Abstract
Sandhoff disease is a prototypical lysosomal storage disorder in which a heritable deficiency of a lysosomal enzyme, β-hexosaminidase, results in the storage of the enzyme's substrates in lysosomes. As with many of the other lysosomal storage diseases, neurodegeneration is a prominent feature. Although the cellular and molecular pathways that underlie the neurodegenerative process are not yet fully understood, macrophage/microglial-mediated inflammation has been suggested as one possible mechanism. We now show that the expanded macrophage/microglial population in the CNS of Sandhoff disease mice is compounded by the infiltration of cells from the periphery. Coincident with the cellular infiltration was an increased expression of macrophage-inflammatory protein 1α (MIP-1α), a leukocyte chemokine, in astrocytes. Deletion of MIP-1α expression resulted in a substantial decrease in infiltration and macrophage/microglial-associated pathology together with neuronal apoptosis in Sandhoff disease mice. These mice without MIP-1α showed improved neurologic status and a longer lifespan. The results indicate that the pathogenesis of Sandhoff disease involves an increase in MIP-1α that induces monocytes to infiltrate the CNS, expand the activated macrophage/microglial population, and trigger apoptosis of neurons, resulting in a rapid neurodegenerative course.
Sandhoff disease is lysosomal storage disorder caused by a deficiency of β-hexosaminidases A and B (1). In patient's cells, glycosphingolipids, glycosoaminoglycans, and oligosaccharides containing terminal β-linked N-acetylglucosamine or N-acetylgalatosamine residues (the substrates of the β-hexosaminidases) accumulate in lysosomes. Massive substrate storage is particularly evident in neurons, where gangliosides, a series of the glycosphingolipids, are synthesized in relatively high amounts (2). As a consequence, the CNS is primarily affected. In the most severely affected infantile patients, the onset of symptoms begins in early infancy and includes motor function disturbances, seizure, visual loss, and deafness. Death usually occurs by 2–5 years of age. Tay–Sachs disease, caused by a deficiency of only β-hexosaminidase A, is biochemically and clinically similar to Sandhoff disease. Collectively, Tay–Sachs and Sandhoff diseases are known as GM2 gangliosidoses. Less severe, later-onset forms of these diseases due to partial enzyme deficiencies are also known. In these late-onset conditions manifestations related to neurodegeneration are prominent as well.
Many of the >40 lysosomal storage disorders involve neurodegeneration as a prominent feature (3, 4). Therapies for lysosomal storage diseases that affect the CNS have been generally unsuccessful because of the difficulty of delivering therapeutics across the blood–brain barrier (5). In addition, a basic lack of knowledge of the underlying neurodegenerative mechanisms in lysosomal storage diseases has hampered the development of therapies.
Macrophage/microglia-mediated inflammatory responses have been invoked as neurodegenerative mechanisms in storage diseases (6–9). In Sandhoff disease model mice, expansion and activation of the macrophage/microglia population were found to occur before detectable neuronal apoptosis and the onset of a rapid decline in neurologic condition (6, 7). It was hypothesized that neuronal damage produces macrophage/microglia activation and expansion, possibly through monocyte infiltration, and, thereby, initiates a cascade of neuronal apoptosis resulting in rapid neurodegeneration (6, 10).
We have investigated whether cells from the periphery are recruited to enter the CNS during Sandhoff disease to supplement the macrophage/microglial population and whether such an infiltration exacerbates the neurodegenerative disease course. We show that macrophages, indeed, are recruited into CNS during the disease and, more importantly, represent a significant mechanism that underlies neurodegeneration in this lysosomal storage disease.
Methods
Generation of Hexb-/-MIP-1α-/- Double Knockout Mice. Sandhoff disease model mice (Hexb-/-) on a 129/Sv background were generated in our laboratory as described (11). Hexb genotypes were identified by PCR using genomic DNA extracted from mouse tails. Macrophage-inflammatory protein (MIP)-1α-deficient (MIP-1α-/-) and GFP transgenic mice on the C57BL/6 background were purchased from The Jackson Laboratory and were characterized in refs. 12–14.
A male MIP-1α-/- mouse was crossed to female Hexb+/- mice to generate doubly heterozygous (MIP-1α+/-Hexb+/-) mice. These doubly heterozygous mice were mated with each other to generate doubly null (Hexb-/-MIP-1α-/-) mice. To more efficiently generate Hexb-/- mice with two normal MIP-1α alleles (Hexb-/-MIP-1α+/+), with a single active MIP-1α allele (Hexb-/-MIP-1α+/-), and with two null MIP-1α alleles (Hexb-/-MIP-1α-/-), we also bred the doubly heterozygous mice with Hexb-/-MIP-1α+/- mice and Hexb-/-MIP-1α+/+ mice. Comparisons were made among the offspring of the same mating pairs to minimize the influence of genetic background. The genotype of the MIP-1α locus was determined by PCR from tail DNA using two pairs of primers. The pair of primers used for detecting the wild-type allele are located within exon 1 (sense, 5′-ATGAAGGTCTCCACCACTGC-3′) and exon 2 (antisense, 5′-AGTCAACGATGAATTGGCG-3′). The pair of primers for detecting the mutant allele are located within the neo gene (sense, 5′-TAAAGCATGCTCCAGACT-3′) and exon 2 (antisense, 5′-CAAAGGCTGCTGGTTTCAAA-3′). The wild-type primers yielded an ≈700-bp fragment, and the mutant primers yielded an ≈100-bp fragment. Combination of the PCR-based procedures for both Hexb and MIP-α alleles allowed unequivocal diagnosis of all of the nine possible genotypes.
In Vivo Tracking of Cells Entering the CNS. Three Hexb+/+ mice; three each of Hexb-/- mice at 2, 3, and 4 months of age; and three Hexb-/- mice null for the MIP-1α gene at 4 months of age received an i.v. or i.p. injection (0.5 mg/g of body weight) of 20% horseradish peroxidase (HRP IV, Sigma-Aldrich). Alternatively, the mutant mice received an injection of 5 × 106 cells isolated from the spleens of GFP transgenic mice. Two days later, the tissues were fixed and sectioned. The HRP-labeled cells were detected by diaminobenzidine and hydrogen peroxide with or without nickel enhancement. GFP-positive cells were detected by immunostaining with an antibody directed to GFP.
Quantitative Gene Expression Assay by Real-Time PCR. The TaqMan PreDeveloped and Assays-on-Demand Gene Expression Assays for murine MIP-1α, tumor necrosis factor α (TNF-α), Mac-1 α-subunit, and GAPDH (PE Applied Biosystems) were used. Total RNA from brain and spinal cord was isolated by using TRI reagent according to the manufacturer's protocol (Invitrogen). Total RNA (1–2 μg) was reverse-transcribed by using a Super-Script II RT Kit (Invitrogen). Real-time PCR was performed on an Prism 7700 Sequence Detection System (PE Applied Biosystems) as described (6). Each PCR amplification was performed in duplicate. Each genotype and time point represents the determination from three to five mice.
Histopathology and Immunohistochemistry. Three to five Hexb-/- MIP-1α+/+, Hexb-/-MIP-1α-/-, and Hexb+/+MIP-1α+/+ mice at 4 months of age were used for this study. The mice were anesthetized and perfused transcardially with 0.9% NaCl followed by cold buffered 4% paraformaldehyde. The brain and spinal cord were removed and immersed in the same fixative at 4°C for 4 h. Then they were transferred to buffered 20% sucrose overnight at 4°C. Each half of the brain and a segment of the spinal cord were processed for paraffin sections. The other half of the brain and spinal cord were cut to 40-μm-thick sections with a vibratome or cryostat.
For immunohistochemistry, the sections were pretreated with 10% normal rabbit or goat serum with 0.2% Triton X-100 and then incubated overnight with goat anti-MIP-1α (1:200; Santa Cruz Biotechnology), mouse anti-Mac-1 (1:500; Serotec), rabbit anti-GFP (1:200; Pharmingen), or rabbit anti-glial fibrillary acidic protein (GFAP) (1:200; DAKO). After extensive washing in PBS, sections were incubated for 1 h in biotinylated secondary antibodies (Vector Laboratories). The immunoreactions were then visualized by using Vectastain ABC reagent and diaminobenzidine substrate (Vector Laboratories). For double labeling of HRP-positive cells with Mac-1 antibody, the sections were incubated with a diaminobenzidine and nickel mix solution (Vector Laboratories) to develop a gray-black stain. After extensive washing, the sections were treated with H2O2 and then processed for Mac-1 immunostaining. For immunofluorescent staining, after incubation with primary antibodies, sections were incubated for 1 h with species-specific affinity FITC- and tetramethylrhodamine B isothiocyanate (TRITC)-conjugated rabbit and goat IgG (Sigma-Aldrich). The sections were mounted and analyzed by using Leica DMR microscope equipped with FITC and TRITC filters.
In Situ Apoptosis Detection. The terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) method was used for detection of apoptotic cells using the TdT-FragEL kit (Oncogene Science) following the manufacturer's instructions. Brief ly, sections from three each of Hexb-/-MIP-1α+/+, Hexb-/-MIP-1α-/-, and Hexb+/+MIP-1α+/+ mice at 4 months of age were deparaffinized, rehydrated, and pretreated with proteinase K for 20 min. After blocking for endogenous peroxidase activity with 2% H2O2 in methanol, the sections were treated with equilibration buffer and incubated with terminal deoxynucleotidyl transferase enzyme at 37°C for 1 h. Then the sections were incubated with a peroxidase–streptavidin conjugate and reacted with diaminobenzidine containing 0.01% H2O2. The sections were counterstained with methyl green. All washes were performed with Tris-buffered saline.
Behavioral Testing. Two tests were used to ascertain neurological function, rotorod, and righting reflex (15). For the rotorod test, motor performance was evaluated weekly from 10 weeks of age on a rotorod (model 7650, Ugo Basile, Varese, Italy) at an initial speed of 4 rpm; the acceleration was increased 2 rpm in every 1 min. If the mice remained on the rod for 350 sec, then the test was completed and scored as 350 sec. If mice fell in <10 sec, then they were given a second trial. The righting reflex was tested at 17 and 19 weeks old by turning the mouse on its back on a flat stainless steel surface. Latency for the mouse to turn itself upright was recorded. Mice not righting themselves within 10 sec were turned right-side up and assigned a score of 10.
Data Analysis. Statistical comparisons were made by Student's t test. P < 0.05 was considered significant.
Results
Infiltration of Peripheral Monocytes/Macrophages into the CNS of Hexb-/- Mice. We had previously shown that the activated macrophage/microglial population expands in the CNS of Sandhoff disease mice over the course of the disease during their ≈4.5-month lifespan (6). To determine whether cells entering from the periphery were contributing to the increase in CNS macrophage/microglia, wild-type (Hexb+/+) and Sandhoff disease mice (Hexb-/-) were injected with a solution containing HRP to label peripheral phagocytic cells. In the Hexb+/+ mice, HRP-labeled cells could be detected only at the leptomeninges and choroid plexus and not in the parenchyma of the CNS (data not shown). In Hexb-/- mice at 2 months of age, the result was similar to Hexb+/+ mice (data not shown). At 3 months of age, HRP-labeled cells could be detected closely associated with vessels in the thalamus, brainstem, and spinal cord (Fig. 1 a and b). At both 3 and 4 months of age, during their acute decline in neurologic function, HRP-labeled cells could be found in the parenchyma of the brain of the Hexb-/- mice (Fig. 1c). These HRP-labeled cells were positive for Mac-1, a marker for monocytes/macrophages (Fig. 1d). Similar results were observed after adoptive transfer of splenocytes expressing GFP into Hexb-/-, mice confirming that cells were migrating into the CNS from the periphery.
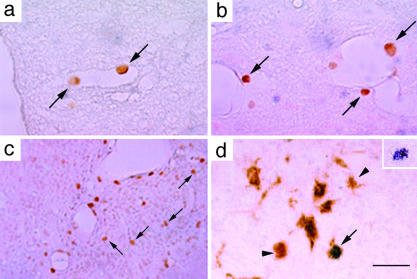
Fig. 1.
Localization and identification of HRP-labeled cells in Hexb-/- mice. In 3-month-old Hexb-/- mice, HRP-labeled cells were detected attached to the vascular wall in vessels in the spinal cord (a) and perivascular regions in the spinal cord (b). (c) In a 4-month-old Hexb-/- mouse, HRP-labeled cells were localized in the parenchyma of the thalamic nucleus. (d) An HRP-labeled cell in the thalamic nucleus of a 4-month-old Hexb-/- mouse was identified as Mac-1-positive (arrow). Arrowheads show Mac-1-positive cells. (Inset) A nickel-enhanced HRP-labeled cell. (Bars: 100 μmin a and b, 220 μmin c, and 60 μm in d.)
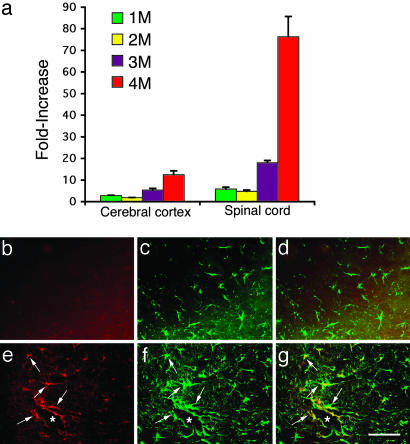
Up-Regulation of MIP-1α Expression. We next examined whether the expression of MIP-1α (16), a CC chemokine that directs leukocyte extravasation, was up-regulated in the CNS of Hexb-/- mice. The relative expression of the MIP-1α gene was determined by real-time PCR in the cerebral cortex and spinal cord of Hexb+/+ and Hexb-/- mice at 1, 2, 3, and 4 months of age (Fig. 2a). The results revealed a gradual increase in MIP-1α expression in both the cerebral cortex and the spinal cord. At 1 and 2 months of age, a slight increase of MIP-1α was detected in both cerebral cortex and spinal cord; however, by 3 and 4 months of age, MIP-1α mRNA levels were elevated by 5- and 12-fold, respectively, in the cerebral cortex relative to Hexb+/+ mice. In the spinal cord, MIP-1α mRNA expression was elevated to 18-fold at 3 months and to 75-fold at 4 months of age when compared with controls.
Fig. 2.
Expression of MIP-1α in Hexb+/+ and Hexb-/- mice. (a) The time course of relative MIP-1α mRNA expression levels in the cerebral cortex and spinal cord of Hexb-/- mice compared with age-matched Hexb+/+ mice. (b–g) Immunofluorescent staining of the spinal cord with MIP-1α and GFAP antibodies. (b–d) Control (Hexb+/+) sections. (e–g) Hexb-/- sections. b and e show MIP-1α immunostaining. c and f show GFAP immunostaining. d and g show double immunostaining for MIP-1α and GFAP. Note that the MIP-1α and GFAP double-positive astrocytes (arrows in e–g) are closely associated with vessels (*). (Bar: 60 μm.)
To determine which cells produce MIP-1α, double immuno-fluorescent staining was performed (Fig. 2). The MIP-1α-positive cells were found closely associated with vessels and with a morphology similar to astrocytes (Fig. 2e). The sections were also stained with GFAP, a specific marker for astrocytes, which demonstrated that these MIP-1α-positive cells were indeed astrocytes with hypertrophied cell soma and abundant processes (Fig. 2 f and g). MIP-1α-positive cells were not detected in Hexb+/+ mice (Fig. 2 b and d). In contrast to the activated morphology of astrocytes in the Hexb-/- mice, the GFAP-positive astrocytes in the control mice had small soma and fine processes (Fig. 2c).
Deletion of MIP-1α Improves the Clinical Course of Sandhoff Disease Mice. To determine whether MIP-1α expression is an important factor in the neuropathogenesis of Sandhoff disease, we generated Hexb-/- mice carrying either one or two mutant MIP-1α alleles together with control mice of the genotype Hexb-/-MIP-1α+/+. The lifespan, body weight, and neurological function of each group of mice was monitored (Figs. 3 and 4). Hexb-/-MIP-1α+/+ mice displayed the prototypic disease course described for the Sandhoff disease model on the mixed 129/Sv × C57BL/6 background (17, 18) (Fig. 3). The mice showed neurological disturbances beginning at ≈13 weeks and weight loss beginning at 15 weeks of age. All mice died or were incapacitated and killed by 19 weeks of age. Their average lifespan was 121.1 ± 2.5 days (n = 10). The group of Hexb-/- mice with a single active MIP-1α allele showed a similar but slightly delayed disease course; their average of lifespan was 138 ± 3.1 days (n = 10), which was significantly longer than Hexb-/-MIP-1α+/+ mice (P = 0.00016). The double knockout mice (Hexb-/-MIP-1α-/-) showed by far the mildest neurological symptoms and longest lifespans. They continued to maintain their body weight until ≈19 weeks and survived up to 175 days of age. Their average lifespan was 156.1 ± 2.7 days (n = 15) (Fig. 4 a and b).
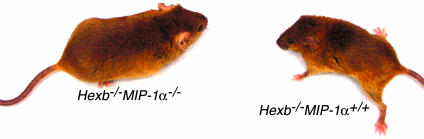
Fig. 3.
Preserved clinical condition of Sandhoff disease mice deficient for MIP-1α was clearly noted at 4 months of age compared with littermates. Hexb-/- mice with different MIP-1α genotypes are indicated.
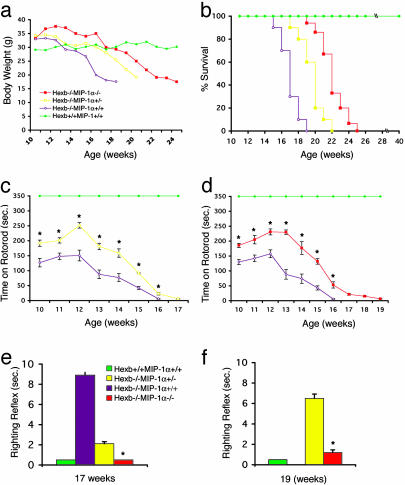
Fig. 4.
Body weight, lifespan, and behavioral testing. (a) The body weight in Hexb-/-MIP-1α-/- and Hexb-/-MIP-1α+/- mice was higher at and after 15 weeks of age compared with Hexb-/-MIP-1α+/+ mice. The data are means from three to five mice for each genotype and data point. (b) Survival of the Hexb-/- mice with different MIP-1α backgrounds. Each group contained 10–15 mice. (c and d) Mean (±SEM) time to fall off the rotorod. Both Hexb-/-MIP-1α+/- and Hexb-/-MIP-1α-/- mice had significantly better (*) performances at all time points tested. The comparison was made only among the littermates. At all time points in c, n = 4 Hexb-/-MIP-1α+/+ mice and 4 Hexb-/-MIP-1α+/- mice. At all time points in d, n = 3 Hexb-/-MIP-1α+/+ mice and 3 Hexb-/-MIP-1α-/- mice. The righting reflex was tested at 17 (e) and 19 (f) weeks of age; n = 4 in each group and time point. Hexb-/-MIP-1α-/- mice had a significantly improved (*) righting reflex compared with other groups. All Hexb-/-MIP-1α+/+ mice were dead by 19 weeks old.
To monitor motor function, two behavioral tasks, rotorod and righting reflex, were performed among the different genotypes (17, 19). The rotorod test to measure motor coordination and balance showed that the performance of Hexb-/-MIP-1α+/- and Hexb-/-MIP-1α-/- mice each were significantly better than that of Hexb-/-MIP-1α+/+ mice over the course of the testing period (P < 0.0001) (Fig. 4 c and d). All Hexb+/+MIP-1α+/+ mice tested remained on the rotorod >350 sec. There was a significantly improved righting ref lex in Hexb-/-MIP-1α+/- and Hexb-/-MIP-1α-/- mice compared with Hexb-/-MIP-1α+/+ mice at 17 weeks of age (both P < 0.01) (Fig. 4e). The results were similar at 19 weeks of age although the Hexb-/-MIP-1α+/+ mice had succumbed to their disease.
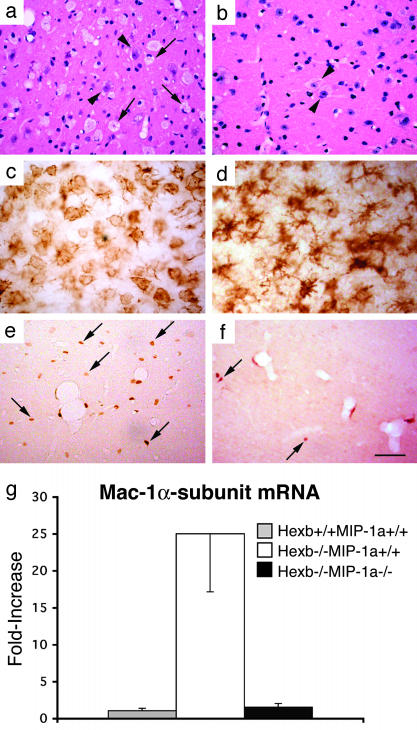
Improved Pathology and Decreased Apoptotic Neuronal Death in Sandhoff Disease Mice Without MIP-1α. We examined double mutant mice to determine whether the deletion of MIP-1α altered the CNS pathology of the Hexb-/- mice. At 4 months of age, many round, foamy cells (Fig. 5a), characteristic of macrophages, were found in the thalamus, brainstem, and spinal cord of Hexb-/-MIP-1α+/+ mice. In these same CNS regions, there were abundant amoeboid, Mac-1-positive brain macrophages (Fig. 5c). These features were not seen in Hexb+/+MIP-1α+/+ mice (data not shown). In Hexb-/-MIP-1α-/- mice, at 4 months of age, the appearance of both the foamy cells and the amoeboid, Mac-1-positive brain macrophages were substantially reduced compared with the Hexb-/-MIP-1α+/+ mice (Fig. 5 c and d) even though the neuronal storage pathology was similar (Fig. 5 a and b).
Fig. 5.
Lessened macrophage/microglia-associated pathology in Hexb-/-MIP-1α-/- mice compared with Hexb-/-MIP-1α+/+ mice. Hematoxylin/eosin and Mac-1 immunostaining show numerous foamy cells (a) and ameboid brain macrophages (c) in the thalamic nuclei of 4-month-old Hexb-/-MIP-1α+/+ mice. In the thalamic nuclei of 4-month-old Hexb-/-MIP-1α-/- mice foamy cells were rare (b) and ramified microglia were detected (d) instead of amoeboid brain macrophages. The arrows indicate foamy cells in the Hexb-/-MIP-1α+/+ mice, and the arrowheads indicate neurons with storage in both genotypes. (e and f) Reduction of HRP-labeled cells (arrow) in 4-month-old Hexb-/-MIP-1α-/- mice (f) compared with age-matched Hexb-/-MIP-1α+/+ mice (e). (g) Down-regulation of Mac-1α-subunit mRNA expression in the spinal cord of Hexb-/-MIP-1α-/- mice at 4 months old compared with age-matched Hexb-/-MIP-1α+/+ mice. (Bars: 20 μmin a–d and 120 μmin e and f.)
The HRP cell tracking assay revealed fewer cells infiltrating the CNS of the Hexb-/-MIP-1α-/- mice at 4 months of age (Fig. 5f) compared with Hexb-/-MIP-1α+/+ mice (Fig. 5e). We quantitatively measured the extent of macrophage/microglial expansion and activation by determining the mRNA expression level of Mac-1α-subunit, a macrophage-specific gene, in the CNS. In the spinal cord of 4-month-old Hexb-/-MIP-1α+/+ mice, the Mac-1α-subunit mRNA was elevated 25-fold compared with Hexb+/+MIP-1α+/+ mice. In contrast, it was elevated just 2-fold in Hexb-/-MIP-1α-/- mice (Fig. 5g). Thus, the deletion of MIP-1α reduced the expression of a macrophage-specific gene by >90% in the CNS of Hexb-/- mice.
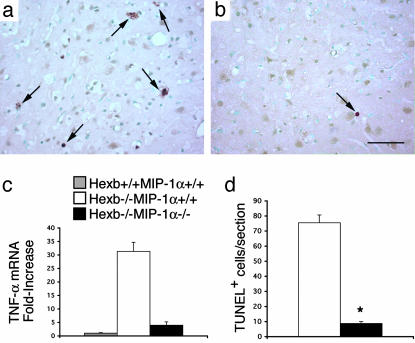
Finally, we determined whether the deletion of MIP-1α could alter neuronal apoptosis in Hexb-/- mice. In agreement with previous results (6, 20), abundant TUNEL-positive cells were detected in the thalamus, brainstem, and spinal cord (Fig. 6 a and d) in 4-month-old Hexb-/-MIP-1α+/+ mice. However, in Hexb-/-MIP-1α-/- mice at 4 months of age, TUNEL-positive cells were nearly absent (Fig. 6 b and d). Expression of the mRNA for the proinflammatory cytokine TNF-α was highly elevated in the spinal cord of Hexb-/-MIP-1α+/+ mice as had been shown in ref. 6 (see Fig. 6c). However, when MIP-1α was deleted, spinal cord TNF-α mRNA was reduced by 87%.
Fig. 6.
Reduction of TUNEL-positive cells (arrows) in Hexb-/-MIP-1α-/- mice at 4 months old (b) compared with age-matched Hexb-/-MIP-1α+/+ mice (a). (d) TUNEL-positive cells were counted (*, P < 0.05). Data are means ± SEM (n = 3–5). (c) The TNF-α mRNA expression level was reduced in the spinal cord of Hexb-/-MIP-1α-/- mice compared with Hexb-/-MIP-1α+/+ mice at 4 months old. (Bar: 40 μm.)
Discussion
CNS inflammation has been detected in several lysosomal storage diseases with neurologic impairment. In addition to Sandhoff and Tay–Sachs diseases (6), activated macrophage/microglia have been identified in the CNS of mice with GM1 gangliosidosis (7), mucopolysaccharidoses I and IIIB (8), metachromatic leukodystrophy (21), and Krabbe disease (9, 22). These findings raised the possibility that inflammation plays a role in neurodegeneration, as has been suggested for diseases such as HIV-associated dementia, Alzheimer's disease, and prion diseases (23–25). A critical question has been the extent to which macrophage/microglial-mediated inflammation contributes to the neurodegeneration in these disorders. In this study we have provided direct evidence that peripheral cells cross the blood–brain barrier and expand the macrophage/microglial population, and that this process accelerates the neurodegeneration in Sandhoff disease.
Our earlier study documented a progressive increase in activated macrophage/microglia in the CNS in the Sandhoff disease mouse (6). In 2-month-old mice activated macrophages/microglia could be detected around blood vessels, and by 3 and 4 months substantial numbers of activated cells were detected in the parenchyma of the spinal cord and brain. However, we were uncertain whether blood monocytes contribute to the expansion of the activated macrophage/microglia population by entering the CNS from the periphery. To answer this question we labeled peripheral cells with HRP. The distribution of HRP+ cells mirrored the distribution of activated macrophages/microglia seen in ref. 6. Early in the disease labeled cells were detected adhering to the endothelium in the CNS, and at latter times HRP-labeled macrophages were found within the parenchyma of the brain and spinal cord. These results suggested that, during the disease process, a signal is generated that recruits blood monocytes to traverse the endothelium and migrate into the parenchyma of the brain.
We hypothesized that a macrophage chemotactic factor may be responsible for recruitment of blood monocytes into the Sandhoff disease brain. MIP-1α is a CC-type chemokine that has been shown to direct leukocyte extravasation in multiple sclerosis and other neuroinflammatory conditions (16, 26, 27). We found that the level of MIP-1α mRNA increased dramatically in the CNS during the progression of Sandhoff disease. Substantial elevations in the MIP-1α mRNA levels were detected at 3 and 4 months of age, the time when cells were found infiltrating the CNS.
We further investigated the role of MIP-1α in the pathogenesis of Sandhoff disease by cross-breeding the mice carrying mutant MIP-1α and Hexb alleles to produce doubly null mice. These double-mutant mice showed a significantly delayed disease course as evidenced by an ≈30% longer lifespan and a significant preservation of neurologic function compared with Sandhoff mice carrying one or two wild-type MIP-1α alleles. Histologic evaluation of the doubly mutant mice revealed markedly improved pathology. In particular, there was a reduction in the numbers of abnormal macrophages/microglia in the CNS, which is likely be a consequence of lowered recruitment of monocytes from the periphery. The Sandhoff mice lacking MIP-1α also demonstrated substantially reduced neuronal apoptosis at 4 months of age, providing an explanation for their preserved neurological condition and longer lifespan. These results demonstrate that MIP-1α is a critical factor in the neuropathogenesis of Sandhoff disease mice. They also raise the possibility that MIP-1α has a key role in the pathogenesis of other lysosomal storage diseases with macrophage/microglial-mediated inflammation.
The mechanism by which the MIP-1α signal is conveyed across the blood–brain barrier remains to be determined. An intriguing finding was that MIP-1α production was most prominent in astrocytes located around blood vessels. Astrocytes sense and respond to neuronal damage and can be in direct cell-to-cell contact with both vascular endothelial cells and neurons (28, 29). One possibility is that astrocytes act as a conduit by directly sensing damaged neurons in Sandhoff disease. In response, the astrocytes produce MIP-1α, the chemokine to the abluminal surface of endothelial cells, which contain receptors for MIP-1α (30). MIP-1α receptor stimulation may induce the activation of the endothelium with the up-regulation of adhesion molecules, allowing monocytes to adhere and extravasate across the blood–brain barrier. The macrophages, having entered the CNS, may then trigger neuronal apoptosis. Our results point to the chemokine, MIP-1α, and its receptor, CCR5, as possible targets for therapies to slow the rapid neurodegenerative course in the disorder.
Acknowledgments
We thank Katie Prothero and Jessica Ellis for assistance with genotyping and behavioral tests and Cynthia Tifft and Kiyomi Mizugishi for helpful discussions.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MIP, macrophage-inflammatory protein; TNF-α, tumor necrosis factor α; HRP, horseradish peroxidase; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling; GFAP, glial fibrillary acidic protein.
References
- 1.Gravel, R. A., Kabak, M. M., Proia, R. L., Sandhoff, K., Suzuki, K. & Suzuki, K. (2001) in The Metabolic and Molecular Basis of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Valle, D., Sly, W. S., Childs, B., Kinzler, K. W. & Vogelstein, B. (McGraw–Hill, New York), Vol. 3, pp. 3827-3876. [Google Scholar]
- 2.Kolter, T., Proia, R. L. & Sandhoff, K. (2002) J. Biol. Chem. 277, 25859-25862. [DOI] [PubMed] [Google Scholar]
- 3.Neufeld, E. F. (1991) Annu. Rev. Biochem. 60, 257-280. [DOI] [PubMed] [Google Scholar]
- 4.Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (2001) in The Metabolic and Molecular Basis of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Valle, D., Sly, W. S., Childs, B., Kinzler, K. W. & Vogelstein, B. (McGraw-Hill, New York), pp. 3371-3896.
- 5.Sly, W. S. & Vogler, C. (2002) Proc. Natl. Acad. Sci. USA 99, 5760-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wada, R., Tifft, C. J. & Proia, R. L. (2000) Proc. Natl. Acad. Sci. USA 97, 10954-10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeyakumar, M., Thomas, R., Elliot-Smith, E., Smith, D. A., van der Spoel, A. C., d'Azzo, A., Perry, V. H., Butters, T. D., Dwek, R. A. & Platt, F. M. (2003) Brain 126, 974-987. [DOI] [PubMed] [Google Scholar]
- 8.Ohmi, K., Greenberg, D. S., Rajavel, K. S., Ryazantsev, S., Li, H. H. & Neufeld, E. F. (2003) Proc. Natl. Acad. Sci. USA 100, 1902-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu, Y. P., Matsuda, J., Kubota, A. & Suzuki, K. (2000) J. Neuropathol. Exp. Neurol. 59, 628-639. [DOI] [PubMed] [Google Scholar]
- 10.Myerowitz, R., Lawson, D., Mizukami, H., Mi, Y., Tifft, C. J. & Proia, R. L. (2002) Hum. Mol. Genet. 11, 1343-1350. [DOI] [PubMed] [Google Scholar]
- 11.Sango, K., McDonald, M. P., Crawley, J. N., Mack, M. L., Tifft, C. J., Skop, E., Starr, C. M., Hoffmann, A., Sandhoff, K., Suzuki, K. & Proia, R. L. (1996) Nat. Genet. 14, 348-352. [DOI] [PubMed] [Google Scholar]
- 12.Cook, D. N., Beck, M. A., Coffman, T. M., Kirby, S. L., Sheridan, J. F., Pragnell, I. B. & Smithies, O. (1995) Science 269, 1583-1585. [DOI] [PubMed] [Google Scholar]
- 13.Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. (1997) FEBS Lett. 407, 313-319. [DOI] [PubMed] [Google Scholar]
- 14.McMahon, E. J., Cook, D. N., Suzuki, K. & Matsushima, G. K. (2001) J. Immunol. 167, 2964-2971. [DOI] [PubMed] [Google Scholar]
- 15.Norflus, F., Tifft, C. J., McDonald, M. P., Goldstein, G., Crawley, J. N., Hoffmann, A., Sandhoff, K., Suzuki, K. & Proia, R. L. (1998) J. Clin. Invest. 101, 1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menten, P., Wuyts, A. & Van Damme, J. (2002) Cytokine Growth Factor Rev. 13, 455-481. [DOI] [PubMed] [Google Scholar]
- 17.Liu, Y., Wada, R., Kawai, H., Sango, K., Deng, C., Tai, T., McDonald, M. P., Araujo, K., Crawley, J. N., Bierfreund, U., et al. (1999) J. Clin. Invest. 103, 497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeyakumar, M., Butters, T. D., Cortina-Borja, M., Hunnam, V., Proia, R. L., Perry, V. H., Dwek, R. A. & Platt, F. M. (1999) Proc. Natl. Acad. Sci. USA 96, 6388-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sango, K., Yamanaka, S., Hoffmann, A., Okuda, Y., Grinberg, A., Westphal, H., McDonald, M. P., Crawley, J. N., Sandhoff, K., Suzuki, K., et al. (1995) Nat. Genet. 11, 170-176. [DOI] [PubMed] [Google Scholar]
- 20.Huang, J. Q., Trasler, J. M., Igdoura, S., Michaud, J., Hanal, N. & Gravel, R. A. (1997) Hum. Mol. Genet. 6, 1879-1885. [DOI] [PubMed] [Google Scholar]
- 21.Hess, B., Saftig, P., Hartmann, D., Coenen, R., Lullmann-Rauch, R., Goebel, H. H., Evers, M., von Figura, K., D'Hooge, R., Nagels, G., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 14821-14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno, M., Komiyama, A., Martin, P. M. & Suzuki, K. (1993) Brain Res. 602, 268-274. [DOI] [PubMed] [Google Scholar]
- 23.Baker, C. A. & Manuelidis, L. (2003) Proc. Natl. Acad. Sci. USA 100, 675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persidsky, Y. & Gendelman, H. E. (2003) J. Leukocyte Biol. 74, 691-701. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Scarano, F. & Baltuch, G. (1999) Annu. Rev. Neurosci. 22, 219-240. [DOI] [PubMed] [Google Scholar]
- 26.Balashov, K. E., Rottman, J. B., Weiner, H. L. & Hancock, W. W. (1999) Proc. Natl. Acad. Sci. USA 96, 6873-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baggiolini, M. (1998) Nature 392, 565-568. [DOI] [PubMed] [Google Scholar]
- 28.Nedergaard, M., Ransom, B. & Goldman, S. A. (2003) Trends Neurosci. 26, 523-530. [DOI] [PubMed] [Google Scholar]
- 29.Ransom, B., Behar, T. & Nedergaard, M. (2003) Trends Neurosci. 26, 520-522. [DOI] [PubMed] [Google Scholar]
- 30.Andjelkovic, A. V. & Pachter, J. S. (2000) J. Neurochem. 75, 1898-1906. [DOI] [PubMed] [Google Scholar]