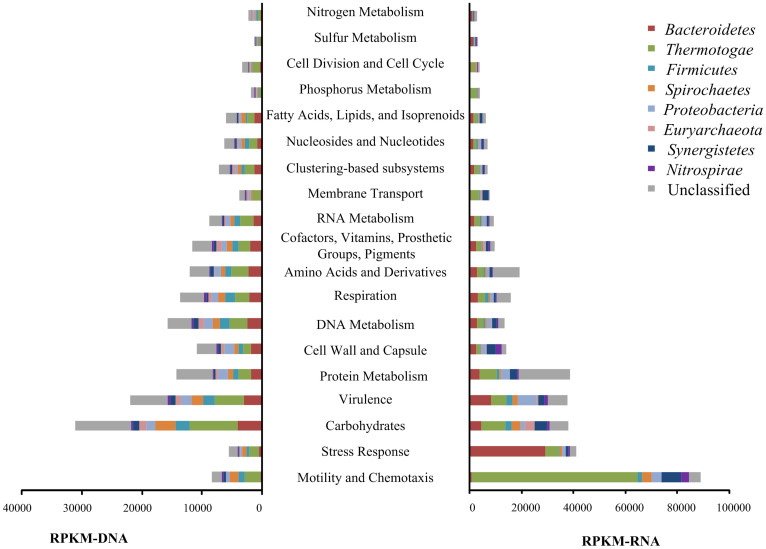
Several interesting phenomena were observed in the transcriptional pattern of SEED subsystems. First, the two subsystems Motility and Chemotaxis and Stress Response showed substantial transcription (Figure 3, right). Two microbial groups played major roles in these two subsystems, Thermotogae in Motility and Chemotaxis and Bacteroidetes in Stress Response (Figure 3, right). Thermotogales is also actively involved in cellulose hydrolysis through the expression of a wide range of beta-glycosidases (Figure 4). Extraordinary transcriptional activities (with MRPKM > 100) were observed for genes (alkyl hydroperoxide reductase [AhpC] and ferric ion binding domain) involved in cell protection against reactive oxygen species in Bacteroidetes.
Figure 3. Distribution of dominating orders in the expression and genomic profile of SEED subsystems in the thermophilic cellulose-degrading consortium.
Left figure: The relative abundance of various subsystems represented by RPKM-DNA, Right figure: The overall transcriptional activities of different subsystems indicated by RPKM-RNA. The subsystems were sorted from bottom to top according to their corresponding transcription activities.
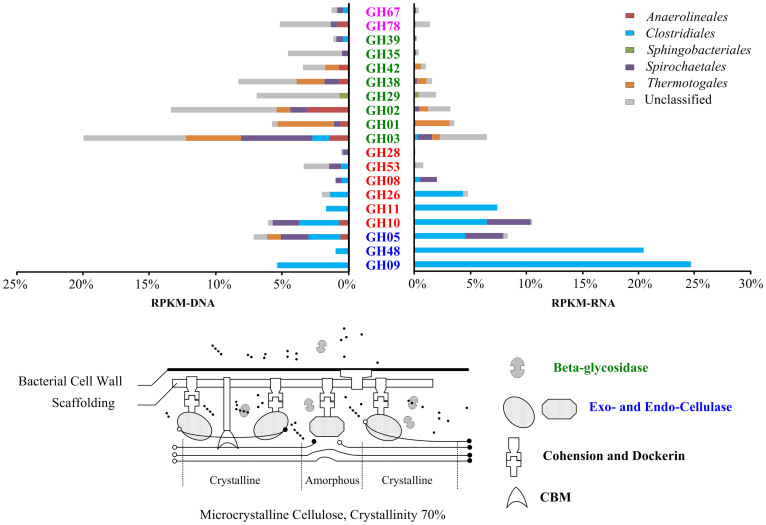
Figure 4. Distribution of various orders in the expression and genomic profiles of GH families related to lignocellulose decomposition in the thermophilic cellulose-degrading consortium.
Left figure: The relative abundance of various GH families as presented by RPKM-DNA, Right figure: The overall transcriptional activities of different GH families as indicated by RPKM-RNA. The GH families were sorted according to their corresponding transcriptional activities with labels colored according their functions in cellulose deconstruction, as defined by Pope et al.3: blue, Cellulases; orange, Endohemicellulases; green, Oligosaccharide-degrading enzymes; pink, Debranching enzymes. RPKM-RNA and RPKM-DNA values were compared as the percentage of all cellulose-hydrolysis related genes. Based on the attached-hydrolysis model of cellulosic biomass as proposed by Lynd, L. et al.1, the anaerobic hydrolysis process of microcrystalline cellulose was illustrated in the flowchart below, with the involved enzymes colored accordingly.
Expression of CAGs
Among the expressed CAGs, metabolic acclimation towards cellulosic substrate deconstruction was evident when enzyme families related to cellulose decomposition showed 1.5 times higher transcriptional activity than those related to other steps in carbohydrate metabolism. Furthermore, enzymes catalyzing cellulose hydrolysis (the blue-labeled cellulase families in Figure 4, right) are highly active despite their comparably lower prevalence in the metagenome (Figure 4 left). In contrast to the simplicity of cellulases, the thermophilic populations maintain the substantial genetic redundancy of oligosaccharide-degrading enzymes from 8 families (Figure 4 left). The excessive genetic diversity of oligosaccharide-degrading genes is regarded as crucial for cellulolytic consortia to maintain metabolic flexibility15.
As expected, Clostridiales play an essential role in cellulose chain breakdown by expressing a large proportion of the active exocellulases (GH05) and all of the endocellulases (GH48 and GH09). The most active endocellulases in Clostridiales (GH09 and GH48) are adjacent to genes encoding the heat shock proteins of the sigma factor and HSP60, respectively (Figure S4). Additionally, the overrepresentation of cohesin and dockerin, proteins that are commonly involved in the formation of lignocellulolytic multi-enzyme complexes (cellulosomes)16 (Figure S5), indicated the vital role of cellulase proximity via cellulosome complexes of Clostridiales in initializing cellulose hydrolysis in this thermophilic consortium. Unlike active cellulase and hemicellulase expression, a transcriptional and genetic deficit of oligosaccharide-degrading enzymes, especially the typical beta-glycosidases of GH01, GH02 and GH03, were noted in this Clostridiales population (Figure 4 left). Spirochaetales and Thermotogales species are the other two active cellulose degraders in the thermophilic consortium that produce GH05 exocellulases and a wide range of beta-glycosidases. Another interesting transcriptional phenomenon occurred despite the general absence of hemicellulose substrates in this consortium, which is solely fed hexose-based polysaccharides (microcrystalline cellulose): hemicellulases (red-labeled GH families in Figure 4) of both Clostridiales and Spirochaetales exhibited noticeable transcriptional activities.
Expression of genes involved in methanogenesis
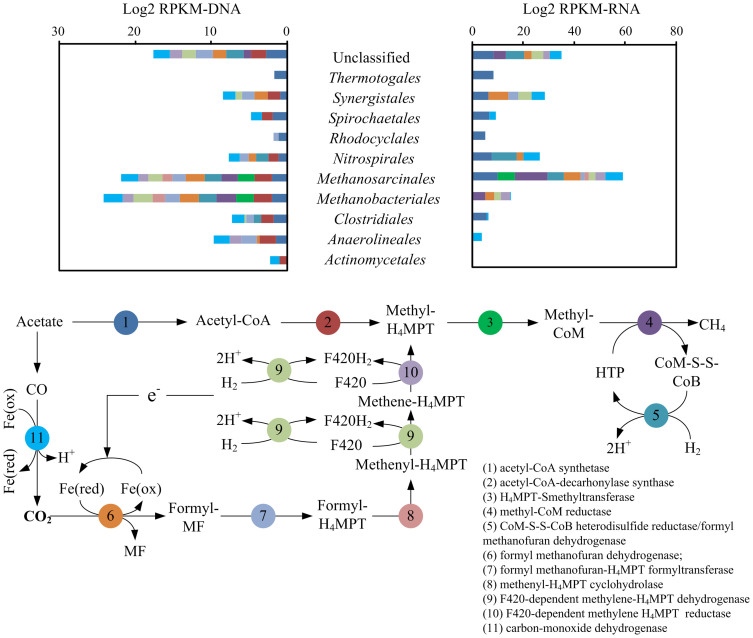
The transcriptional activities of genes involved in the thermophilic methanogenesis process were investigated in a similar manner based on RPKM-RNA and MRPKM. Genes involved in methanogenesis belong to the two archaeal orders Methanobacteriales and Methanosarcinales, with Methanobacteriales being the most prevalent by 60% (Figure 5, left). However, genes from the aceticlastic Methanosarcinales showed remarkably higher transcriptional activities (a six-fold higher MRPKM value) compared to those of the hydrogenotrophic Methanobacteriales. The overall aceticlastic pathway was three times more active than the hydrogenotrophic methanogenesis pathway in terms of the transcription of characteristic functional enzymes for each pathway (excluding mutual enzymes shared by hydrogenotrophic and aceticlastic pathways as represented by steps 3, 4 and 5 in Figure 5).
Figure 5. Transcriptional activities (right) and genomic prevalence (left) of genes in the methanogenesis pathway.
Genes were classified into various phylogenetic orders, as shown in the top figure, and colored according to their functions in the methanogenesis process (as adopted from the KEGG Methane Metabolism pathway), as shown in the flowchart (bottom figure).
Discussion
The technical reproducibility of NGS-based metatranscriptomic sequencing is a topic lacking wide recognition. At this early stage of applying RNA-seq to metatranscriptomes, it is common to see frontier research without an emphasis on replication, especially for technical replicates17,18,19. After an extensive literature survey, we found only one previous work that addressed the issue of technical replicates. Tsementzi, D. et al.20 noticed that the variability in technical replicates was almost as large as it was in the biological replicates. Their findings highlighted the presence of noticeable variation between mRNA technical replicates. In contrast, extensive studies have shown that the NGS-based transcriptome of a single organ (stem cells21, liver and kidneys22) or single species (yeast23) is highly replicable with little technical variation. Therefore, we speculate that inadequate sequencing coverage other than RNA extraction contributes primarily to the variation between metatranscriptomic technical replicates as observed in this study (Table S1), and the enormous amount of mRNA molecules, at approximately 8 × 1023 microbial mRNA molecules per liter of reactor sludge with volatile suspended solids of 800 mg/L (~200 mRNA molecules per bacterial cell24) makes it economically difficult to obtain the sequencing depth required to ensure the representation of the entire metatranscriptional profile25. The scale of variation observed in this study was comparable to that reported by Tsementzi, D. et al.20. Therefore even the large NGS-based metatranscriptional libraries constructed here (3.8 million mRNA sequences after filtering ribosomal and small RNA content out of 29.5 million Illumina reads for each replicated RNA library, Figure S2) could at best provide a snapshot of the major activities of the community at a particular time point. Additionally, we picked the peak of biogas production to ensure active microbial metabolism in the microbiota because based on the long-term monitoring the peak biogas formation occurred almost simultaneously with the highest cellulose uptake rate in the sequencing batch reactor (SBR) cycle.
Because of the annotation difficulty caused by the short reads of Illumina sequencing, we compared the distribution and function of different phylotypes within the community at the order level. The adaptation of the thermophilic microbial consortium to cellulose yielded simplified communities in which members of Anaerolineas, Clostridiales, Bacteroidales and Thermotogales (listed in order of dominance) were the most prevalent populations. The consortium showed notable microbial diversity with 700 species (Shannon Index of 6.7) (Figure S6), which was comparable to that of the cow rumen (approximately 1000 OTUs5) and the termite hindgut (800 OTUs and Shannon Index of 5.0511). Compared to our previous community analysis at 120 days26, a significant decline in the Clostridiales population and the consequent increase in Bacteroidales and Anaerolineas was noted in the SBR long-term operation at 545 days (this study). Eichorst, M. et al.27 observed a similar community shift from Firmicutes to a novel Bacteroidetes population in an aerobic thermophilic microbiome that was adapted to microcrystalline cellulose, and they argued that the gradual accumulation of solubilized cellulose after initial hydrolysis was the reason for this trend. In contrast, our annotation-based comprehensive protein database (NCBI nr database) did not disclose the direct metabolism advantage of Bacteroidales or Anaerolineas growing on soluble oligosaccharides (Figure 4). However, since unknown proteins (proteins that cannot be taxonomically classified), especially the beta-glycosidase of GH02 and GH03, played important roles in the oligosaccharide metabolism of the community, we cannot conclude that Bacteroidales or Anaerolineas are unimportant to oligosaccharide consumption. Instead, we observed the strong transcription of putative Sus-like polysaccharide utilization loci (PULs) (with RPKM-RNA comparable to GH09 observed in Clostridiales) in an unclassified order of the Bacteroidetes phylum following the identification protocol proposed by Rosewarne, C. et al.28. This phenomenon not only consolidated the involvement of Sus-like PULs from Bacteroidetes in cellulose-hydrolysis28,29 but also helped, in part, to reveal that the accumulation of the Bacteroidetes population in this cellulolytic consortia may actually be related to its special cellulose hydrolysis capability. Additionally, we noticed the strong transcription of genes involved in cell protection against oxygen species in Bacteroidales (Figure 3), indicating the ability of this population to grow in a facultative manner. We speculate that temporary oxygen exposure during sample preparation might cause the strong oxidative stress resistance observed in Bacteroidetes.
Clostridiales play vital roles in cellulose hydrolysis via cellulosome complexes. The proximity of endocellulases to heat shock protein explains the transcriptional advantage of these cellulases in response to high temperatures (Figure S4). The consistency of this genetic arrangement with that of Clostridium clariflavum DSM 19732 circumscribed the phylogenetic origin of the active Clostridiales as a branching strain of this species. Thermotogales only exhibited an exo-cutting capacity towards the exposed chains produced by endocellulases of Clostridiales, suggesting their dependency on Clostridiales for carbohydrate metabolism. In return, by expressing beta-glycosidases, both Spirochaetales and Thermotogales could facilitate the microbial uptake of tetrasaccharides and cellobiose, the accumulation of which will otherwise cause strong inhibition on Clostridiales cellulases such as GH48. This synergistic mechanism could explain the earlier observed cellulose degradation enhancement in Clostridium thermocellum when the Spirochaeta phylotypes were present within its environment30 and help to shed light on the ubiquitous presence of symbiotic Spirochetes in the gut of diverse termites11,31.
Because the consortium was absolutely predominated by Clostridiales at 120 days of enrichment, which constituted up to 70% of the community, it is reasonable to speculate that the Clostridiales populations contributed the majority of both the hydrolysis and beta-sugar metabolism of the community at this earlier time point. In contrast, a major contribution of beta-glycosidase activity was observed in Thermotogales and Spirochaetales populations within 545 days in the metatranscriptome, suggesting that synergistic effects between cellulose-hydrolyzing Clostridiales and beta-sugar-consuming Thermotogales and Spirochaetales play a critical role over the long-term in the SBR. However, further validation of this hypothesis is required to reveal the dynamics of population involvement in cellulose bioconversion. Additionally, we observed a strong mobility via bacterial flagella in Thermotogales that was consistent with the general cell motility reported in the cellulolytic members of this order32,33,34. This increase in the transcription of genes involved in cell motility highlights the importance of physical cell movement in facilitating the capture and breakdown of beta-sugars in Thermotogales.
Despite the general absence of hemicellulose substrate, a moderate transcription level of hemicellulases (labeled red in Figure 4) in Clostridiales and Spirochaetales was observed. Aside from the rare possibility of being housekeeping genes with expressions that are unaffected by experimental conditions, co-transcription with cellulases was more likely to be the machinery coordinating the hemicellulase regulation of Clostridiales and Spirochaetales populations. The lack of gene clusters for cellulases and hemicellulases35 had long been regarded as the genetic barrier for these two enzymes to co-transcribe in Clostridiales until the recent discovery of celC–glyR3–licA co-transcription in Clostridium thermocellum36. However, this co-transcriptional regulation has never been observed for strains in Spirochaetales before. In the present study, we did not observe genetic clusters consisting of cellulase and hemicellulase on the scaffolds of either Clostridiales or Spirochaetales. Thus, the observed co-transcription pattern of hemicellulases and cellulases in these two populations could serve to consolidate the co-transcription machinery that coordinates hemicellulase activities in these populations.
Despite the fact that acetate is always the major intermediate product, the prevalence of exclusively hydrogenotrophic Methanobacteriales over aceticlastic methanogens (primarily Methanosarcinales) is common in the cellulose-based methanization system6,37,38. Previously, the presence of over-competing hydrogenotrophic populations was interpreted as the influence of the inhibitory effect of environmental factors on the activity of the aceticlastic methanogens. Such factors include a high level of ammonia or volatile fatty acids (VFAs), extreme pH values or elevated temperatures39,40,41,42. However, our results may overturn this hypothesis because the less prevalent aceticlastic population showed significantly higher overall transcriptional activity in methanogenesis than its hydrogenotrophic counterpart. These findings suggested that the minority of aceticlastic methanogens is not necessarily associated with a repressed metabolism. Instead, we speculate that other overlooked factors, such as a slower growth rate of aceticlastic methanogens, may actually shape the Methanobacteriales-dominated distribution of methanogens in the thermophilic cellulose methanization consortium. This finding also indicates the weakness of studying methanogenesis pathways based on the phylogenetic prevalence of representative methanogens.
Hydrogenotrophic Methanobacteriales often co-exist with syntrophic acetate-oxidizing bacteria (SAOB), which facilitate the fermentation of acetate to hydrogen and carbon dioxide. Some researchers claimed that enhanced acetate oxidation by SAOB is crucial for maintaining the effectiveness of this hydrogen-utilizing methanogenesis pathway43,44,45; however, the attempt to enhance hydrogenotrophic methanogenesis via the bio-augmentation of the SAOB population was unsuccessful43. Therefore, the synergistic mechanism between Methanobacteriales and syntrophic bacteria remains unclear. Unfortunately, owing to the limited identification of thermophilic SAOB and the lack of known enzymes that are specific to its acetate oxidizing pathway, active SAOB populations could not be clearly identified. However, the active symbiotic involvement of Nitrospirales in methanogenesis suggested that the population actively transcribed CoM-S-S-CoB heterodisulfide reductase, which regenerated coenzyme M and coenzyme B after the final methanogenesis reaction (Step 4 in Figure 5). This population also exhibited visible activity towards sulfate reduction, which consumes hydrogen for electrons and thus provides a thermodynamically favorable environment for acetate oxidation to take place44,45.
By combining NGS-based metatranscriptomics and metagenomics, the present study provides initial transcriptional insights into the expressed biological functions during thermophilic cellulosic biomass methanization. Novel information on phylogeny and the functions of the 40,000 active genes identified in the metatranscriptome highlight the importance of complementary interactions between microbial groups (Thermotogales, Spirochaetales, and unclassified order of Bacteroidetes and Clostridiales) for efficient cellulose hydrolysis. More importantly, we observed stronger transcriptional activities in genes that were involved in aceticlastic methanogenesis pathways when the aceticlastic Methanosarcinales are less dominant than their hydrogenotrophic counterparts, Methanobacteriales. This finding contradicts the earlier hypothesis on the repressed activity of aceticlastic methanogens and suggests that the less prevalent aceticlastic populations could play more important roles in cellulose methanogenesis than previously expected. More intensive biological and technical replication is required to reveal whether this is a general pattern in similar systems. Further metatranscriptomic investigation of aceticlastic methanogenesis activity in lignocellulosic biomass methanogenesis systems, especially in the large-scale digesters, could help to better explain the ecological contributions of different methanogens during these processes and eventually provide practical guidelines for microbial manipulation in cellulose decomposition.
Methods
Thermophilic cellulose methanization consortium
Anaerobic digestion sludge (ADS) collected from a local wastewater treatment plant (Shek Wu Hui Sewage Treatment Plant, Hong Kong) was enriched in a sequential batch reactor (SBR, working volume of 800 ml) at 55°C, with microcrystalline cellulose (20 μm, Sigma, USA) as the primary substrate at a loading concentration of 2 g/L, and glucose was used as a co-substrate at a COD ratio of 10:1. Glucose was added to maintain the effectiveness of cellulose conversion in long-term operation; this effect from the monosugar co-substrate was discussed in detail in our previous study46. The pH of the SBR at the beginning was approximately pH 7.0 and was automatically maintained above pH 6.0 throughout the fermentation. Each batch was suspended when no more gas was produced. After 545 days of enrichment, the batch cycle time was stabilized at approximately 47 h. The adapted culture showed a cellulose uptake rate of 1.2 g/(L·d). Based on the continuous monitoring of SBR performance, we noticed that the peak biogas formation also corresponded to the maximum cellulose uptake speed in the SBR cycle. To ensure the active transcriptional status of genes encoding reactive enzymes that were involved in both cellulose hydrolysis and methanogenesis processes, sludge samples were collected from the SBR at peak gas formation, which occurred approximately 24 h after cellulose dosing.
DNA extraction
Two replicates of genomic DNA were individually extracted from 4 ml of sludge slurry (equal to an approximately 500-μg dry weight sample) from the thermophilic consortium with a FastDNA SPIN Kit for Soil (MP Biomedicals, LLC, Illkirch, France). The concentrations of extracted DNA replicates were 214.2 ng/μl with a 260/280 ratio of 1.88 and 203.7 ng/μl with a 260/280 ratio of 1.87 as quantified by a Nanodrop spectrophotometer (ND-1000, USA). Additionally, the quality of the extracted DNA and RNA was verified by electrophoresis (Figure S7).
Total RNA isolation and cDNA synthesis
At the same time point as the above DNA samples, total RNA isolation was performed with a TRIzol Plus RNA Purification Kit (Lift Technologies, USA) immediately after sampling. In brief, a 12-ml (roughly equal to a 1500-μg dry weight sample) consortium sample was centrifuged at 13,000 ×g for 2 min at 4°C. Replicates of the biomass pellets were subject to independent cell lysis and total RNA precipitation according to the manufacturer's instructions. Next, genomic DNA was removed with an Amplification Grade DNase I Kit (Sigma, USA). RNA qualification was performed by electrophoresis (Figure S7). The integrity of the extracted RNA was checked with a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The extracted RNA replicates had RNA integrity numbers of 6.7 and 6.6, respectively, on a scale of 1–10, with 10 representing no degradation (Figure S8). First-strand and second-strand cDNA were synthesized with the Superscript III First-Strand Synthesis SuperMix (Invitrogen, CA, USA) and the Second-Strand cDNA Synthesis Kit, respectively (BeyoTime, Jiangsu, China).
DNA, cDNA library construction and sequencing
Independent libraries of ~180 bp were prepared for replicated DNA and cDNA samples by following the manufacturer's instructions (Illumina). A sequencing depth of 2.8 G was applied to each metagenomic and metatranscriptomic replicate (Table S5).
Quality control (QC) of metagenomic and metatranscriptomic datasets
Metagenomic and metatranscriptomic raw reads derived from the Illumina Hiseq2000 platform were filtered for quality by discarding any read with 0.1% unknown nucleotides or with 50% nucleotides with a quality score lower than 20. Next, reads in the DNA dataset were checked for artificial duplicates that were produced during sequencing. The quality control for artificial duplicate reads were removed by a self-written script following the MG-RAST47 artifact identification protocol (See Table S5 for detailed dataset information after QC).
Bioinformatic analysis
Reproducibility analysis
Replicated libraries in the metagenomic dataset (named DNA datasets) and a metatranscriptomic dataset (named RNA datasets) were individually submitted to the MG-RAST server (Table S5 for accession number). Taxonomy annotation was performed using the Best Hit Classification Algorithm against the GenBank and SSU databases provided by the server. The cutoffs for searching against the SSU database had an E-value of 1E-20, a similarity of 60% and an alignment length of 50 bp. Cutoffs for searching against the GenBank database had an E-value cutoff of 1E-5, a similarity cutoff of 60% and an alignment length cutoff of 15 amino acids. Functional annotation was conducted against SEED subsystems using an E-value cutoff of 1E-5 and a hierarchical classification algorithm. The annotation based on the MG-RAST server was used for the reproducibility test.
rRNA subtraction and annotation
The analysis workflow is illustrated in Figure 1. First, rRNA sequences were separated from mRNA sequences in the post-QC RNA dataset by riboPicker48. Sequences encoding rRNA genes within the DNA dataset (named rDNA sequences) were simultaneously isolated from the protein-coding sequences (named coding DNA sequences) in the DNA dataset by BLAST49 against the Silva SSU (version 11.1)50 database with an E-value cutoff of 1E-20.
To reveal the community structure, rDNA sequences were aligned to reference ribosomal RNA sequences in the Greengene 13_5 database12. The aligned rDNA reads (87% of all rDNA reads) were clustered using the Uclust_ref algorithm51 adopted by QIIME 1.7.052 into operational taxonomic units (OTUs), with a similarity cutoff of 0.97.
Assembly of metagenomic and metatranscriptomic datasets
The artifact-filtered sequences of two replicated DNA datasets were assembled together using IDBA_UD53 with combined kmer sizes of 20, 40, 60, 80 and 100. Scaffolds longer than 300 bp were subjected to open reading frame (ORFs) prediction by MetaGeneMark54. To ensure the annotation accuracy of the derived ORFs, only ORFs longer than 300 bp were kept for expression analysis.
ORF annotation
ORFs were searched against the nr database using Rapsearch55 at an E-value cutoff of 1E-5. The search results were parsed by MEGAN4 to assign taxa (with the LCA algorithm) and KEGG/SEED-subsystem functions. Next, the amino acid sequences of the predicted ORFs were screened against the PfamA database version 26.056 by Pfam_scan (E-value cutoff of 1E-4)5 for particular glycoside hydrolase (GH) families and carbohydrate binding modules (CBM) as classified by the Carbohydrate Active enZyme (CAZy) database57.
Quantifying transcriptional activity
For quantification, the coding DNA and mRNA sequences were mapped back to ORFs with Bowtie58 allowing two mismatches over the whole sequence length. Based on the Bowtie alignment, RSEM59 was used to compute the RPKM (Reads Per Kilobase of transcript per Million mapped reads)60 values for DNA and RNA datasets (hereafter, RPKM-DNA and RPKM-RNA as defined in Equation S1 and S2). These two values were used as measurements for the relative abundance of a gene in the metagenome and the overall transcriptional activity of a gene in the metatranscriptome. Given the uneven richness of various genes in the metagenome, the overall transcriptional activity of each gene must be normalized by its relative abundance in the microbial community to reveal its absolute transcriptional activity. As a result, MRPKM (short for Metatranscriptomic RPKM) was proposed in the present study to evaluate the absolute transcriptional activity (which is equivalent to the expression level) of each gene in the metatranscriptome. MRRKM could be calculated as the ratio of RPKM-RNA to RPKM-DNA.
In the present study, the RPKM-DNA value sums for genes assigned to any taxa were used to reveal the prevalence of that population in the community, and the transcriptional activities of different populations were compared on the basis of the average MRPKM values of genes assigned. Additionally, the overall transcriptional activities of metabolic pathways were evaluated based on the sum of RPKM-RNA values for the genes involved.
Author Contributions
Y.X. conducted the experiments, analyzed the data and drafted the manuscript. Y.W., T.J. and H.Z. conducted the experiments. T.Z. and H.F. designed the experiments and contributed to the manuscript editing. All authors reviewed the manuscript.
Supplementary Material
supplementary information
Acknowledgments
The authors wish to thank Hong Kong ITF (GHP/015/11SZ) and Hong Kong GRF (HKU 712509E) for their financial support of this study. Yu Xia and Yubo Wang wish to thank The University of Hong Kong for their postgraduate studentships.
References
- Lynd L. R., Weimer P. J., Van Zyl W. H. & Pretorius I. S. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. et al. Omics-based interpretation of synergism in a soil-derived cellulose-degrading microbial community. Sci. Rep. 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope P. B. et al. Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc. Natl. Acad. Sci. 107, 14793–14798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke F. et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565 (2007). [DOI] [PubMed] [Google Scholar]
- Hess M. et al. Metagenomic Discovery of Biomass-Degrading Genes and Genomes from Cow Rumen. Science 331, 463–467 (2011). [DOI] [PubMed] [Google Scholar]
- Hanreich A. et al. Metagenome and metaproteome analyses of microbial communities in mesophilic biogas-producing anaerobic batch fermentations indicate concerted plant carbohydrate degradation. Syst. Appl. Microbiol. 36, 330–338 (2013). [DOI] [PubMed] [Google Scholar]
- D'haeseleer P. et al. Proteogenomic Analysis of a Thermophilic Bacterial Consortium Adapted to Deconstruct Switchgrass. PLoS ONE 8, e68465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü F. et al. Metaproteomics of cellulose methanisation under thermophilic conditions reveals a surprisingly high proteolytic activity. ISME J. 8, 88–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E. A. et al. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. U. S. A. 111, E2329–2338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth P. et al. Metatranscriptomics of the Human Oral Microbiome during Health and Disease. mBio 5, e01012–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. et al. Comparative Metagenomic and Metatranscriptomic Analysis of Hindgut Paunch Microbiota in Wood- and Dung-Feeding Higher Termites. PLoS ONE 8, e61126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K. & Zhang T. Metagenomic and Metatranscriptomic Analysis of Microbial Community Structure and Gene Expression of Activated Sludge. PLoS ONE 7, e38183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfert T., Csaba G., Zimmer R. & Friedel C. C. Mining RNA–Seq Data for Infections and Contaminations. PLoS ONE 8, e73071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister E. B. et al. Mesophilic and Thermophilic Conditions Select for Unique but Highly Parallel Microbial Communities to Perform Carboxylate Platform Biomass Conversion. Plos One 7, e39689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M. T. et al. Abundance and Diversity of Dockerin-Containing Proteins in the Fiber-Degrading Rumen Bacterium, Ruminococcus flavefaciens FD-1. PLoS ONE 5, e12476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt C. G. et al. Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting a microbial mat in a geothermal spring. ISME J. 7, 1775–1789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewski M. et al. Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. J. Biotechnol. 158, 248–258 (2012). [DOI] [PubMed] [Google Scholar]
- Luo F. et al. Metatranscriptome of an Anaerobic Benzene-Degrading Nitrate-Reducing Enrichment Culture Reveals Involvement of Carboxylation in Benzene Ring Activation. Appl. Environ. Microbiol. AEM.00717–14 (2014). 10.1128/AEM.00717-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsementzi D., Poretsky R., Rodriguez-R L. M., Luo C. & Konstantinidis K. T. Evaluation of metatranscriptomic protocols and application to the study of freshwater microbial communities. Environ. Microbiol. Rep. (2014) 10.1111/1758-2229.12180. [DOI] [PubMed] [Google Scholar]
- Cloonan N. et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods 5, 613–619 (2008). [DOI] [PubMed] [Google Scholar]
- Marioni J. C., Mason C. E., Mane S. M., Stephens M. & Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U. et al. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science 320, 1344–1349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford S. M., Sharma S., Rinta-Kanto J. M. & Moran M. A. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 5, 461–472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley J. C. & Ye Y. Metagenomics: Facts and Artifacts, and Computational Challenges. J. Comput. Sci. Technol. 25, 71–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Ju F., Fang H. H. P. & Zhang T. Mining of Novel Thermo-Stable Cellulolytic Genes from a Thermophilic Cellulose-Degrading Consortium by Metagenomics. PLoS ONE 8, e53779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichorst S. A. et al. Community dynamics of cellulose-adapted thermophilic bacterial consortia. Environ. Microbiol. 15, 2573–2587 (2013). [DOI] [PubMed] [Google Scholar]
- Rosewarne C. P., Pope P. B., Cheung J. L. & Morrison M. Analysis of the bovine rumen microbiome reveals a diversity of Sus-like polysaccharide utilization loci from the bacterial phylum Bacteroidetes. J. Ind. Microbiol. Biotechnol. 41, 601–606 (2014). [DOI] [PubMed] [Google Scholar]
- Mackenzie A. K. et al. Two SusD-Like Proteins Encoded within a Polysaccharide Utilization Locus of an Uncultured Ruminant Bacteroidetes Phylotype Bind Strongly to Cellulose. Appl. Environ. Microbiol. 78, 5935–5937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroeder M., Leschine S. B. & Canale-Parola E. Spirochaeta caldaria sp. nov., a thermophilic bacterium that enhances cellulose degradation by Clostridium thermocellum. Arch. Microbiol. 161, 17–24 (1994). [Google Scholar]
- Ohkuma M., Iida T. & Kudo T. Phylogenetic relationships of symbiotic spirochetes in the gut of diverse termites. FEMS Microbiol. Lett. 181, 123–129 (1999). [DOI] [PubMed] [Google Scholar]
- Wery N. et al. Marinitoga camini gen. nov., sp. nov., a rod-shaped bacterium belonging to the order Thermotogales, isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 51, 495–504 (2001). [DOI] [PubMed] [Google Scholar]
- Huber R., Woese C. R., Langworthy T. A., Kristjansson J. K. & Stetter K. O. Fervidobacterium islandicum sp. nov., a new extremely thermophilic eubacterium belonging to the ‘Thermotogales'. Arch. Microbiol. 154, 105–111 (1990). [Google Scholar]
- Podosokorskaya O. a. et al. Fervidobacterium riparium sp. nov., a thermophilic anaerobic cellulolytic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 61, 2697–701 (2011). [DOI] [PubMed] [Google Scholar]
- Guglielmi G. & Béguin P. Cellulase and hemicellulase genes of Clostridium thermocellum from five independent collections contain few overlaps and are widely scattered across the chromosome. FEMS Microbiol. Lett. 161, 209–215 (1998). [DOI] [PubMed] [Google Scholar]
- Newcomb M., Millen J., Chen C.-Y. & Wu J. H. D. Co-transcription of the celC gene cluster in Clostridium thermocellum. Appl. Microbiol. Biotechnol. 90, 625–634 (2011). [DOI] [PubMed] [Google Scholar]
- Munk B., Bauer C., Gronauer A. & Lebuhn M. Population dynamics of methanogens during acidification of biogas fermenters fed with maize silage. Eng. Life Sci. 10, 496–508 (2010). [Google Scholar]
- Bauer C., Korthals M., Gronauer A. & Lebuhn M. Methanogens in biogas production from renewable resources – a novel molecular population analysis approach. Water Sci. Technol. 58, 1433 (2008). [DOI] [PubMed] [Google Scholar]
- Schnurer A. & Nordberg A. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci. Technol. 57, 735–740 (2008). [DOI] [PubMed] [Google Scholar]
- Koster I. W. & Lettinga G. The influence of ammonium-nitrogen on the specific activity of pelletized methanogenic sludge. Agric. Wastes 9, 205–216 (1984). [Google Scholar]
- Karakashev D., Batstone D. J., Trably E. & Angelidaki I. Acetate Oxidation Is the Dominant Methanogenic Pathway from Acetate in the Absence of Methanosaetaceae. Appl. Environ. Microbiol. 72, 5138–5141 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakashev D., Batstone D. J. & Angelidaki I. Influence of Environmental Conditions on Methanogenic Compositions in Anaerobic Biogas Reactors. Appl. Environ. Microbiol. 71, 331–338 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerholm M., Levén L. & Schnürer A. Bioaugmentation of syntrophic acetate-oxidizing culture in biogas reactors exposed to increasing levels of ammonia. Appl. Environ. Microbiol. 78, 7619–7625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori S. Syntrophic Acetate-Oxidizing Microbes in Methanogenic Environments. Microbes Environ. 23, 118–127 (2008). [DOI] [PubMed] [Google Scholar]
- Montoya L., Celis L. B., Gallegos-García M., Razo-Flores E. & Alpuche-Solís Á. G. Consortium diversity of a sulfate-reducing biofilm developed at acidic pH influent conditions in a down-flow fluidized bed reactor: Consortium diversity of a sulfate-reducing biofilm. Eng. Life Sci. 13, 302–311 (2013). [Google Scholar]
- Xia Y., Cai L., Zhang T. & Fang H. H. P. Effects of substrate loading and co-substrates on thermophilic anaerobic conversion of microcrystalline cellulose and microbial communities revealed using high-throughput sequencing. Int. J. Hydrog. Energy 37, 13652–13659 (2012). [Google Scholar]
- Meyer F. et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Lim Y. W. & Edwards R. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinforma. Oxf. Engl. 28, 433–435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Leung H. C. M., Yiu S. M. & Chin F. Y. L. Meta-IDBA: a de Novo assembler for metagenomic data. Bioinformatics 27, i94–i101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Lomsadze A. & Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38, e132–e132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Choi J.-H. & Tang H. RAPSearch: a fast protein similarity search tool for short reads. BMC Bioinformatics 12, 159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information